Abstract
The active vitamin D metabolite 1α,25-dihydroxyvitamin D3 [1α,25(OH)2D3] has wide but not fully understood antitumor activity. A previous transcriptomic analysis of 1α,25(OH)2D3 action on human colon cancer cells revealed cystatin D (CST5), which encodes an inhibitor of several cysteine proteases of the cathepsin family, as a candidate target gene. Here we report that 1α,25(OH)2D3 induced vitamin D receptor (VDR) binding to, and activation of, the CST5 promoter and increased CST5 RNA and protein levels in human colon cancer cells. In cells lacking endogenous cystatin D, ectopic cystatin D expression inhibited both proliferation in vitro and xenograft tumor growth in vivo. Furthermore, cystatin D inhibited migration and anchorage-independent growth, antagonized the Wnt/β-catenin signaling pathway, and repressed c-MYC expression. Cystatin D repressed expression of the epithelial-mesenchymal transition inducers SNAI1, SNAI2, ZEB1, and ZEB2 and, conversely, induced E-cadherin and other adhesion proteins. CST5 knockdown using shRNA abrogated the antiproliferative effect of 1α,25(OH)2D3, attenuated E-cadherin expression, and increased c-MYC expression. In human colorectal tumors, expression of cystatin D correlated with expression of VDR and E-cadherin, and loss of cystatin D correlated with poor tumor differentiation. Based on these data, we propose that CST5 has tumor suppressor activity that may contribute to the antitumoral action of 1α,25(OH)2D3 in colon cancer.
Introduction
There is an increasing interest in the active vitamin D metabolite 1α,25-dihydroxyvitamin D3 [1α,25(OH)2D3] and its analogs as preventive and therapeutic anticancer agents, and a number of clinical trials are presently underway (1–4). Epidemiological and experimental data in cultured cells and in animal models indicate their beneficial effect against colon cancer through the inhibition of cell proliferation and invasion and the induction of pro-apoptotic and pro-differentiation activities (5–7). Global transcriptomic studies have led to the identification of a few 1α,25(OH)2D3 target genes, including some that encode cell-cycle regulators (p21WAF1/CIP1, p27KIP1, several cyclins), the gene that encodes the crucial intercellular adhesion and invasion suppressor E-cadherin, and several that encode cytoskeletal proteins (8–11). In addition, 1α,25(OH)2D3 exerts an indirect gene-regulatory effect through the antagonism of the Wnt/β-catenin signaling pathway, which is aberrantly activated in most human colon cancers. By inducing a rapid interaction of its receptor (vitamin D receptor [VDR]) and β-catenin and the subsequent nuclear export of β-catenin, 1α,25(OH)2D3 opposes the regulation of β-catenin/TCF complexes over a large number of genes in colon carcinoma cells (12, 13).
Cystatin D is a member of the cystatin superfamily of endogenous inhibitors of endosomal/lysosomal cysteine proteases. Cystatins have also been reported to play a role in other diverse biological processes, including cell proliferation, differentiation, survival, and migration and interleukin and nitric oxide production (ref. 14 and refs. therein). Cystatin D has a more restricted pattern of tissue expression and a narrower inhibitory profile than other cystatins: it inhibits cathepsin S, H, and L but not cathepsin B (15, 16). The role of proteases in cancer has been recognized for a long time, while that of cysteine cathepsins in particular as major players in tumor progression has recently been proposed (17, 18). In addition to the proteolytic cleavage of matrix components, adhesion proteins, and other proteases in a cascade fashion (18), some cathepsins have been implicated in novel mechanisms of cell transformation such as the control of cell cycle in the nucleus and the resistance to chemotherapy (14, 19).
Based on results of a transcriptomic analysis performed in human SW480-ADH colon cancer cells that indicated an increase in the level of cystatin D (CST5) RNA upon treatment with 1α,25(OH)2D3 (9), we have investigated the regulation and biological activity of this gene in colon cancer. In this study, we present evidence of the direct transcriptional regulation of the human CST5 gene by 1α,25(OH)2D3 in colon cancer cells. Our results also show that cystatin D profoundly affects the cell phenotype, inhibiting proliferation and migration and increasing cell adhesiveness. These effects are, at least partially, due to the repression of the c-MYC oncogene and the transcriptional activity of β-catenin and to the induction of E-cadherin. Conversely, CST5 knockdown by shRNA abolished the inhibition of cell proliferation by 1α,25(OH)2D3 and had the opposite effect of 1α,25(OH)2D3 on several target genes. Interestingly, mutant cystatin D proteins with reduced antiproteolytic activity preserve the antiproliferative but not the cell migration-inhibitory effects. Moreover, expression of CST5 in human colon carcinoma cells drastically reduces their tumorigenic potential in immunodeficient mice. Remarkably, analysis of human samples revealed that cystatin D expression is lost in poorly differentiated colon cancers. Furthermore, a strong correlation exists between the expression of cystatin D and that of VDR and E-cadherin proteins in tumors. Together, these data point to a role for CST5 as a tumor suppressor gene.
Results
1α,25(OH)2D3 increases cystatin D protein expression by direct activation of its gene promoter.
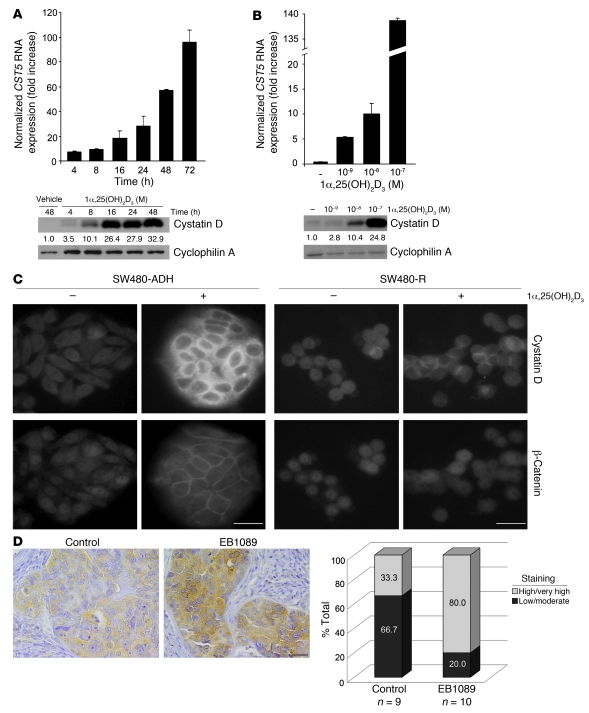
To validate the upregulation of CST5 by 1α,25(OH)2D3 suggested by a transcriptomic study in SW480-ADH cells (3.7-fold increase at 4 hours after treatment) (9), we performed kinetics analyses of the cellular RNA and protein content after addition of 1α,25(OH)2D3. 1α,25(OH)2D3 (10–7 M) gradually increased CST5 RNA and protein levels, which were about 60- and 30-fold greater, respectively, at 48 hours after treatment (Figure 1A). This effect was dose dependent (Figure 1B) and specific, as no induction of cystatin C was found (data not shown). The amount of secreted cystatin D protein in the medium increased slightly at 24–48 hours after treatment (2-fold; data not shown). The upregulation of CST5 by 1α,25(OH)2D3 was also found in other colon cancer cell lines, such as LS174T, Caco-2, and Colo205, and this correlated well with the activation of a consensus response element (Supplemental Figure 1, A and B; supplemental material available online with this article; doi: 10.1172/JCI37205DS1). The increase in cystatin D protein was confirmed by immunofluorescence studies, which also showed a predominant localization in the cytoplasm of SW480-ADH cells treated with 1α,25(OH)2D3 (Figure 1C). The relocation of β-catenin from the nucleus and cytosol toward the plasma membrane was used as control for 1α,25(OH)2D3 activity. No cystatin D was detected in SW480-R cells, which contain very low VDR levels and do not respond to 1,25(OH)2D3 (Figure 1C). To analyze whether the regulation of cystatin D expression takes place in vivo, we performed immunohistochemistry analyses of tumors generated by SW480-ADH cells in immunodeficient mice that were treated with EB1089, a less calcemic 1α,25(OH)2D3 analog (20). In line with in vitro data, EB1089 treatment increased cystatin D expression (Figure 1D).
Figure 1. 1α,25(OH)2D3 induces cystatin D expression.
