Abstract
Many eukaryotic transcription factors are bimodal in their regulatory properties and can both repress and activate expression of their target genes. These divergent transcriptional properties are conferred through recruitment of auxiliary proteins, denoted coactivators and corepressors. Repression plays a particularly critical role in the functions of the nuclear receptors, a large family of ligand-regulated transcription factors involved in metazoan development, differentiation, reproduction, and homeostasis. The SMRT corepressor interacts directly with nuclear receptors and serves, in turn, as a platform for the assembly of a larger corepressor complex. We report here that SMRT is expressed in cells by alternative mRNA splicing to yield two distinct variants or isoforms. We designate these isoforms SMRTα and SMRTτ and demonstrate that these isoforms have significantly different affinities for different nuclear receptors. These isoforms are evolutionarily conserved and are expressed in a tissue-specific manner. Our results suggest that differential mRNA splicing serves to customize corepressor function in different cells, allowing the transcriptional properties of nuclear receptors to be adapted to different contexts.
Nuclear receptors are transcription factors that play multiple roles in metazoan development and physiology (1–6). Nuclear receptors operate by binding to specific promoter elements on DNA and by modulating transcription of adjacent target genes in response to hormone ligand (3, 7–9). The nuclear receptors include, among others, the thyroid hormone receptors (TRs),1 the retinoic acid receptors (RARs), and the retinoid × receptors (RXRs) (3–5, 7, 10, 11). Each of these receptors localizes to the nucleus and binds to DNA in both the absence and presence of hormone ligand. These receptors can repress transcription of their target genes in the absence of hormone, but activate target gene transcription upon binding to hormone agonist (3, 7–9, 12, 13). This bimodal transcriptional regulation is accomplished through a hormone-regulated exchange of a corepressor complex, found on the nuclear receptor in the absence of hormone, for a coactivator complex recruited in the presence of hormone agonist (14). Corepressor and coactivator protein complexes regulate transcription through direct interaction with the basal transcription machinery and through modification of chromatin structure (15).
Both activation and repression are essential for correct receptor function. For example, RAR-mediated repression is required for appropriate anterior/posterior segregation in vertebrates, and disruption leads to aberrant head formation during murine development (16). TR-mediated repression is required for correct Xenopus larval development, and abrogation of repression leads to premature metamorphosis (17). Aberrations in the regulation of repression can result in human disease. For example, resistance to thyroid hormone syndrome, an inherited endocrine disorder, has been mapped to mutations in TRs that disrupt the hormone-driven release of corepressor (18–23); similarly, mutant RARs that fail to release corepressor correctly in response to hormone ligand play a causal role in human acute promyelocytic leukemia (24–28).
SMRT (silencing mediator of retinoic acid and thyroid hormone receptors) and its paralog, N-CoR (nuclear receptor corepressor), are central mediators of transcriptional repression by TRs, RARs, and RXRs (29–32). SMRT and N-CoR make direct contact with their nuclear receptor partners and serve, in turn, as platforms for the recruitment of additional components of a larger corepressor complex that includes histone deacetylases, TBL1, TBLR1, and GPS2 (33–37). A series of at least four “repression domains” within the N-terminal portion of SMRT and N-CoR (denoted RD1 to RD4) serve as docking surfaces for these additional corepressor subunits, whereas a series of more C-terminal receptor interaction domains (denoted S1 and S2 in SMRT and N1 to N3 in N-CoR) mediate contacts with the nuclear receptors (see Fig. 1A) (38–42). In the absence of hormone, conserved “CoRNR box” amino acid motifs, located within each of these receptor interaction domains, tether to a hydrophobic groove on the surface of the unliganded nuclear receptors (38–42). In the presence of hormone agonist, a conformational change in the C-terminal helix 12 of the nuclear receptors occludes this corepressor docking surface, causing release of SMRT or N-CoR, release of the remainder of the corepressor complex, and derepression of target gene expression (43–47). This repositioning of helix 12 by agonist simultaneously forms a new surface that recruits coactivators, thereby conferring transcriptional activation (43, 46). Many nuclear receptors bind to DNA as protein dimers, and it is believed that two CoRNR motifs within a single SMRT or N-CoR are employed in tethering these corepressors to the two receptors that compose the dimer (41, 42, 48, 49). Previous work has suggested that RAR preferentially interacts with the S2 domain, RXR preferentially interacts with the S1 domain, and TR can interact with both the S1 and S2 domains (31, 38, 42, 50).
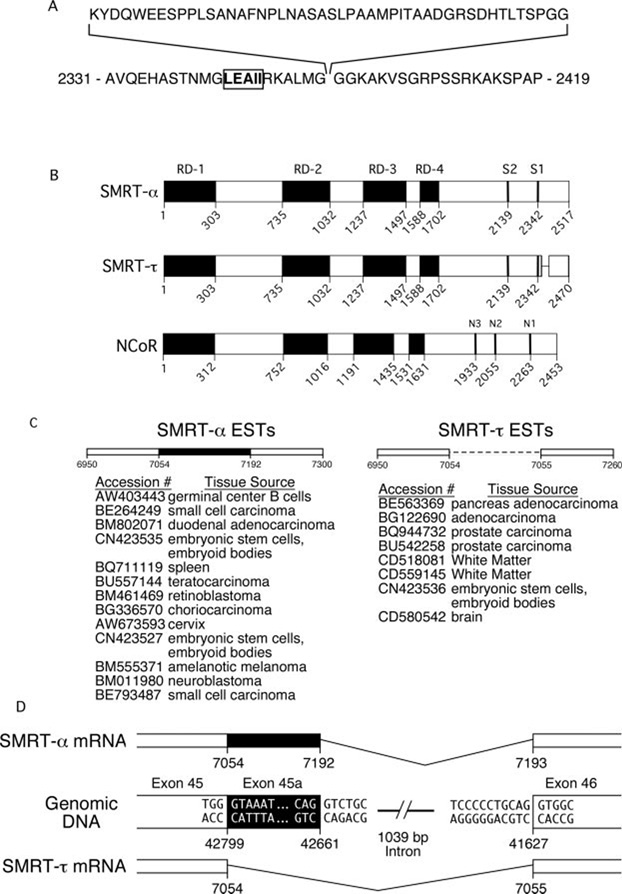
FIG. 1. Schematic representation of SMRT isoforms.