(A) Kinetics of CST5 RNA (top) and protein (bottom) induction by 1α,25(OH)2D3. SW480-ADH cells were incubated with 1α,25(OH)2D3 (10–7 M) or vehicle for the indicated times, and the levels of CST5 RNA and protein were measured by quantitative RT-PCR (top) or Western blotting (bottom) as explained in Methods. Normalized mean values and SD obtained in 3 independent experiments are shown. (B) Dose-curve induction of CST5 RNA (top) and protein (bottom) by 1α,25(OH)2D3. Numbers between the blots in A and B correspond to mean of the fold increase values obtained in 2 experiments. (C) Immunofluorescence analysis of cystatin D induction. Images of SW480-ADH and SW480-R cells treated with 1α,25(OH)2D3 (10–7 M) or vehicle for 48 hours. The relocalization of β-catenin was analyzed as control of 1α,25(OH)2D3 action. Scale bars: 20 μm. (D) The 1α,25(OH)2D3 analog EB1089 induces cystatin D expression in vivo. Left: Immunohistochemical analysis of cystatin D expression in tumors generated by SW480-ADH cells in immunodeficient mice that were treated with EB1089 (10–7 M) or placebo. Scale bar: 200 μm. Right: Quantification of cystatin D expression by estimation of staining intensity as described in Methods. The number of samples analyzed per group and the percentage corresponding to each level of cystatin D staining are shown.
To assess the functionality of the cystatin D protein induced by 1α,25(OH)2D3, we studied the activity of its cathepsin targets in SW480-ADH cells. A series of enzymatic assays showed that cathepsin L activity in extracts from 1α,25(OH)2D3-treated cells was lower than in those of vehicle-treated cells (Supplemental Figure 2A). Total cathepsin activity also decreased, whereas that of cathepsin B was unchanged by 1α,25(OH)2D3 (Supplemental Figure 2A). Recombinant cystatin D protein was used as control. No changes in total extracellular cathepsin activity were found (data not shown).
We next examined the mechanism of cystatin D induction. The finding that the increase in CST5 RNA was abrogated by the transcription inhibitor actinomycin D but not significantly by the translation inhibitor cycloheximide indicated a direct transcriptional effect of 1α,25(OH)2D3 (Figure 2A). The in silico analysis of the CST5 gene promoter allowed the identification of several putative VDR binding hemisites (consensus RGKTCA) that could be grouped in 4 (A–D) regions (Figure 2B). Upon cloning of the –1,867/+262 region, transactivation assays in SW480-ADH cells using different fragments of the promoter showed that most of the activation depended on the cluster of sites in region B (–650/–262) (Figure 2C). The same result was obtained in VDR-deficient HEK293T cells when they were cotransfected with an exogenous VDR (Figure 2C, right). In addition, experiments using the SW480-R cells transfected with either wild-type VDR or the transcription activation–deficient VDRΔAF2 mutant assessed the requirement of a functional VDR for the activation of the CST5 promoter by 1α,25(OH)2D3 (Figure 2D). This activation was also observed in 2 other human colon cancer cells lines (LS174T and HCT116) (Supplemental Figure 1C). ChIP assays showed that 1α,25(OH)2D3 induces binding of VDR to the –840/–571 fragment (region B) of the promoter in vivo (Figure 2E). No binding to region A was detected (data not shown). Additionally, 1α,25(OH)2D3 promoted the release of the SMRT corepressor from the promoter and an increase in histone H4 acetylation, demonstrating the direct activation of the CST5 promoter (Figure 2E). The CYP24 gene, which is highly responsive to 1α,25(OH)2D3, was used as control (Figure 2E). Together, these results show that CST5 is a direct transcriptional target of 1α,25(OH)2D3.
Figure 2. 1α,25(OH)2D3 directly activates the CST5 promoter.
(A) Quantitative RT-PCR analysis of CST5 RNA expression in SW480-ADH cells treated with 1α,25(OH)2D3 or vehicle that were pretreated for 30 minutes with actinomycin D (2 μg/ml) or cycloheximide (8 μg/ml) as indicated. (B) Scheme of the human CST5 gene promoter showing putative VDR binding sites grouped in regions A–D. (C) Activation of CST5 promoter constructs by 1α,25(OH)2D3 in SW480-ADH cells and in HEK293T cells cotransfected with an expression vector for wild-type VDR. Twenty-four hours after transfection, cells were treated with vehicle or 1α,25(OH)2D3 (10–7 M) for an additional 48 hours. The empty pGL3 vector was used as control. (D) The activation of the CST5 promoter by 1α,25(OH)2D3 requires a transcriptionally competent VDR. SW480-R cells were cotransfected with the promoter construct pGL3-1867 or pGL3-251 and either the wild-type VDR or the mutant ΔAF2-VDR (VDRΔAF2), or an empty vector. The cells were then treated with 1α,25(OH)2D3 (10–7 M) or vehicle for 48 hours. As control, the consensus 4xVDRE-DR3-Tk-Luc (VDRE) reporter construct was cotransfected with the wild-type or mutant VDR. Values correspond to promoter induction by 1α,25(OH)2D3 in 3 independent experiments done in triplicate. (E) 1α,25(OH)2D3 induces VDR binding to and an active chromatin conformation of the CST5 promoter in vivo. ChIP assay showing the induction by 1α,25(OH)2D3 of VDR binding, SMRT corepressor release, and increased histone H4 acetylation (AcH4) of the CST5 gene promoter in SW480-ADH cells. The CYP24 gene was used as control. The promoter regions studied are indicated.
Cystatin D inhibits proliferation, migration, and anchorage-independent growth of cultured colon cancer cells and their tumorigenic potential in vivo.
To examine the effects of cystatin D upregulation, we expressed ectopically human cystatin D cDNA in SW480-ADH, LS174T, and HCT116 colon cancer cell lines, which have undetectable or low endogenous levels of this protease inhibitor. Expression of the exogenous cystatin D protein in transfected clones following antibiotic selection was analyzed by Western blotting: in all cases, the level was comparable to that of 1α,25(OH)2D3-treated SW480-ADH cells (Supplemental Figure 3). The activity of exogenous cystatin D was assessed in enzymatic assays (data not shown). Exogenous cystatin D and 1α,25(OH)2D3 decreased SW480-ADH cell proliferation comparably and their combination had an additive effect (Figure 3A). Ectopic cystatin D expression enhanced the increase in E-cadherin (CDH1) and CYP24 RNA and in E-cadherin protein levels by 1α,25(OH)2D3 (Figure 3, B and C). Furthermore, exogenous cystatin D reduced c-MYC expression in the absence or presence of 1α,25(OH)2D3. No significant changes were found for the mesenchymal marker LEF-1. Notably, exogenous cystatin D decreased SNAI1 RNA and protein expression (Figure 3D).
Figure 3. Ectopic cystatin D expression inhibits proliferation and alters gene expression in SW480-ADH cells.
(A) Cystatin D inhibits cell proliferation. (B) Quantitative RT-PCR analysis showing the level of CYP24, CDH1, and LEF1 RNA in cystatin D–expressing cells. (C) Western blot analysis showing changes in E-cadherin, c-MYC, and LEF-1 proteins in cystatin D–expressing cells. (D) Cystatin D inhibits SNAI1 expression. Quantitative RT-PCR (left) and Western blot (right) analyses showing reduced SNAI1 RNA and protein levels in cystatin D–expressing cells. Numbers between the blots in C and D correspond to mean of the fold increase values obtained in 2 experiments. *P < 0.05, **P < 0.01, ***P < 0.001. The Western blot in D shows noncontiguous lanes run on the same gel.
To further investigate its effects in SW480-ADH cells, we knocked down CST5 by stable shRNA expression. CST5 downregulation, confirmed by Western blot analysis, reduced the phenotypic change induced by 1α,25(OH)2D3 (Figure 4A). In addition, it abrogated the antiproliferative effect of 1α,25(OH)2D3 (Figure 4B). Moreover, and in contrast to the effect of CST5 overexpression, CST5 knockdown decreased CDH1 and CYP24 RNA and E-cadherin protein expression and increased that of c-MYC protein (Figure 4, C and D). Likewise, CST5 knockdown increased LEF-1 (Figure 4, C and D) and SNAIL1 (Figure 4E) expression.
Figure 4. CST5 silencing affects SW480-ADH cell proliferation and gene expression.
(A) Phase-contrast images of control shRNA and CST5 shRNA cells that were treated with 10–7 M 1α,25(OH)2D3 or vehicle for 48 hours. Western blot analysis showing decreased level of cystatin D protein in CST5 shRNA cells. Scale bar: 20 μm. (B) CST5 knockdown abrogates the inhibition of cell proliferation by 1α,25(OH)2D3. Control shRNA and CST5 shRNA cells were treated with vehicle or 1α,25(OH)2D3 for the indicated times. Proliferation of cells in 1α,25(OH)2D3- versus vehicle-treated cultures is shown. (C) Quantitative RT-PCR analysis showing decreased CDH1 and CYP24 and increased LEF1 RNA expression in CST5 shRNA cells. (D) Western blot analysis showing changes in E-cadherin, c-MYC, and LEF-1 proteins in CST5 shRNA cells. (E) CST5 knockdown increases the expression of SNAI1 RNA (left) and protein (right). Numbers between the blots in A, D, and E correspond to mean of the fold increase values obtained in 2 experiments.*P < 0.05, **P < 0.01, ***P < 0.001. The Western blot in E shows noncontiguous lanes run on the same gel.
Exogenous cystatin D also inhibited the proliferation of HCT116 (Figure 5A) and LS174T (Supplemental Figure 4) cells. Conversely, neither recombinant cystatin D protein added to cultured medium nor the nonpermeable E-64 pan-cathepsin inhibitor altered the proliferation of these 2 cell lines, suggesting that this effect requires intracellular mechanisms (Supplemental Figure 5). We studied 2 in vitro parameters of cell transformation: migration and anchorage-independent growth. Cystatin D expression reduced migration of HCT116 cells (SW480-ADH and LS174T cells lack basal migration capacity) in Boyden chamber assays (Figure 5B). Likewise, cystatin D inhibited the growth of HCT116 cells in semisolid agar (Figure 5C).
Figure 5. Ectopic cystatin D expression inhibits proliferation, migration, and anchorage-independent growth of HCT116 cells.
(A) Cystatin D inhibits cell proliferation. (B) Cystatin D inhibits cell migration. HCT116 cells transfected with empty vector (Mock) or stably expressing CST5 were seeded in triplicate on Transwell filters, and 24 hours later migratory cells that had attached to the lower surface of filters were counted. Representative images and a quantification of data from 3 independent experiments are shown. Original magnification, ×200. (C) Cystatin D inhibits anchorage-independent cell growth. Images of foci grown in semisolid agar and quantification are shown. Original magnification, ×63. ***P < 0.001.
Next, we examined the antitumoral effects of cystatin D in vivo. To this end, we inoculated immunodeficient mice subcutaneously with HCT116 or LS174T cells expressing or not expressing ectopic cystatin D. In both cases, the number and size of tumors generated were drastically inhibited by cystatin D expression (Figure 6).
Figure 6. Cystatin D inhibits tumor growth in vivo.
Immunodeficient mice were injected subcutaneously with (A) mock-transfected HCT116 cells or one of 2 clones of HCT116 cells transfected with CST5 (clone 9 or 20); or (B) mock-transfected LS147 T cells or one of 2 clones of LS147T cells transfected with CST5 (clone 9 or 11). The volume of the tumors generated was measured during the indicated period. Number of mice developing tumors at the end of the evaluation period: HCT116, mock: 25 of 25, CST5: 5 of 26; LS174T, mock: 9 of 9, CST5: 9 of 21.
Cystatin D induces intercellular adhesion proteins and inhibits genes promoting epithelial-mesenchymal transition.
Cells expressing exogenous cystatin D exhibited a strong adhesive phenotype (Figure 7A, left). Accordingly, immunofluorescence and confocal microscopy analysis showed that cystatin D–expressing HCT116 cells had increased levels of the adherens junction E-cadherin and p120-catenin and of the tight junction occludin proteins (Figure 7A, right). This was confirmed by Western blot analysis (Figure 7B). Furthermore, cystatin D–expressing cells contained higher levels of CDH1 RNA than control cells (Figure 7C). As CDH1 and OCLD are targets of transcriptional inhibition by genes promoting epithelial-mesenchymal transition (EMT), we studied whether cystatin D could modulate these genes. Indeed, cystatin D–expressing cells expressed lower levels of SNAI2, ZEB1, and ZEB2 RNA than empty vector–transfected (mock) cells (Figure 7D). In contrast, only a weak reduction in SNAI1 and no change in TWIST expression were found.
Figure 7. Cystatin D induces an adhesive phenotype and represses genes involved in EMT.
(A) Ectopic cystatin D induces intercellular adhesion. Phase-contrast (left) and immunofluorescence and confocal microscope images (right) of control (Mock) and cystatin D–expressing HCT116 cells. Expression of E-cadherin, p120-catenin, and occludin proteins was analyzed using specific antibodies. Scale bars: 10 μm (left) and 20 μm (right). (B) Western blot analysis showing the induction of adhesion proteins by cystatin D in HCT116 cells. Numbers between the blots correspond to mean of the fold increase values obtained in 3 experiments. (C) Quantitative RT-PCR analysis showing increased levels of CDH1 RNA in cystatin D–expressing HCT116 cells. (D) Cystatin D represses EMT genes. Quantitative RT-PCR analysis of the expression of the EMT genes SNAI1, SNAI2/SLUG, TWIST, ZEB1, and ZEB2 in mock and cystatin D–expressing HCT116 cells.
Cystatin D extends the cell cycle and inhibits Wnt/β-catenin signaling and the c-MYC oncogene.
To explore the mechanism of the inhibition of cell proliferation by cystatin D, we first performed flow cytometry analyses. Cystatin D–expressing cells displayed slower entry into cell cycle upon synchronization than control cells, as shown by the smaller proportion of cells in G2/M phase at 4 hours after release of the cell-cycle blockade (Figure 8A). This explains their lower proliferation rate. Cystatin D–expressing cells showed a decreased level of c-MYC protein, a key cell-cycle regulator (Figure 8B). Consistently, 2 c-MYC promoter constructs were less active in cystatin D–expressing cells than in control cells (Figure 8C). As the aberrant activation of the Wnt/β-catenin pathway that causes the induction of c-MYC and other proliferation and invasion genes is a hallmark of colon cancer (21, 22), we examined its activity in cystatin D–expressing cells. Analogously to 1α,25(OH)2D3 treatment (12, 13), exogenous cystatin D inhibited the transcriptional activity of β-catenin/TCF complexes, the downstream effector of the Wnt pathway (Figure 8D). This did not result from a general inhibitory effect on transcription, as the Notch pathway (evaluated by using a NICD reporter construct) was not affected by cystatin D expression (data not shown). We conclude that the antiproliferative action of cystatin D is at least in part mediated by the repression of the c-MYC oncogene, which in turn is probably mediated by the inhibitory effect on the Wnt/β-catenin pathway.
Figure 8. Cystatin D extends the cell cycle and inhibits of c-MYC expression and β-catenin/TCF transcriptional activity.
(A) Cystatin D decelerates cell-cycle entry. Mock and cystatin D–expressing HCT116 cell clones were stimulated with 10% serum to enter into cell cycle following synchronization in G1 phase using double block with thymidine and mimosine as described in Methods, and the proportion of cells in each phase was estimated by flow cytometry. Asynchronously growing cells were used for comparison (black profile). Data from a representative experiment of 3 performed are shown. The bottom graph represents the mean percentage of cells present in each phase of the cell cycle (G1 in black, S in gray, and G2/M in white) 4 hours after release of the blockade. (B) Western blot analysis showing the decrease in c-MYC protein content in cells expressing exogenous cystatin D following the release of cell-cycle blockade. Numbers between the blots correspond to mean of the fold increase values obtained in 3 experiments. (C) The human c-MYC gene promoter is less active in cystatin D–expressing cells. Mock and 2 cystatin D–expressing HCT116 cell clones were transfected with either of 2 constructs of the c-MYC promoter, and luciferase activity was measured 48 hours later. (D) β-Catenin/TCF transcriptional activity is reduced in cystatin D–expressing cells. Mock and 2 cystatin D–expressing HCT116 cell clones were transfected with the wild-type TOP-Flash and mutant FOP-Flash reporter plasmids, and the TOP/FOP ratio of luciferase activity was measured 48 hours later.
Cystatin D proteins with reduced antiproteolytic activity maintain the antiproliferative but not the migration-inhibitory effect.