A, the amino acid sequence of the SMRTα exon and surrounding sequences. The CoRNR box sequence of the SMRT S1 domain is indicated in boldface and boxed. The numbers indicate the amino acid positions in SMRTα. B, schematic alignment of the domains of SMRTα and SMRTτ as well as N-CoR. Black boxes indicate the repression domains, and vertical bars indicate the positions of the CoRNR box sequences within each receptor interaction domain. The amino acid positions for each isoform are indicated. C, EST clones that contain sequences identical to either SMRTα or SMRTτ and the library tissue sources for each EST. D, schematic representation of the alternative mRNA splicing events that give rise to SMRTα and SMRTτ. The nucleotide positions within the open reading frames of SMRTα and SMRTτ are indicated. Nucleotide numbers within the human genome chromosome 12 HS12_9912 segment sequence are also indicated (GenBank™/EBI accession number NT_009755).
Many nuclear receptors are expressed as a series of interrelated isotypes or isoforms. For example, two genetic loci in vertebrates encode TRs (denoted TRα and TRβ); three genetic loci encode RARs (denoted RARα, RARβ, and RARγ); and three genetic loci encode RXRs (denoted RXRα, RXRβ, and RXRγ) (51–55). Alternative mRNA splicing and promoter utilization result in further diversification of the receptors that are produced from a given locus (56–60). These various nuclear receptor isotypes are expressed in tissue- and development-specific patterns, display distinct interactions with corepressors and coactivators, and exhibit distinct transcriptional properties (50, 61). N-CoR and SMRT can similarly be considered isotypes of one another, thereby paralleling the multiple isotypes found in their nuclear receptor partners. We report here a further extension of this concept by demonstrating that SMRT is itself expressed by alternative mRNA splicing to generate at least two distinct isoforms (denoted SMRTα and SMRTτ) that are expressed at different levels in different tissues (see Fig. 1A). SMRTα contains 47 amino acids in its C-terminal domain that are absent from SMRTτ (30, 31). The SMRTα-specific 47 amino acids map only 5 residues away from the S1 CoRNR box. As a consequence, although both SMRT isoforms interact nearly equally with RARα, they differ significantly in the ability to interact with and mediate repression by different isoforms of TR. We conclude that the receptor interaction properties of SMRT can be modified by alternative mRNA splicing and that, as a result, the repression properties of different nuclear receptors are likely to differ in different cell and tissue contexts. These observations also help reconcile apparent discrepancies in the literature as to the relative affinity of TRs for N-CoR versus SMRT (17, 35, 42, 48, 62, 63).
EXPERIMENTAL PROCEDURES
Plasmids
PCR was used to introduce BamHI and XhoI restriction sites at the ends of DNA fragments representing the S1 domain (amino acids 2313–2517), the S2 domain (amino acids 2077–2312), or both the S1 and S2 domains (amino acids 2077–2517) of SMRTα (GenBankTM/EBI accession number AF113003) or the corresponding fragments of SMRTτ. These PCR products were cloned into the corresponding BamHI and XhoI sites in pGEX-KG (64). For expression in vitro or in transfected mammalian cells, these receptor interaction subdomains of SMRT were transferred from the pGEX plasmids into a modified version of pSG5 (Stratagene, La Jolla, CA) containing an N-terminal Mycepitope tag. The mammalian expression plasmids pSG5-TRα1, pSG5-TRβ1, and pSG5-Gal4DBD-TRβ1HBD (where DBD is DNA-binding domain and HBD is hormone-binding domain; amino acids 177–461) were described previously (65), as were the twice reiterated chicken lysozyme F2 element- and Gal4 17-mer-luciferase reporter vectors (66).
Protein Expression and Purification
Glutathione S-transferase (GST) fusion proteins of the various SMRT receptor interaction domains were expressed in Escherichia coli strain BL21, purified by binding to glutathione-agarose (43, 64), and recovered by elution in two changes of 20 mm glutathione and 100 mm Tris-Cl (pH 8.0) for 30 min at 4 °C. Protein concentrations were determined by SDS-PAGE (67), Coomassie Blue staining (68), and scanning using an Alpha Innotech FluorChem 8900 densitometer running AlphaEaseFC Version 4.0.1 software (Alpha Innotech Corp., San Leandro, CA). Ovalbumin was analyzed in parallel as a protein concentration standard. Native TRα1, TRβ1, RARα, and RXRα were expressed in Sf9 insect cells using a recombinant baculovirus system and were prepared as described previously (69).
Computational DNA Sequence Analysis
BLAST alignments of SMRTα and SMRTτ sequences were performed using the software on the National Center for Biotechnology Information web site (available at www.ncbi.nlm.nih.gov/BLAST/) (70) and the public domain expressed sequence tag (EST) and human genomic sequence data bases. Pairwise sequence alignments were performed using the Align algorithm in Biology Workbench Version 3.2 at the San Diego Supercomputer Center (available at workbench.sdsc.edu/) (71–73).
Protein-Protein and Protein-DNA Interaction Assays
The ability of various SMRT isoforms and subdomains to bind to nuclear receptors in the presence of DNA was assayed in vitro by a electrophoretic mobility shift/supershift assay as described previously (62). Briefly, TR homodimers, RAR homodimers, RXR homodimers, or the corresponding heterodimers were formed on 32P-radiolabeled DNA probes and incubated with a range of purified GST or GST-SMRT protein concentrations. The resulting protein·DNA complexes were resolved by native gel electrophoresis and visualized using a Storm 840 PhosphorImager. The ability of the SMRT constructs to retard (“supershift”) the mobility of the nuclear receptor·DNA complexes was quantified using the PhosphorImager. Apparent dissociation constants were calculated using Prism Version 4.0 software (GraphPad Software, San Diego, CA) to fit the equation Y = Bmax·X/(Kapp + X); Bmax was constrained to be the same for both SMRTα and SMRTτ in all cases. Relative affinities are expressed as the ratio of SMRTτ Kapp to SMRTα Kapp.