To examine whether the antiproteolytic activity of cystatin D is necessary for its newly identified effects on cancer cells, we generated by PCR-mediated mutagenesis 2 mutant forms of this protease inhibitor. One of these cystatin D mutants had a Gly replacing Trp at position 108 (CystD W108G), while the other carried this mutation as well as a deletion of the first 12 amino acids (CystD W108G/Δ1–12). Mutations were verified by sequencing, and the loss of cysteine protease activity was checked by transfection of the mutant CST5 constructs in Cos-7 cells (Supplemental Figure 2B). To evaluate the activity of these mutant cystatin D proteins, we first expressed them in HCT116 colorectal carcinoma cells (Figure 9A) and then studied their effects on cell proliferation. Similar to the results obtained with wild-type cystatin D, both mutant proteins inhibited cell proliferation (Figure 9B). However, and in contrast to the above findings with wild-type cystatin D, neither of the 2 mutant proteins decreased cell migration (Figure 9C). In addition, we studied the transcriptional effect of both cystatin D mutants on selected target genes. As can be seen in Figure 9D, and also in contrast to wild-type cystatin D, the 2 mutant proteins decreased E-cadherin, while they did not alter p120-catenin expression. However, in line with their antiproliferative effect and like the wild-type protein, cystatin D mutants decreased the level of c-MYC (Figure 9D). Consistent with the decrease in E-cadherin protein, and in contrast to wild-type cystatin D, the 2 mutant cystatin D proteins reduced the RNA level of CDH1 and slightly increased that of SNAI1 (Figure 9, E and F). Moreover, they failed to inhibit ZEB1 RNA expression and were less efficient than wild-type cystatin D in the case of ZEB2 (Figure 9F). Together, these results indicate that the inhibition of cysteine proteases is only partially responsible for the effects of cystatin D in colon cancer cells.
Figure 9. Cystatin D mutant proteins with reduced antiproteolytic activity distinctly affect cell proliferation, migration, and gene expression.
(A) Western blot analysis showing the expression of CystD W108G and CystD W108G/Δ1–12 proteins in SW480-ADH cells. (B) CystD W108G and CystD W108G/Δ1–12 have the same antiproliferative effect as wild-type cystatin D. (C) CystD W108G and CystD W108G/Δ1–12 lack migration-inhibitory activity. (D) Effects of CystD W108G and CystD W108G/Δ1–12 on target genes. Western blot analysis showing E-cadherin, p120-catenin, and c-MYC protein expression in cells expressing mutant cystatin D proteins. Numbers between the blots in A and D correspond to mean of the fold increase values obtained in 3 experiments. (E and F) Quantitative RT-PCR analysis showing the RNA levels of CDH1 and of SNAI1, ZEB1, and ZEB2 in cells expressing either wild-type or mutant cystatin D proteins. ***P < 0.001.
Cystatin D expression decreases in human colorectal tumorigenesis, in good correlation with tumor dedifferentiation and VDR and E-cadherin loss.
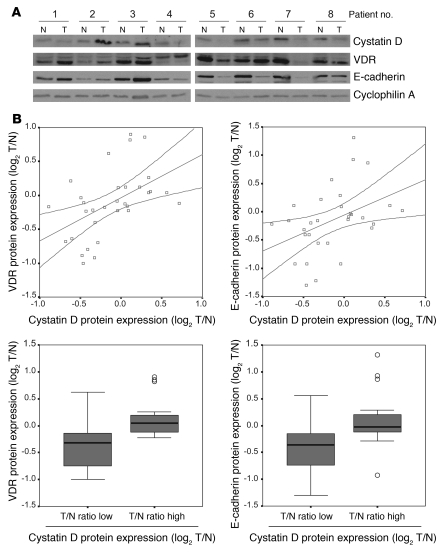
To investigate the relevance of our results in human colon cancer, we first studied the expression of cystatin D protein in tissue microarrays containing tumor and adjacent normal tissue samples. We found a progressive loss of cystatin D expression that correlated with tumor dedifferentiation: the strong staining in normal tissue and polyps decreased significantly in well- and moderately differentiated carcinomas and was absent in a large proportion of poorly differentiated tumors (Figure 10, A, top, and B). In line with previous studies (23), VDR expression also decreased associated with tumor progression (Figure 10A, bottom). These results were confirmed by Western blot analysis of a series of 32 matched normal and tumor samples. We defined overexpression and reduction as changes of at least 2-fold in protein level (Figure 11A). In 40.6% (13 of 32) cases, the level of cystatin D was lower in tumor than in normal tissue. Additionally, we found a strong direct correlation between the expression of cystatin D and VDR (Spearman correlation coefficient, r = 0.562; P = 0.001), supporting that the regulation of cystatin D expression by 1α,25(OH)2D3 observed in cultured cells and xenografts may also take place in human colon cancer (Figure 11B). Moreover, the expression of cystatin D also correlated with that of the E-cadherin (Spearman correlation coefficient, r = 0.492; P = 0.005), which, like VDR, is a marker of differentiation of human colorectal tumors (23) (Figure 11B). Box plot analysis of VDR and E-cadherin expression relative to that of cystatin D confirmed their correlation in the colorectal cancer series (Kruskal-Wallis test, P = 0.001 and P = 0.006, respectively).
Figure 10. Cystatin D expression is downregulated during human colon cancer progression.
(A) Immunohistochemical analysis of cystatin D and VDR expression in tissue microarrays. Counterstaining was with hematoxylin. Representative slices of nontumoral tissue and of well-, moderately, and poorly differentiated (differ.) carcinomas. Scale bar: 200 μm. (B) Quantification of cystatin D expression by estimation of staining intensity as described in Methods. The number of samples analyzed per group and the percentage corresponding to each level of cystatin D staining are shown.
Figure 11. Cystatin D expression directly correlates with that of VDR and E-cadherin in colon carcinomas.
(A) Representative Western blot showing the expression of cystatin D, VDR, and E-cadherin proteins in a series of matched normal (N) and tumoral (T) human colon tissues. (B) Top: Relationship between the log2 tumor versus normal (T/N) ratio of normalized protein levels of cystatin D and VDR or E-cadherin. Among the 32 sample pairs, the T/N ratio of cystatin D expression was less than 0.5 in 13 samples; between 0.5 and 2 in 14 samples; and greater than 2 in 5 samples. Bottom: Box plot of the log2 T/N ratio of normalized VDR or E-cadherin expression in samples with high or low cystatin D levels in the colorectal cancer series. Boxes include values in the 25%–75% interval; internal lines represent the median; the outliers (circles) of the VDR and E-cadherin expression are indicated.
Discussion
Natural and synthetic vitamin D compounds are increasingly studied as anticancer agents (1–4). In colon cancer, previous data from us and others have shown that 1α,25(OH)2D3 regulates the proliferation and phenotype of colon carcinoma cells through the transcriptional control of a number of target genes and antagonism of the Wnt/β-catenin signaling pathway (9, 12). These results contribute to explain the higher susceptibility to colon cancer caused by vitamin D deficiency in animal models and the results of epidemiological and clinical studies that indicate antitumoral action of vitamin D in humans (2–4, 24). Here we report that 1α,25(OH)2D3 is a strong direct inducer of CST5 in human colon cancer cells and that this reported inhibitor of members of the cathepsin family of proteases drastically inhibits the tumorigenic phenotype of colon carcinoma cells in vitro and in vivo.
Results obtained in human patients strongly indicate that CST5 gene is downregulated during colon tumorigenesis associated with tumor dedifferentiation. The correlation between the cystatin D expression and VDR protein levels in colon biopsy samples supports a role for 1α,25(OH)2D3 in the regulation of CST5 in the organism. Remarkably, VDR expression is associated with cell differentiation, absence of node involvement, and favorable prognosis in colorectal cancer (23, 25–27), which is in line with the loss of cystatin D in poorly differentiated tumors. Also the direct correlation between the expression of cystatin D and E-cadherin, which is a marker and crucial regulator of the epithelial phenotype and invasion, further supports its relation with tumor differentiation.
Lysosomal cysteine proteases have been implicated in multiple steps during tumor progression, including early steps of immortalization and transformation, intermediate steps of tumor invasion and angiogenesis, and late steps of metastasis and drug resistance (28). Thus, cathepsins B and S seem to contribute to the angiogenic switching and basement membrane degradation in early preneoplastic lesions. Likewise, cathepsins B, C, L, and Z potentiate the release and activation of pro-growth factors, thus favoring tumor growth. On the other hand, cathepsins H and L contribute to invasive growth either through degradation of basement membrane or extracellular matrix components or proteolysis of specific target proteins on the cell surface (28, 29). All these protumorigenic properties of cathepsins are balanced by the activity of the different members of the cystatin family of protease inhibitors. However, in recent years, cystatins have been proposed to play important roles in tumor progression apparently unrelated to their cathepsin-inhibitory action, and numerous studies involving cystatins C and E/M have described this dual function. Specifically, cystatin C has been proposed as a TGF-β receptor antagonist (30), whereas cystatin E/M has been widely studied because of its role as tumor suppressor mainly in breast cancer (14, 31). Our results derived from the functional analysis of mutant cystatin D proteins with reduced antiproteolytic activity indicate that cystatin D exerts its antimigratory effects through cathepsin inhibition, while its antiproliferative effect on colon cancer cells seems to be independent of this activity.
The antiproliferative action of cystatin D in vitro and the antitumor growth in vivo on colon cancer cells is most probably linked to the repression of c-MYC oncogene and the interference with the Wnt signaling pathway, 2 effects that mimic and putatively mediate at least in part 1α,25(OH)2D3 action in these cells (12). Remarkably, the suppression of c-MYC overexpression is sufficient to cause sustained tumor regression in several model systems, and a threshold level of c-MYC protein is required for tumor maintenance (ref. 32 and refs. therein). Moreover, the cystatin D–mediated increase in intercellular adhesion caused by the induction of E-cadherin and other adhesive proteins must strongly contribute to the reversion of the transformed phenotype and the inhibition of cell proliferation by 1α,25(OH)2D3. A plausible explanation for these effects is the downregulation of the EMT genes SNAI1, SNAI2, ZEB1, and ZEB2, known repressors of CDH1, OCLD, and related genes (33). Alternatively, the recent description by Weinberg’s group (34, 35) that E-cadherin loss in breast cancer cells promotes cancer stem cell–like properties and metastasis through the induction of EMT suggests that cystatin D may primarily affect E-cadherin.
The number and importance of the actions of cystatin D and its strong, rapid, and direct transcriptional regulation by 1α,25(OH)2D3 indicate that cystatin D is an important mediator of 1α,25(OH)2D3 action in colon cancer cells in vitro and, according to the results with tumor biopsies, also exerts putative protective effects in humans. This is further emphasized by the results obtained after expressing an exogenous cystatin D or by downregulation of the endogenous CST5 gene by means of shRNA. Previously, cystatin D expression had been detected only in saliva and tears (36). The loss of cystatin D expression during colon tumorigenesis associated to dedifferentiation supports a role for this protein in the control of cell phenotype in vivo and its status as a candidate tumor suppressor. Analogously, cystatin M has recently been shown to have antitumoral effects and to be downregulated in breast cancer (14, 37). The finding that addition to cells of recombinant cystatin D protein did not reproduce the effects of endogenous cystatin D indicates that its antitumor effects are mostly exerted intracellularly. The exact molecular mechanism by which cystatin D exerts its actions is unclear. One possibility is based on its antiproteolytic activity. Thus, cystatin D may regulate E-cadherin integrity or the processing of one or more transcription factors controlling cell proliferation and/or E-cadherin expression. In line with this, the cystatin D target cathepsin L has been reported to cleave E-cadherin in vitro and possibly in RT2 mice (17). In addition, cathepsin L controls mouse 3T3 and human breast cancer cell proliferation within the nucleus by modulating the proteolytic processing of the CCAAT-displacement protein/cut homeobox (CDP/Cux) transcription factor (19). In human colon cancer cells, however, we did not detect any of the E-cadherin cleaved fragments (64 and 30–35 kDa) that are generated by cathepsin L. Likewise, we found no changes in the expression of CDP/Cux polypeptides associated with cystatin D expression (data not shown). These differences may be due to species- and/or cell type–specific activity of cathepsin L. Together with the observed increase in the level of CDH1 RNA in cystatin D–expressing cells, these results favor a role for cystatin D in the regulation of CDH1 RNA transcription or stability. A second possibility is that cystatin D has effects unrelated to the inhibition of cathepsins by interacting with other noncharacterized proteins and so modulating distinct processes. One illustrative example in this regard is that of the tissue inhibitor of metalloproteinases–2 (TIMP2), which inhibits mitogenesis and angiogenesis at least in part by metalloproteinase-independent mechanisms (38). The finding that mutant cystatin D proteins with reduced antiproteolytic activity inhibit cell proliferation but not cell migration makes plausible that both cathepsin inhibition–dependent and –independent mechanisms are responsible for cystatin D antitumoral activity. Notably, 2 other cystatins, A and E, have been shown to be regulated by 1α,25(OH)2D3 in keratinocytes and squamous cell carcinoma cells, respectively, although no related functional studies have been reported yet (39, 40).
Together, our findings reveal an unpredicted activity of CST5 as a tumor suppressor. Furthermore, our results illustrate what we believe to be a novel mechanism of the anticancer action of the most active vitamin D metabolite and provide a rationale for its preventive and therapeutic use against colon cancer.
Methods
Cell culture, constructs, and transfections.
SW480-ADH and SW480-R cells (derived from the SW480 human colon cancer cell line by limit dilution analysis; ref. 12), HCT116 and LS174T cells were cultured in DMEM plus 10% FBS (Invitrogen). Treatment with 1α,25(OH)2D3 was performed in DMEM supplemented with charcoal-treated serum to remove liposoluble hormones. The cells were treated with 10–7 M 1α,25(OH)2D3 or the corresponding vehicle/ethanol for the times indicated. Several fragments of the CST5 promoter (–1,867/+262, –1,128/+262, –650/+262, and –251/+262) were amplified by PCR using genomic DNA from SW480-ADH cells as template and the following primers: 5′-ACGCGTCCGCAGGATCACCTTCAG-3′, 5′-ACGCGTCACAGGTGTGGACAAAGTGG-3′, 5′-ACGCGTTCCAGGAGCTTCCTCTTCCT-3′, 5′-ACGCGTGAATCCAGAGTGAGCCAAGC-3′, and 5′-AGATCTTGTACTCGCTGATGGCAAAG-3′. The resulting products were cloned into the pCRII-TOPO vector (Invitrogen) and sequenced. Subsequently, they were subcloned as MluI/BglII fragments into the promotorless pGL3basic Firefly luciferase expression vector (Promega). The 4xVDRE-DR3-Tk-Luc construct was provided by C. Carlberg (University of Kuopio, Kuopio, Finland). The CDH1 promoter activity was studied using the -987-TK-Luc construct (12). To study β-catenin/TCF transcriptional activity, we used the TOP-Flash and FOP-Flash plasmids containing multimerized wild-type (CCTTTGATC) or mutated (CCTTTGGCC) TCF/LEF-1 binding sites upstream of a minimal c-fos promoter driving luciferase gene expression (41) (a gift from H. Clevers, Hubrecht Institute and University Medical Center, Utrecht, The Netherlands). The expression vector for the truncated VDR lacking the 11 carboxyterminal amino acids (ΔAF2) was donated by Ana Aranda of Insituto de Investigaciones Biomédicas, Madrid. The P1P2myc-Luc and P2myc-Luc plasmids containing fragments of the human c-MYC promoter (42) were provided by J. León (Universidad de Cantabria, Santander, Spain). For transient transactivation assays, cells cultured in 24-well dishes were transfected in triplicate using the jetPEI reagent (PolyPlus Transfection). Reporter assays were carried out 48 hours after transfection by measuring Firefly (Luc) and Renilla reniformis luciferase (Rluc) activities separately using the Dual Luciferase reagent kit (Promega) and a Lumat LB 9507 luminometer (Berthold Technologies). Luc activity was normalized to the Rluc activity. SW480-ADH, HCT116, and LS174T cells stably expressing cystatin D were generated by transfection with either pcDNA3.1-CST5 or pcDNA3.1 plasmids (control, mock), followed by selection with 0.5 mg/ml (SW480-ADH and LS174T) or 2 mg/ml (HCT116) G418 (Sigma-Aldrich) for 2 weeks. pcDNA3.1-CST5 plasmid was obtained by subcloning the full-length human CST5 cDNA from the pEMBL19 plasmid (36) by digestion with EcoRI and XbaI.
Gene silencing.
To knock down CST5, we infected cells with lentiviral particles containing a U6 promoter driving an shRNA targeting the respective RNA. MISSION shRNA lentiviral particles against human CST5 or scramble negative control (Sigma-Aldrich) were used. Control cells were infected with lentivirus bearing a nontargeting shRNA that activates the RISC complex and the RNAi pathway but that contains at least 5 mismatched nucleotides compared with any human gene (clone SHC002; Sigma-Aldrich).
Generation of CST5 mutants.
Point mutation and deletion of CST5 was carried out using QuickChange Site-Directed Mutagenesis Kit (Stratagene) according to the manufacturer’s guidelines. For the single CST5 W108G mutant, we used the pcDNA3.1-CST5 plasmid as template and the sense oligonucleotide 5′-CCAGATCAATGAAGTTCCCGGGGGAGGATAAAATTTCCATTC-3′. For the double CST5 W108G/Δ1–12, mutant, we used the CST5 W108G mutant as template and the sense oligonucleotide 5′-CCTTGATGGTGGCCGTGGCCGGCATCCATGCCACAGACCCTC-3′. Mutations were verified by sequencing.
Cell proliferation and migration assays.