Subcellular Localization of SMRTα and SMRTτ
Green fluorescent protein (GFP) fusions of full-length SMRTαand SMRTτwere generated by ligation of the appropriate corepressor coding regions into a pCMV-GFP vector.2 These constructs were introduced into CV-1 cells (1.0 × 105/well in a 6-well plate) using the Effectene transfection reagent (QIAGEN Inc., Valencia, CA) according to the manufacturer’s protocol. The cells were fixed 48 h after transfection in acetone/methanol (1:1) for 10 min. The cells (on glass coverslips) were then stained with 50 ng/ml 4′,6-diamidino-2-phenylindole to visualize nuclei, mounted on slides using 25 µl of Vectashield (Vector Laboratories, Burlingame, CA), and sealed with fingernail polish. The slides were visualized using a Nikon Microphot epifluorescence microscope. Digital images were captured with a Nikon Cool Pix 4500 digital camera.
Dominant-negative Derepression Assay
CV-1 cells (3 × 105/well) were plated in 24-well culture plates and incubated overnight in a humidified 5% CO2 atmosphere at 37 °C in Dulbecco’s modified Eagle’s medium (Invitrogen) containing 10% fetal bovine serum. The medium was then replaced with Dulbecco’s modified Eagle’s medium plus 10% fetal bovine serum (hormone-depleted); a total of 250 ng of plasmid DNA was introduced per well using the Effectene transfection reagent according to the manufacturer’s protocol. The DNA mixture included 50 ng of the appropriate luciferase reporter plasmid, 50 ng of either plasmid pSG5-TRβ1 or pSG5-Gal4DBD-TRβ1HBD, 50 ng of pCH110-β-galactosidase vector (employed as an internal transfection control), and 0–100 ng of pSG5Myc-SMRTα(S1/S2) or pSG5Myc-SMRTτ(S1/S2). Total plasmid DNA was adjusted by addition of an empty pSG5 vector so as to be equal in all samples. After an additional 24-h incubation at 37 °C, the cells were harvested, lysed, and analyzed for luciferase activity using the Promega luciferase assay system and for β-galactosidase activity as described previously (43). Duplicate transfections were immunoblotted with anti-Myc antibody (Gamma1 Laboratories, Lexington, KY). The immunoblot was visualized using horseradish peroxidase-conjugated anti-mouse IgG secondary antibody and ChemiGlow chemiluminescence substrate (Alpha Innotech Corp.). Images of the immunoblot were captured using an Alpha Innotech FluorChem 8900 densitometer and quantified using AlphaEaseFC software.
Reverse Transcription-PCR
Organs were harvested from 6-week-old male C57/BL6 mice and quick-frozen on dry ice before brief storage at −80 °C. Mouse organs were homogenized in TRIzol reagent (Invitrogen) using a T 8 ULTRA-TURRAX homogenizer (IKA Works, Inc., Wilmington, NC); total RNA was prepared according to the manufacturer’s protocol. cDNA was synthesized using random hexamer primers, 4 µg of total RNA, and avian myeloblastosis virus reverse transcriptase (Promega) as described previously (74). cDNAs corresponding to SMRTα and SMRTτ were selectively amplified by PCR using a common SMRTα/τ-Up primer (5′-caccggaacaggccttatgacc-3′) and a common SMRTα/τ-Down primer (5′-ggttgtaggggaatggcgtgg-3′). PCR was carried out for the number of cycles given in each figure legend using the following cycling parameters: 94 °C for 30 s, 62 °C for 45 s, and 72 °C for 1 min. SMRTα and SMRTτ mRNAs generated products of 442 and 301 bp, respectively, which were resolved on a 2% agarose gel (0.5× Tris borate/EDTA running buffer, 100 V for 75 min), visualized by ethidium bromide staining, and quantified using an Alpha Innotech FluorChem 8900 densitometer and AlphaEaseFC software. The identity of each PCR product was confirmed by DNA sequencing. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) cDNA (GenBank™/EBI accession number NM_008084) was amplified using the same cycle parameters described for SMRT and oligonucleotides GAPDH-Up (5′-gctgaacgggaagctcactgg-3′) and GAPDH-Down (5′-gcctgcttcaccaccttcttgatg-3′), producing a 125-bp product. Both the SMRT and GAPDH primers span introns such that genomic DNA would produce significantly larger DNA products; however, no evidence of genomic DNA contamination was observed.
RESULTS
Two Distinct Forms of SMRT That Differ in Their C-terminal Receptor Interaction Domains Are Expressed
Inspection of the known SMRT cDNA sequences revealed an interesting heterogeneity in the C-terminal corepressor domain, defined by the inclusion or exclusion of a 47-codon sequence immediately flanking the S1 receptor interaction domain (Fig. 1, A and B) (30, 31, 75, 76). To examine this phenomenon in more detail, we searched the expressed sequence tag data base (dbEST) for the 141-nucleotide sequence that encodes these additional 47 amino acids and, separately, for the 100-nucleotide sequence that represents the junction sequence created in the absence of this insert. In both cases, we found multiple entries containing sequence identical to the query sequences (Fig. 1C). Both forms of SMRT were found in ESTs from multiple species and tissue types, suggesting that both SMRT variants are expressed in a variety of contexts. We then analyzed the corresponding SMRT sequences in the human genome. A region on chromosome 12q24 proved identical to the 47-codon insert found in the SMRT variant cDNA; comparison of the EST and genomic sequences indicated that this SMRT heterogeneity was likely the product of alternative splicing, with the longer form arising from the use of an alternative 5′-donor site (Fig. 1D). The published human SMRTα sequence (also referred to as SMRTe) contains the 47-amino acid sequence (75, 76). The published SMRT cDNAs originally denoted TRAC-1 and TRAC-2 lack the 47-codon insert (31). For nomenclature purposes, we therefore refer to SMRT variants that contain the 47-amino-acid region as SMRTα and those that lack it as SMRTτ. Intriguingly, a 47-amino acid segment related to the insert found in SMRTα is present in the N-CoR paralog (15.2% amino acid identity and 54.3% similarity) (70, 72); in contrast to SMRT, however, there is no evidence for a τ-like N-CoR splice variant in the published sequence or EST data base.