To measured proliferation, the cells (8 × 103) were seeded in 24-well plates and treated for up to 6 days with 10–7 M of 1α,25(OH)2D3 or vehicle. Living cells were counted after trypsinization. Alternatively, we used [3-(4,5-dimethythiazol-2-yl)-2,5-diphenyl] tetrazolium bromide (MTT) assays, which were performed according to the manufacturer’s instructions (Roche Diagnostics). All experiments were performed in quadruplicate.
For migration assays, the cells were seeded on 8.0-μm-pore Transwells (Corning). After 24 hours incubation, cells attached to the lower surface of the filter were stained using Diff-Quik reagents (Dade Behring) and counted (10 fields/Transwell). Recombinant human cystatin D protein was from R&D Systems.
Anchorage-independent growth assays.
HCT116 cells (5 × 103) transfected with an empty vector or pcDNA3-CST5 were trypsinized and suspended in 1.5 ml of 0.35% Difco Noble agar (BD) in DMEM containing 10% FBS. The agar-cell mixture was plated on top of a bottom layer of 0.5% agar (1.5 ml). The experiment was performed in triplicate for each cell line. After 2 weeks, viable colonies larger than 50 μm were scored.
Cathepsin L activity assays.
Cathepsin L activity was assessed using fluorogenic substrate Z-Phe-Arg-AMC (benzyloxycarbonyl-Phe-Arg-7-amino-4-methylcoumarin) (Bachem) in the presence of E-64 (Sigma-Aldrich), a broad spectrum inhibitor of cysteine proteases, and CA-074 (Bachem), a specific inhibitor of cathepsin B, by using the Inubushi method (43) with some modifications. Briefly, whole-cell extracts were prepared by lysis in cathepsin L lysis buffer (400 mM sodium phosphate buffer pH 6, 75 mM NaCl, 4 mM EDTA, 0.25% Triton X-100), incubated 1 hour on ice, and homogenized by sonication. Twenty micromoles of substrate was incubated with the whole-cell extracts in the cathepsin assay buffer (100 mM sodium acetate buffer pH 5.5, 1 mM EDTA, 2 mM DTT) in the presence of 50 μmol E-64 or CA-074 at 37°C for 10 minutes. The amount of 7-amino-4-methylcoumarin liberated from the substrate was monitored fluorometrically with excitation at 370 nm and emission at 480 nm. Total cysteine peptidase activity was determined as the difference between the total activity and the background activity of the non-cysteine peptidases determined by using E-64. Cathepsin L–like activity was measured by inhibiting cathepsin B activity with CA-074.
Western blotting.
Whole-cell extracts were prepared by washing the monolayers twice in PBS and subjected to cell lysis by incubation in RIPA buffer (150 mM NaCl, 1.5 mM MgCl2, 10 mM NaF, 10% glycerol, 4 mM EDTA, 1% Triton X-100, 0.1% SDS, 1% sodium deoxycholate, 50 mM HEPES pH 7.4) plus phosphatase- and protease-inhibitor mixture (25 mM β-glycerophosphate, 1 mM Na3VO4, 1 mM PMSF, 10 μg/ml leupeptin, 10 μg/ml aprotinin) for 15 minutes on ice, followed by centrifugation at 15,700 g for 10 minutes at 4°C. Protein concentration was measured using the Bio-Rad DC protein assay kit. Analysis of cell lysates or immunoprecipitates was performed by electrophoresis in SDS gels and protein transfer to Immobilon P membranes (Millipore). The membranes were incubated with the appropriate primary and secondary HRP-conjugated antibodies, and the antibody binding was visualized using the ECL detection system (Amersham, GE Healthcare). We used rabbit polyclonal antibodies generated against cystatin D (36), occludin (Zymed, Invitrogen), and cyclophilin A (Upstate, Millipore); mouse monoclonal antibodies against E-cadherin, p120-catenin (BD Biosciences), and c-MYC (Santa Cruz Biotechnology Inc.); rat monoclonal antibody against VDR (Chemicon); and goat polyclonal antibody against β-actin and lamin B (Santa Cruz Biotechnology Inc.). The anti-Snail1 antibody was provided by K.-F. Becker (Technische Universität München, Munich, Germany). Secondary antibodies used were HPR-conjugated anti-rabbit IgG (H+L) (MP Biomedicals), anti-mouse IgG (H+L) (Promega), and anti-rat IgG and anti-goat IgG (Santa Cruz Biotechnology Inc.).
Immunofluorescence and confocal microscopy.
Cells were rinsed once in PBS and fixed in 3.7% formaldehyde for 10 minutes at room temperature. The cells were permeabilized in 0.2% Triton X-100 for 10 minutes at room temperature. Nonspecific sites were blocked by incubation with PBS containing 1% Difco Skim Milk (BD) for 10 minutes at room temperature before cells were incubated with the primary antibodies against cystatin D, E-cadherin, p120-catenin, and occludin, diluted (1:100) in PBS for 1 hour at 37°C. After 4 washes in PBS, the cells were incubated with the secondary antibodies Alexa Fluor 488 donkey anti-mouse or Alexa Fluor 488 goat anti-rabbit (Molecular Probes, Invitrogen) for 45 minutes at room temperature, washed 3 times in PBS, and mounted in VECTASHIELD (Vector Laboratories). Propidium iodide staining was done for 10 minutes at room temperature and followed by 4 washes in PBS. Images were acquired with an Olympus DP70 digital camera mounted on a Zeiss Axiophot microscope equipped with epifluorescence (Figure 1), and confocal images were captured with a Leica TCS SP2 confocal microscope (Figure 7). For double labeling experiments, images of the same confocal plane were generated and superimposed. Phase-contrast images were captured with a Leica DC300 digital camera mounted on an inverted Leitz Labovert FS Microscope. All images were processed using Adobe Photoshop CS2 software.
Immunohistochemistry.
Human tissues were obtained as formalin-fixed, paraffin-embedded tissue sections from the archives of the tumor bank of Hospital Universitario Central de Asturias. They remained anonymous according to guidelines approved by the hospital’s Research Ethics Board. Tissue arrays containing a total of 51 samples including 3 replicates representing different locations were used to evaluate cystatin D and VDR expression according to the differentiation status. After dewaxing and rehydrating, samples were blocked in 15% goat serum and then incubated overnight at 4°C with anti–cystatin D (1:1,000) or anti-VDR antibody (1:150). Visualization of specific interactions was monitored by using the EnVision HRP System (Dako) according to the manufacturer’s instructions, and the staining was completed by incubation with diaminobenzidine colorimetric reagent (Dako), followed by counterstaining with hematoxylin. Finally, the slides were dehydrated and mounted. Controls included samples that were incubated with a preimmune serum. Normal parotid tissue was used as positive control for cystatin D. Protein expression was graded independently by 2 observers as very high (+++), high (++), moderate (+), low (+/–), or negative (–), depending on the level of epithelial staining.
Flow cytometry.
Cells were synchronized by 17 hours incubation with 2.5 mM thymidine (Sigma-Aldrich) in McCoy’s 5A medium. Later, they were washed twice in PBS and incubated in normal medium for an additional 6 hours at 37°C to release them from the cell-cycle blockade. To increase the proportion of G1-arrested cells, we then treated cultures with 0.5 mM l-mimosine (Sigma-Aldrich) for 20 hours. At various time points, cells were washed in PBS containing 50 mM EDTA, trypsinized, resuspended in 1 ml PBS/EDTA, and fixed by addition of 3 ml ice-cold 100% ethanol and left overnight at 4°C. Fixed cells were then pelleted and washed in 1 ml PBS/EDTA, and DNA was stained with 0.025 mg/ml propidium iodide (Sigma-Aldrich) in PBS/EDTA containing 0.05% NP40 and 5 ng/μl RNase A. The fraction of the population in each phase of the cell cycle was determined as a function of DNA content using the FACScan flow cytometer FC 500 MPL (Beckman Coulter) equipped with MXP software.
ChIP assays.
ChIP assays were carried out as reported previously (44) using specific CST5 or CYP24 primers: CST5 (–840/–571), 5′-CCACAGTGACGCTTGGTCTA-3′ (forward) and 5′-GTCTGGGCAATAGAGCCGTA-3′ (reverse); negative control, 5′-ATCTCCCAGAGAGCAAAGCA-3′ (forward) and 5′-GAATCCAGAGTGAGCCAAGC-3′ (reverse); CYP24, 5′-CGTTTCCTCCTGTCCCTCTC-3′ (forward) and 5′-TGCCTTCCTGGGGGTTATCTC-3′ (reverse). Antibodies against VDR, SMRT (Santa Cruz Biotechnology Inc.), and histone H4 (acetyl K12) (Upstate), and rabbit IgG (Santa Cruz Biotechnology Inc.) were used. The sensitivity of PCR amplification was evaluated on serial dilutions of total DNA collected after sonication (input fraction).
Real-time RT-PCR.
Cellular RNA levels of CST5, SNAI1, SNAI2/SLUG, TWIST, ZEB1, ZEB2, CDH1, and LEF1 were quantified by real-time RT-PCR using the primers listed in Supplemental Table 1. The level of CYP24 RNA was measured in relation to that of 18S rRNA using the comparative Ct method and RNA TaqMan probes (Applied Biosystems). Values were normalized versus the geometric average of 3 control housekeeping genes (TATA-binding protein, TBP; succinate dehydrogenase complex subunit A, SDHA; and ubiquitin C, UBC) as described previously (45). For the synthesis of the first strand of cDNA, 400 ng of total RNA was retrotranscribed using the Gold RNA PCR Core Kit (Applied Biosystems) according to the manufacturer’s instructions. Random hexamers were used for cDNA synthesis. The relative concentration of the target and the reference genes was calculated by interpolation on a standard curve for each gene generated with a serial dilution of cDNA obtained from SW480-ADH cells. The reaction was performed in a LightCycler apparatus using the LightCycler-FastStart DNA MasterPLUS SYBR Green I Kit (Roche). Thermal cycling for all genes was initiated with a denaturation step of 95°C for 10 minutes and consisted of 40 cycles (denaturation at 95°C for 10 seconds, specific annealing temperatures shown in Supplemental Table 1 for 5 seconds, and elongation at 72°C for 5 seconds). At the end of the PCR cycles, melting curve analyses were performed, as well as electrophoresis of the products on nondenaturing 8% polyacrylamide gels, followed by sequencing, in order to validate the generation of the specific PCR product expected.
Xenograft tumor growth.
We used severely immunodeficient female scid mice obtained from The Jackson Laboratory. The maintenance and handling of animals were as recommended by the European Union (ECC directive 86/609/EEC, November 24, 1986), and all experiments were approved by the Animal Experimentation Committee at the Instituto de Investigaciones Biomédicas, Madrid. Every effort was made to minimize animal suffering and to reduce the number of animals used. Mice were subcutaneously injected with 3 × 106 HCT116 or LS174T cells expressing empty vector (mock) or pcDNA3-CST5 in the left and right flank, respectively. Tumor size was measured 3 times per week by using the ellipsoid volume formula (0.5 × L × W × H), where L, W, and H are the tumor length, width, and height (in cm). Animals were euthanized when their external tumor diameter reached 1.5 cm.
Quantification of cystatin D, VDR, and E-cadherin protein expression in human samples.
All patients gave written informed consent, and the protocol was approved by the Research Ethics Board of the Hospital Universitario Puerta de Hierro, Madrid. Normal and tumor tissue samples were obtained immediately after surgery, snap-frozen in liquid nitrogen, and stored at –80°C until processing. Tissue protein was extracted by pulverizing the samples in liquid nitrogen using a mortar and homogenization using a Potter-Elvehjem apparatus in lysis buffer (50 mM Tris-HCl pH 7.5, 1% NP-40, 0.25% sodium deoxycholate, 150 mM NaCl, 2 mM MgCl2, 1 mM EDTA, 10% glycerol, 0.5 mM DTT, and protease and phosphatase inhibitor mix) on ice. After 20 minutes centrifugation at 15,700 g, the expression level of cystatin D, VDR, and E-cadherin proteins was analyzed by Western blotting.
Statistics.
The data are expressed as mean ± SD unless otherwise specified. Statistical significance was assessed by 2-tailed unpaired Student’s t test. When P was greater than 0.05, the data were considered not significant. All statistical analyses were performed using SPSS 13.0 statistical software. As the tumor/normal tissue (T/N) ratios of VDR and cystatin D expression were not normally distributed (Kolmogorov-Smirnov test, Lilliefors correction), we normalized the data distribution by using log2 for statistical analysis (20). The geometric (rather than the arithmetic) average of the T/N ratio was used. Correlations between protein expression levels were analyzed using the Spearman correlation coefficient. In addition, cystatin D protein expression was divided into 2 groups, low and high expression, according to the median value of this variable, and the expression of VDR and E-cadherin in these 2 groups was represented in a box plot graphic. The comparison between the 2 groups of data was done using the Kruskal-Wallis test.
Supplementary Material
Acknowledgments
We are grateful to R. Bouillon, M. Verstuyf, and J.P. Van de Velde for the 1α,25(OH)2D3, and to the individuals mentioned in Methods for providing plasmids or antibodies. We also thank D. Navarro and T. Martínez for technical assistance, N. Pendás-Franco and M.J. Larriba for help with the migration assays and animal experiments, and Marta S. Pitiot for help with the immunohistochemical studies. This work was supported by the Ministerio de Ciencia e Innovación (SAF2007-60341, SAF2006-00476, ISCIII-RETIC RD06/0020/0009, and RD06/0020/0020), Comunidad de Madrid (S-GEN-0266/2006), and the European Union (MRTN-CT-2005-019496, NucSys and Micoenvimet, FP7). The Instituto Universitario de Oncología and Hospital Universitario Central de Asturias tumor bank are supported by Obra Social Cajastur and Acción Transversal del Cáncer–RTICC.
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Nonstandard abbreviations used: 1α,25(OH)2D3, 1α,25-dihydroxyvitamin D3; EMT, epithelial-mesenchymal transition; VDR, vitamin D receptor; VDRE, vitamin D response element.
Citation for this article: J. Clin. Invest. 119:2343–2358 (2009). doi:10.1172/JCI37205
References
- 1.Deeb K.K., Trump D.L., Johnson C.S. Vitamin D signalling pathways in cancer: potential for anticancer therapeutics. Nat. Rev. Cancer. 2007;7:684–700. doi: 10.1038/nrc2196. [DOI] [PubMed] [Google Scholar]
- 2.Gorham E.D., et al. Optimal vitamin D status for colorectal cancer prevention: a quantitative meta analysis. Am. J. Prev. Med. 2007;32:210–216. doi: 10.1016/j.amepre.2006.11.004. [DOI] [PubMed] [Google Scholar]
- 3.Lappe J.M., Travers-Gustafson D., Davies K.M., Recker R.R., Heaney R.P. Vitamin D and calcium supplementation reduces cancer risk: results of a randomized trial. Am. J. Clin. Nutr. 2007;85:1586–1591. doi: 10.1093/ajcn/85.6.1586. [DOI] [PubMed] [Google Scholar]
- 4.Ng K., et al. Circulating 25-hydroxyvitamin D levels and survival in patients with colorectal cancer. J. Clin. Oncol. 2008;26:2984–2991. doi: 10.1200/JCO.2007.15.1027. [DOI] [PubMed] [Google Scholar]
- 5.Ordóñez-Morán P., et al. Vitamin D and cancer: an update of in vitro and in vivo data. . Front. Biosci. 2005;10:2723–2749. doi: 10.2741/1731. [DOI] [PubMed] [Google Scholar]
- 6.Campbell M.J., Adorini L. The vitamin D receptor as a therapeutic target. Expert Opin. Ther. Targets. 2006;10:735–748. doi: 10.1517/14728222.10.5.735. [DOI] [PubMed] [Google Scholar]
- 7.Eelen G., et al. Mechanism and potential of the growth-inhibitory actions of vitamin D and analogs. Curr. Med. Chem. 2007;14:1893–1910. doi: 10.2174/092986707781058823. [DOI] [PubMed] [Google Scholar]
- 8.Akutsu N., et al. Regulation of gene expression by 1α,25-dihydroxyvitamin D3 and its analog EB1089 under growth-inhibitory conditions in squamous carcinoma cells. . Mol. Endocrinol. 2001;15:1127–1139. doi: 10.1210/me.15.7.1127. [DOI] [PubMed] [Google Scholar]
- 9.Pálmer H.G., et al. Genetic signatures of differentiation induced by 1α,25-dihydroxyvitamin D3 in human colon cancer cells. . Cancer Res. 2003;63:7799–7806. [PubMed] [Google Scholar]