SMRTα and SMRTτ Display Similar Protein Accumulation and Subcellular Localizations in Cells
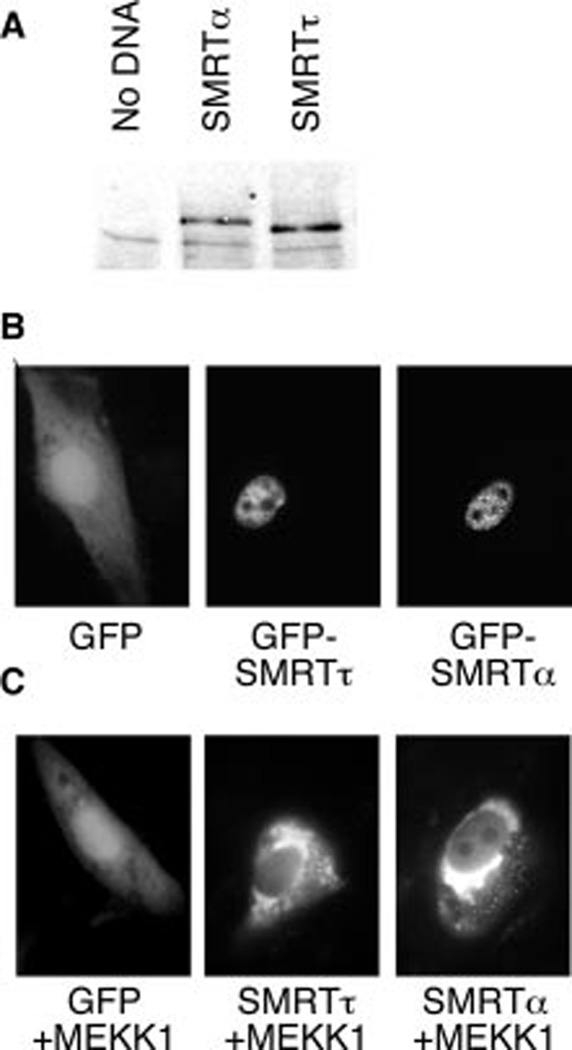
To determine the effect of the 47-amino-acid insert on SMRT function, we created expression vectors for epitope-tagged full-length versions of both the SMRTα and SMRTτ variants and introduced these into CV-1 cells by transfection. Analysis of extracts of these cells by immunoblotting revealed that both the SMRTα and SMRTτ forms were expressed as appropriately sized proteins and accumulated at steady state to similar levels (Fig. 2A). We also created GFP fusions of both SMRT variants and introduced these into CV-1 cells to visualize the subcellular localizations of these proteins. As anticipated, unmodified GFP exhibited a diffuse, primarily cytoplasmic subcellular distribution (Fig. 2B). In contrast, both GFP-SMRTα and GFP-SMRTτ were primarily nuclear in distribution, displaying a diffuse localization over the nucleoplasm that was excluded from the nucleoli (Fig. 2B). Visible within this diffuse nucleoplasmic signal, both SMRTα and SMRTτ also displayed a brighter punctate pattern (Fig. 2B) that has been noted before for SMRTα and that has been proposed to represent clusters of corepressor complexes that also contain HDAC3, TBL1, TBLR1, and GPS2 (15). We have reported previously that SMRTτ responds to the epidermal growth factor receptor/Ras/ MEKK1 cascade signaling by a change from a nuclear to a cytoplasmic distribution in many of the cells (77); SMRTα displayed a similar relocalization in response to co-introduction of an activated MEKK1 construct (Fig. 2C). We conclude that both the SMRTα and SMRTτ variants are synthesized, accumulate, distribute, and respond to growth factor signaling in similar fashions under the conditions tested.
FIG. 2. Protein accumulation and nuclear localization of SMRTα and SMRTτ.
A, Western blot of CV-1 cells either untransfected or transfected with plasmids expressing Myc-tagged full-length SMRTα or Myc-tagged SMRTτ. B, fluorescent micrograph of CV-1 cells transfected with plasmids expressing GFP, GFP-SMRTα, or GFP-SMRTτ. C, change in the subcellular localization of SMRTα or SMRTτ in response to MEKK1.
SMRTα and SMRTτ Diverge in Their Ability to Interact with TR
The 47-amino-acid insert found in the SMRTα splice variant occurs only 5 amino acids C-terminal to the CoRNR box in the S1 receptor interaction domain (Fig. 1A) (41, 42). We tested the ability of each isoform of SMRT to interact with different nuclear receptors. Previous studies have demonstrated that SMRT and N-CoR preferentially interact with nuclear receptors when the latter are bound as dimers to their respective DNA response elements and that DNA binding can influence the relative affinities of these corepressor paralogs for different nuclear receptors (41, 62, 78). We therefore used an electro-phoretic mobility shift assay (EMSA) that permitted us to investigate the interactions of the SMRT corepressors with nuclear receptors in the context of their DNA response elements. A fixed amount of receptor was added to a radiolabeled DNA probe in the presence of increasing amounts of a construct containing the receptor interaction domains of either the SMRTα or SMRTτ isoform (purified as a bacterially produced GST fusion protein). The ability of the SMRT construct to bind to and supershift the receptor·DNA complex was analyzed by native gel electrophoresis and quantified by PhosphorImager analysis.
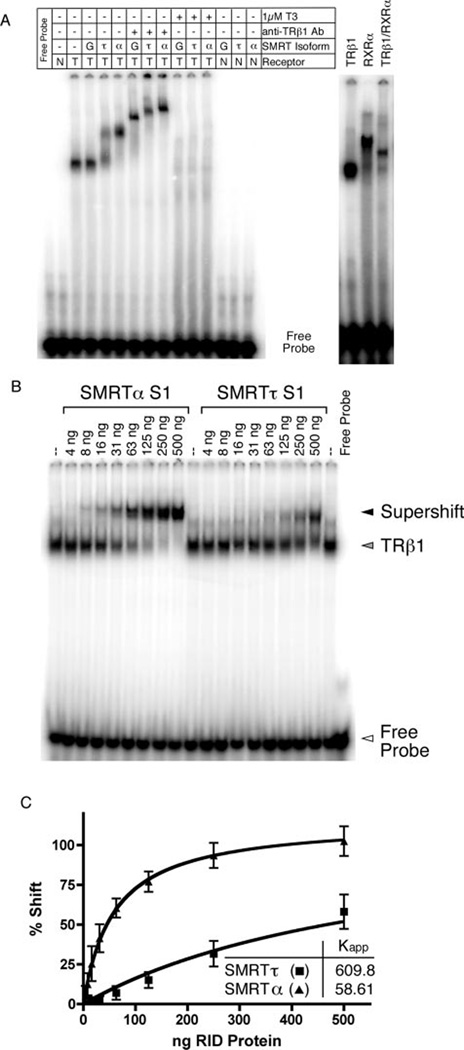
In the absence of the SMRT corepressor, TRβ1 bound to the DR4 probe as a receptor homodimer (Fig. 3A, third lane). This is consistent with prior studies (13, 62, 79, 80) and was confirmed by the following criteria. (a) Little or no protein·DNA complex was observed when using non-recombinant baculovirus/Sf9 extracts with the DR4 probe or when using the TRβ1 preparation with an irrelevant DNA probe or with a DNA probe containing only a single half-site (Fig. 3A, first and second lanes) (data not shown). (b) The TRβ1·DNA complexes were supershifted to a slower mobility by anti-TR antibody or by addition of RXR (which forms a heterodimer with TRs) (Fig. 3A, compare third and fourth lanes with seventh lane and compare sixteenth and eighteenth lanes). (c) The homodimeric TRβ1·DNA complex was destabilized by addition of triiodothyronine (T3) (Fig. 3A, compare third and fourth lanes with tenth lane). Addition of the SMRTτ isoform to the homodimeric TRβ1·DNA complex resulted in formation of a new tertiary complex indicative of an interaction between SMRTτ and the TRβ1 homodimer (Fig. 3A, compare third and fifth lanes). The amount of tertiary complex formed was proportional to the amount of GST-SMRTτ added (Fig. 3B, tenth through eighteenth lanes), and no tertiary complex formation was observed when using non-recombinant GST constructs (Fig. 3A, fourth lane). SMRTτ did not form a complex with the DNA probe in the absence of the nuclear receptor (Fig. 3A, fourteenth lane), and the SMRTτ·TRβ1·DNA complex was further supershifted by anti-TR antibody (Fig. 3A, compare fifth and seventh lanes). Finally, the SMRTτ·TRβ1·DNA complex was disrupted, as expected, by addition of T3 (Fig. 3A, compare fifth and eleventh lanes).
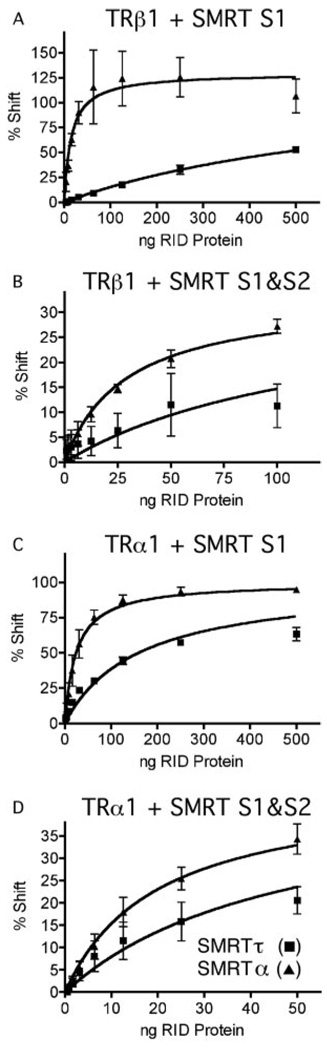
FIG. 3. EMSA interaction between TRβ1 and the SMRT S1 domains.
A, human TRβ1 derived from a recombinant baculovirus/Sf9 cell system (T) or equivalent non-recombinant preparations (N) were incubated with radiolabeled DR4 oligonucleotide DNA and GST (G), GST-SMRTα(S1) (α), or GST-SMRTτ(S1) (τ). Anti-TRβ1 antibody (Ab; catalog no. MA1–215, Affinity BioReagents, Golden, CO), 1 µM T3, and/or human RXRα (from baculovirus/Sf9 preparations) was also added to certain samples as indicated. B, varying amounts of purified GST-SMRTα(S1) or GST-SMRTτ(S1) protein were added to a constant amount of TRβ1 protein and radiolabeled DR4 probe. C, the TRβ1·DNA complexes supershifted to a slower mobility by the SMRTα or SMRTτ S1 domain (i.e. bound by SMRTα or SMRTτ) were quantified relative to the amount of SMRT protein added to each binding reaction. From these data, the apparent dissociation constants for both the SMRTα and SMRTτ S1 domains were determined. The graph represents the mean of n > 3 replicates. Error bars indicate S.E. RID, receptor interaction domain.
Having established the overall validity of the supershift EMSA, we next used it to compare the relative binding of SMRTαand SMRTτto the TRβ1·DNA complex. Notably, the S1 domain of SMRTα displayed a much higher affinity for the TRβ1·DNA complex than did the S1 domain of SMRTτ, manifested as a requirement for significantly less SMRTα S1 construct compared with SMRTτ S1 construct to generate a TRβ1·DNA supershift (Fig. 3, A, compare fifth and sixth lanes; and B, compare first through ninth lanes with tenth through eighteenth lanes). It should be noted that the amounts of SMRTα and SMRTτ constructs were carefully quantified, that the constructs were used as purified preparations at equal concentrations, and that comparable results were obtained using independently isolated preparations (data not shown). As observed with SMRTτ, the SMRTα·TRβ complex could be supershifted by anti-TR antibody (Fig. 3A, compare sixth and ninth lanes) and was dissociated by saturating T3 (compare sixth and twelfth lanes).
Quantification of repeated experiments confirmed the reproducibility of these results (Fig. 3C) and verified that the data fitted a hyperbolic binding equation. Using this model, apparent dissociation constants (Kapp) for the interaction between TRβ1 and SMRTα and SMRTτ were determined as described under “Experimental Procedures”; analogous supershift EMSAs have been used to determine the apparent affinity constants for several other transcription factors, including Sp1 and the lac and trp repressors (81–85). Based on this form of analysis TRβ1 interacted >10-fold more strongly with SMRTα than with SMRTτ (Fig. 3C and Table I). It should be noted that this mathematical representation provided a close fit to the actual data; however, the EMSA method is not a true equilibrium assay, and these numbers represent apparent rather than absolute dissociation constants. Nonetheless, these apparent dissociation constants are a useful means to describe the relative affinity of SMRTα and SMRTτ for different receptors and will be cited, acknowledging their limitations, in the remainder of this work.
TABLE I.