- 10.Swami S., Raghavachari N., Muller U.R., Bao Y.P., Feldman D. Vitamin D growth inhibition of breast cancer cells: gene expression patterns assessed by cDNA microarray. Breast Cancer Res. Treat. 2003;80:49–62. doi: 10.1023/A:1024487118457. [DOI] [PubMed] [Google Scholar]
- 11.Wood R.J., Tchack L., Angelo G., Pratt R.E., Sonna L.A. DNA microarray analysis of vitamin D-induced gene expression in a human colon carcinoma cell line. Physiol. Genomics. 2004;17:122–129. doi: 10.1152/physiolgenomics.00002.2003. [DOI] [PubMed] [Google Scholar]
- 12.Pálmer H.G., et al. Vitamin D3 promotes the differentiation of colon carcinoma cells by the induction of E-cadherin and the inhibition of β-catenin signaling. . J. Cell Biol. 2001;154:369–387. doi: 10.1083/jcb.200102028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shah S., et al. The molecular basis of vitamin D receptor and beta-catenin crossregulation. Mol. Cell. 2006;21:799–809. doi: 10.1016/j.molcel.2006.01.037. [DOI] [PubMed] [Google Scholar]
- 14.Zhang J., et al. Cystatin M: a novel candidate tumor suppressor gene for breast cancer. Cancer Res. 2004;64:6957–6964. doi: 10.1158/0008-5472.CAN-04-0819. [DOI] [PubMed] [Google Scholar]
- 15.Freije J.P., et al. Structure and expression of the gene encoding cystatin D, a novel human cysteine proteinase inhibitor. J. Biol. Chem. 1991;266:20538–20543. [PubMed] [Google Scholar]
- 16.Balbin M., et al. Structural and functional characterization of two allelic variants of human cystatin D sharing a characteristic inhibition spectrum against mammalian cysteine proteinases. . J. Biol. Chem. 1994;269:23156–23162. [PubMed] [Google Scholar]
- 17.Gocheva V., et al. Distinct roles for cysteine cathepsin genes in multistage tumorigenesis. Genes Dev. 2006;20:543–556. doi: 10.1101/gad.1407406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gocheva V., Joyce J.A. Cysteine cathepsins and the cutting edge of cancer invasion. Cell Cycle. 2007;6:60–64. doi: 10.4161/cc.6.1.3669. [DOI] [PubMed] [Google Scholar]
- 19.Goulet B., et al. Increased expression and activity of nuclear cathepsin L in cancer cells suggests a novel mechanism of cell transformation. Mol. Cancer Res. 2007;5:899–907. doi: 10.1158/1541-7786.MCR-07-0160. [DOI] [PubMed] [Google Scholar]
- 20.Pálmer H.G., et al. The transcription factor SNAIL represses vitamin D receptor expression and responsiveness in human colon cancer. Nat. Med. 2004;10:917–919. doi: 10.1038/nm1095. [DOI] [PubMed] [Google Scholar]
- 21.Clevers H. Wnt/beta-catenin signaling in development and disease. Cell. 2006;127:469–480. doi: 10.1016/j.cell.2006.10.018. [DOI] [PubMed] [Google Scholar]
- 22.Segditsas S., Tomlinson I. Colorectal cancer and genetic alterations in the Wnt pathway. Oncogene. 2006;25:7531–7537. doi: 10.1038/sj.onc.1210059. [DOI] [PubMed] [Google Scholar]
- 23.Peña C., et al. E-cadherin and vitamin D receptor regulation by SNAIL and ZEB1 in colon cancer: clinicopathological correlations. Hum. Mol. Genet. 2005;14:3361–3370. doi: 10.1093/hmg/ddi366. [DOI] [PubMed] [Google Scholar]
- 24.Garland C.F., Garland F.C. Do sunlight and vitamin D reduce the likelihood of colon cancer? Int. J. Epidemiol. 2005;35:217–220. doi: 10.1093/ije/dyi229. [DOI] [PubMed] [Google Scholar]
- 25.Vandewalle B., Adenis A., Hornez L., Revillion F., Lefebvre J. 1,25-dihydroxyvitamin D3 receptors in normal and malignant human colorectal tissues. . Cancer Lett. 1994;86:67–73. doi: 10.1016/0304-3835(94)90181-3. [DOI] [PubMed] [Google Scholar]
- 26.Cross H.S., et al. Vitamin D receptor and cytokeratin expression may be progression indicators in human colon cancer. Anticancer Res. 1996;16:2333–2337. [PubMed] [Google Scholar]
- 27.Evans S.R., et al. Vitamin D receptor expression as a predictive marker of biological behavior in human colorectal cancer. Clin. Cancer Res. 1998;4:1591–1595. [PubMed] [Google Scholar]
- 28.Joyce J.A., Hanahan D. Multiple roles for cysteine cathepsins in cancer. Cell Cycle. 2004;3:1516–1619. doi: 10.4161/cc.3.12.1289. [DOI] [PubMed] [Google Scholar]
- 29.Kuester D., Lippert H., Roessner A., Krueger S. The cathepsin family and their role in colorectal cancer. Pathol. Res. Pract. . 2008;204:491–500. doi: 10.1016/j.prp.2008.04.010. [DOI] [PubMed] [Google Scholar]
- 30.Sokol J.P., Schiemann W.P. Cystatin C antagonizes transforming growth factor beta signaling in normal and cancer cells. Mol. Cancer Res. 2004;2:183–195. [PubMed] [Google Scholar]
- 31.Shridhar R., et al. Cystatin M suppresses the malignant phenotype of human MDA-MB-435S cells. Oncogene. 2004;23:2206–2215. doi: 10.1038/sj.onc.1207340. [DOI] [PubMed] [Google Scholar]
- 32.Shachaf C.M., et al. Genomic and proteomic analysis reveals a threshold level of MYC required for tumor maintenance. Cancer Res. 2008;68:5132–5142. doi: 10.1158/0008-5472.CAN-07-6192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Peinado H., Olmeda D., Cano A. Snail, Zeb and bHLH factors in tumour progression: an alliance against the epithelial phenotype? Nat. Rev. Cancer. 2007;7:415–428. doi: 10.1038/nrc2131. [DOI] [PubMed] [Google Scholar]
- 34.Onder T.T., et al. Loss of E-cadherin promotes metastasis via multiple downstream transcriptional pathways. Cancer Res. 2008;68:3645–3654. doi: 10.1158/0008-5472.CAN-07-2938. [DOI] [PubMed] [Google Scholar]
- 35.Mani S.A., et al. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell. 2008;133:704–715. doi: 10.1016/j.cell.2008.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Freije J.P., et al. Human cystatin D. cDNA cloning, characterization of the Escherichia coli expressed inhibitor, and identification of the native protein in saliva. J. Biol. Chem. 1993;268:15737–15744. [PubMed] [Google Scholar]
- 37.Schagdarsurengin U., Pfeifer G.P., Dammann R. Frequent epigenetic inactivation of cystatin M in breast carcinoma. Oncogene. 2007;26:3089–3094. doi: 10.1038/sj.onc.1210107. [DOI] [PubMed] [Google Scholar]
- 38.Stetler-Stevenson W.G. The tumor microenvironment: regulation by MMP-independent effects of tissue inhibitor of metalloproteinases-2. Cancer Metastasis Rev. 2008;27:57–66. doi: 10.1007/s10555-007-9105-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Takahashi H., et al. 1,25-dihydroxyvitamin D(3) increases human cystatin A expression by inhibiting the Raf-1/MEK1/ERK signaling pathway of keratinocytes. Arch. Dermatol. Res. 2003;295:80–87. doi: 10.1007/s00403-003-0396-5. [DOI] [PubMed] [Google Scholar]
- 40.Wang T.T., et al. Large-scale in silico and microarray-based identification of direct 1,25-dihydroxyvitamin D3 target genes. Mol. Endocrinol. 2005;19:2685–2695. doi: 10.1210/me.2005-0106. [DOI] [PubMed] [Google Scholar]
- 41.Korinek V., et al. Constitutive transcriptional activation by a beta-catenin-Tcf complex in APC–/– colon carcinoma. . Science. 1997;275:1784–1787. doi: 10.1126/science.275.5307.1784. [DOI] [PubMed] [Google Scholar]
- 42.Lee T.C., Ziff E.B. Mxi1 is a repressor of the c-Myc promoter and reverses activation by USF. J. Biol. Chem. 1999;274:595–606. doi: 10.1074/jbc.274.2.595. [DOI] [PubMed] [Google Scholar]
- 43.Inubushi T., Kakegawa H., Kishino Y., Katunuma N. Specific assay method for the activities of cathepsin L-type cysteine proteinases. J. Biochem. 1994;116:282–284. doi: 10.1093/oxfordjournals.jbchem.a124520. [DOI] [PubMed] [Google Scholar]
- 44.Ballestar E., et al. Methyl-CpG binding proteins identify novel sites of epigenetic inactivation in human cancer. EMBO J. 2003;22:6335–6345. doi: 10.1093/emboj/cdg604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vandesompele J., et al. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002;3:RESEARCH0034. doi: 10.1186/gb-2002-3-7-research0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.