Affinities of SMRT isoforms for receptors
| SMRT isoforms | Kapp | S.E. | 95% CI |
|---|---|---|---|
| DR4 element | |||
| TRβ1 homodimers | |||
| S1 | |||
| τ | 609.8 | 100.5 | 406.5–813.0 |
| α | 58.61 | 11.17 | 36.01–81.22 |
| S1/S2 | |||
| τ | 230.7 | 25.92 | 178.2–283.3 |
| α | 85.03 | 10.67 | 63.39–106.7 |
| TRα1 homodimers | |||
| S1 | |||
| τ | 213.3 | 35.15 | 142.6–284.0 |
| α | 39.67 | 7.941 | 23.70–55.65 |
| S1/S2 | |||
| τ | 70.10 | 8.863 | 52.13–88.07 |
| α | 32.91 | 4.445 | 23.90–41.92 |
| Chicken lysozyme F2 element | |||
| TRβ1 homodimers | |||
| S1 | |||
| τ | 746.8 | 22.81 | 700.2–793.5 |
| α | 15.56 | 4.047 | 7.286–23.84 |
| S1/S2 | |||
| τ | 126.2 | 34.66 | 55.36–197.1 |
| α | 28.68 | 4.870 | 18.72–38.64 |
| TRα1 homodimers | |||
| S1 | |||
| τ | 158.4 | 13.30 | 130.7–186.1 |
| α | 24.58 | 2.914 | 18.53–30.59 |
| S1/S2 | |||
| τ | 47.52 | 9.925 | 27.62–67.42 |
| α | 19.84 | 3.684 | 12.45–27.23 |
| DR4 element | |||
| RXRα/TRβ1 heterodimers | |||
| S1 | |||
| τ | 542.0 | 44.23 | 447.1–638.8 |
| α | 78.21 | 7.042 | 63.10–93.31 |
| S1/S2 | |||
| τ | 846.5 | 147.4 | 545.0–1148 |
| α | 207.0 | 81.80 | 39.77–374.3 |
| RXRα/TRα1 heterodimers | |||
| S1 | |||
| τ | 1119 | 280.9 | 515.9–1721 |
| α | 574.3 | 162.6 | 225.7–923.0 |
| S1/S2 | |||
| τ | 3454 | 307.3 | 2826–4083 |
| α | 617.3 | 136.4 | 338.5–896.2 |
| DR5 element | |||
| RARα homodimers | |||
| S1 | |||
| τ | 406.8 | 34.33 | 337.8–475.8 |
| α | 127.2 | 13.40 | 100.3–154.2 |
| S2 | |||
| τ/α | 20.17 | 3.730 | 12.58–27.76 |
| S1/S2 | |||
| τ | 19.21 | 3.013 | 3.15–25.27 |
| α | 14.52 | 1.984 | 10.53–18.51 |
| RARα/RXRα heterodimers | |||
| S1 | |||
| τ | 8528 | 3083 | 1993–15064 |
| α | 3476 | 1349 | 615.3–6337 |
| S2 | |||
| τ/α | 411.0 | 74.79 | 228.0–594.0 |
| S1&S2 | |||
| τ | 261.3 | 58.10 | 136.6–385.9 |
| α | 187.9 | 44.22 | 93.10–282.8 |
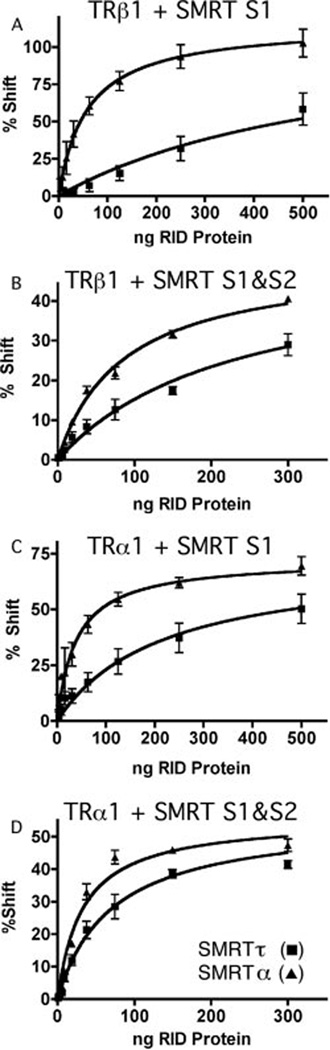
TRs can interact with either the S1 or S2 domain of SMRT, and TR homodimers are believed to be able to contact both the S1 and S2 domains simultaneously. The S2 domain is identical in the SMRTα and SMRTτ isoforms. We therefore also examined the effect of the isoform-specific differences in the S1 domain when tested in combination with the S2 domain. Whereas the difference in the relative affinities of the SMRTα and SMRTτ isoforms for TRβ1 was most readily observed using GST-SMRT constructs limited to the S1 domain (Fig. 4A), it was still easily discernable with GST-SMRT constructs containing both the S1 and S2 domains (Fig. 4B). In the latter assays, TRβ1 had a 2.7-fold greater affinity for the S1 and S2 domains of SMRTα compared with those of SMRTτ.
FIG. 4. Interaction between SMRT receptor interaction domains and TRα1 or TRβ1 on a DR4 DNA element.
Varying amounts of purified GST-SMRTα(S1) or GST-SMRTτ (S1) were added to binding reactions containing TRβ1 (A), or TRα1 (C) together with a radiolabeled DR4 oligonucleotide probe. Alternatively, varying amounts of purified GST-SMRTα(S1/S2) or GST-SMRTτ(S1/S2) were added to binding reactions containing TRβ1 (B) or TRβ1 (D) together with the radiolabeled DR4 oligonucleotide probe. The receptor-DNA complexes supershifted by addition of the SMRT constructs were quantified relative to the amount of SMRT protein added to each binding reaction. Error bars indicate S.E. of three replicate experiments. RID, receptor interaction domain.
TRs are expressed as two different isotypes from two distinct genetic loci: TRβ and TRα. To determine whether the preference displayed by SMRTα for TRβ1 extends to the other TR isotype, we repeated our EMSA experiments using TRα1 in place of TRβ1. TRα1 had a 5.1-fold greater affinity for GST-SMRTα(S1) and a 2.1-fold greater affinity for GST-SMRTα(S1/S2) than for equivalent constructs of SMRTτ (Fig. 4, C and D; and Table I). Controls confirmed that non-recombinant GST failed to super-shift the TRα1·DNA complex and that the SMRT·TRα·DNA complex was dissociated by addition of T3 (data not shown). We conclude that SMRTα exhibits a significantly stronger interaction with TRs than does SMRTτ and that this preference for SMRTα over SMRTτ can be observed for both TRα1 and TRβ1.
The Relative Affinities of SMRTα and SMRTτ for TRs Are Further Influenced by the Nature of the DNA Response Element and by Heterodimer Formation
In addition to the prototypic DR4 element studied above, TRs also interact with and regulate transcription of target genes through divergent repeats separated by a 6-base spacer (referred to as DIV6), such as that found in the lysozyme F2 silencer element. Given that receptor binding to DNA can alter the relative affinity of different nuclear receptors for SMRT versus N-CoR, we determined whether the nature of the DNA response element could also affect the preference of TRs for SMRTα versus SMRTτ. We examined the affinity of TRβ1 and TRα1 for SMRT using a probe containing the DIV6 element. On this element, TRβ1 displayed a 48-fold greater affinity for the SMRTα S1 domain (a 4.4-fold greater affinity for the S1/S2 domains of SMRTα) than for the equivalent SMRTτ construct (Fig. 5, A and B; and Table I). TRα1 also exhibited an increased ability to discriminate between SMRTα and SMRTτ on the DIV6 element. TRα1 had a 6.4-fold greater affinity for the SMRTα S1 domain and an ~2.4-fold greater affinity for the S1/S2 domains of SMRTα than for SMRTτ (Fig. 5, C and D; and Table I). The ability of TRs arrayed on the DIV6 lysozyme F2 element to bind strongly to SMRT is consistent with studies implicating this element in TR-mediated repression in the absence of hormone (e.g. Ref. 62).
FIG. 5. Interaction between SMRT receptor interaction domains and TRα1 or TRβ1 on a DIV6 DNA element.
Varying amounts of purified GST-SMRTα(S1) or GST-SMRTτ(S1) were added to binding reactions containing TRβ1 (A) or TRα1 (C) together with a radiolabeled DIV6 oligonucleotide probe. Alternatively, varying amounts of purified GST-SMRTα(S1/S2) or GST-SMRTτ(S1/S2) were added to binding reactions containing TRβ1 (B) or TRα1 (D) together with the radiolabeled DIV6 oligonucleotide probe. The receptor-DNA complexes supershifted by addition of the SMRT constructs were quantified relative to the amount of SMRT protein added to each binding reaction. Error bars indicate S.E. of two or more replicate experiments. RID, receptor interaction domain.
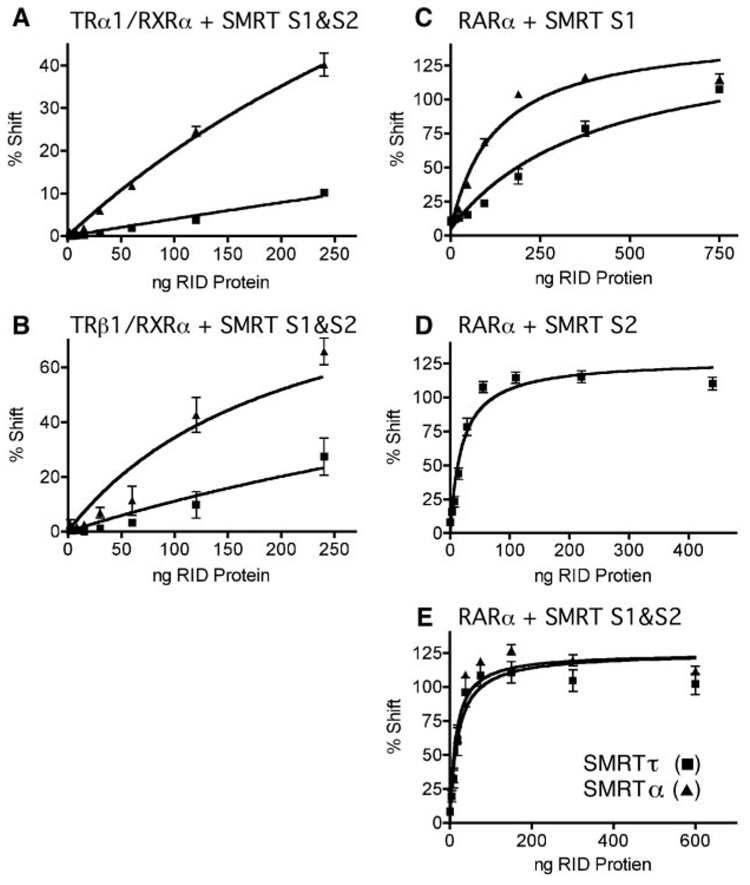
The above experiments were all performed using TR homodimers. We next examined the ability of heterodimers of TR and RXR to recruit SMRTα versus SMRTτ. Notably, heterodimers of TR and RXR have been implicated in transcriptional activation rather than repression (62). Consistent with these previous results, we observed that the SMRTτ(S1/S2) construct interacted with RXRα/TRα heterodimers with significantly less avidity than with TRα homodimers (Fig. 6A). In contrast, the SMRTα(S1/S2) construct not only interacted with TRα homodimers more strongly than did the SMRTτ form, but also interacted strongly with RXRα/TRα heterodimers (Fig. 6A). The ability of the SMRTα construct to interact with heterodimers was somewhat more pronounced for RXRα/TRβ1 than for RXRα/TRβ (Fig. 6B). These results suggest that the nature of the SMRT isoform has significant influence on the ability of the corepressor to be recruited by heterodimeric versus homodimeric versions of TRs.
FIG. 6. Interaction between SMRT receptor interaction domains and RXRα/TR heterodimers or RARα homodimers.
Varying amounts of purified GST-SMRTα(S1/S2) or GST-SMRTτ(S1/S2) were added to binding reactions containing TRα1 and RXRα (A) or TRβ1 and RXRα (B) and a DR4 oligonucleotide probe. Alternatively, varying amounts of purified GST-SMRTα(S1) or GSTSMRT α(S1) (C), GST-SMRT(S2) (D), or GST-SMRTα(S1/S2) or GST-SMRTτ(S1/S2) (E) were added to binding reactions containing RARα and a radiolabeled DR5 oligonucleotide probe. The receptor·DNA complexes supershifted by addition of the SMRT constructs were quantified relative to the amount of SMRT protein added to each binding reaction. Error bars indicate S.E. of three replicate experiments. RID, receptor interaction domain.
Previous reports have suggested that N-CoR has a much greater affinity for TRs than does SMRT, leading to the suggestion that N-CoR serves as the preferred corepressor partner for this receptor in vivo (15). We re-examined this question in light of the enhanced interaction properties of the SMRTα isoform. We compared the avidities of the SMRTα(S1/S2) and N-CoR(N1/N2/N3) constructs for TRα1 and TRβ1. Under these conditions, N-CoR displayed only a 1.8 –2.7-fold higher apparent affinity for TRs than did SMRTα, a much smaller difference than that seen upon comparison of N-CoR and SMRTτ. (data not shown). We suggest that at least some of the prior reports demonstrating much greater discrepancies between N-CoR and SMRT may have employed SMRTτ and that SMRTα has the potential to function as an authentic corepressor partner for TRs, if at a slightly lower efficiency than N-CoR.
Unlike TRα1 and TRβ1, RARα Interacts Nearly Equally with Both the SMRTα and SMRTτ Constructs
TRs can interact with either the S1 or S2 domain of SMRT; in contrast, RARα preferentially interacts with the SMRTτ S2 domain and displays comparatively less binding to the S1 domain of this isoform (31, 41). Consistent with these reports, we observed a significantly weaker interaction of RARα homodimers with the S1 domain of SMRTτ compared with the S2 domain of SMRT (which is invariant in both SMRTα and SMRTτ) (Fig. 6, compare C and D; note the change in scale of the ordinate). Although the interaction of RARα homodimers with the isolated S1 domain of SMRTα was somewhat stronger than that with the isolated S1 domain of SMRTτ (Fig. 6C), this modest SMRT isoform specificity was abolished when the ability of RARα homodimers to interact with the combined S1 and S2 domains of SMRT was assayed (Fig. 6E and Table I). RARα heterodimers formed with RXRα also bound efficiently to the S1 and S2 domains of both SMRTα and SMRTτ (although at some-what reduced levels compared with the corresponding RARα homodimers) (Table I). We conclude that, unlike TRs, there is relatively little effect of this SMRT mRNA splicing event on the ability of the corepressor to bind to RARα.
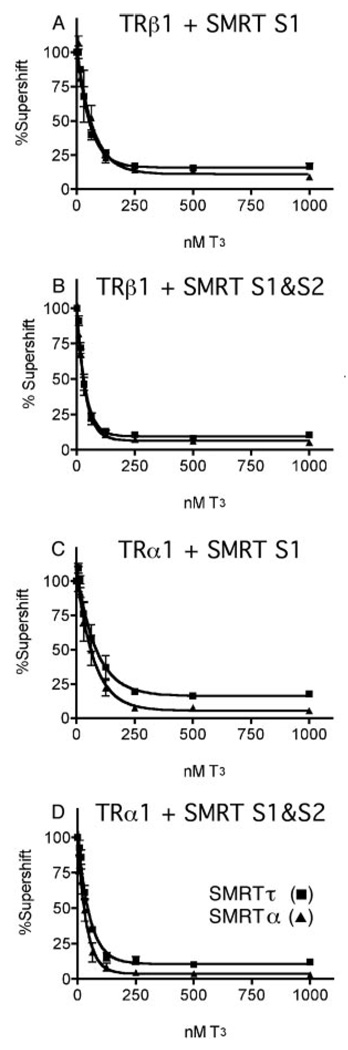
SMRTα and SMRTτ Are Indistinguishable in Their Ability to Be Released from TRs in Response to T3
Given the higher affinity of SMRTα compared with SMRTτ for TRs in the absence of hormone, we wished to determine whether these corepressor isoforms might also differ in their ability to be released from TR in response to T3 agonist. Increasing amounts of T3 (8–100 nm) were added to parallel supershift EMSA reactions, and the amount of SMRT·TR complex was determined and quantified as described above (Fig. 7). No significant difference could be detected in the amount of T3 required to release either isoform of SMRT from either TRα1 or TRβ1 (Fig. 7). Similar results were obtained using a GST pull-down assay in the absence of DNA (data not shown). We conclude that, whereas SMRTα displays a higher affinity for TRs than does SMRTτ in the absence of hormone, this does not affect the T3-induced conformational change in the receptor that is responsible for agonist-driven corepressor release and that both SMRT isoforms are efficiently displaced from TR in response to hormone.
FIG. 7. Disruption of SMRT·TR complexes with thyroid hormone.
A fixed amount of purified GST-SMRTα(S1) or GST-SMRTτ(S1) were added to binding reactions containing TRβ1 (A) or TRα1 (C) together with a radiolabeled DR4 oligonucleotide probe. Alternatively, a fixed amount of purified GST-SMRTα(S1/S2) or GST-SMRTτ(S1/S2) were added to binding reactions containing TRβ1 (B) or TRα1 (D) together with the radiolabeled DR4 probe. Varying amounts of T3 were added to each reaction. The SMRT-TR-DNA complexes formed in the presence of the differing T3 concentrations were quantified. (SMRT-TR-DNA complexes observed in the absence of hormone were defined as 100%.) Error bars for each data point indicate S.E. (n = 3).
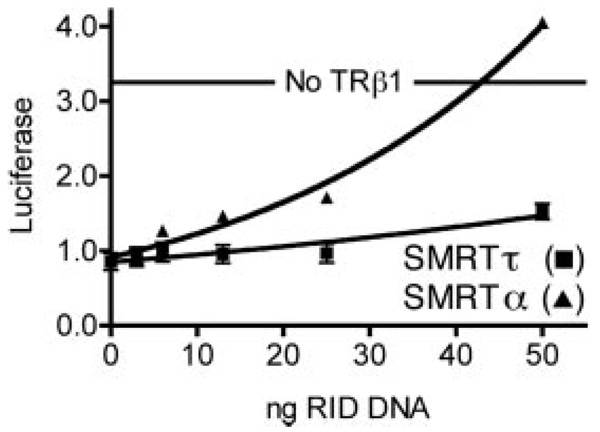
TR-mediated Repression in Vivo Is Preferentially Enhanced by SMRTα Relative to SMRTτ
Given the difference in the ability of TRβ1 to interact with SMRTα versus SMRTτ in vitro, we investigated whether this would manifest itself as a different ability of each SMRT isoform to affect TR-mediated repression. Using native TR constructs, we and others have been unable to demonstrate enhanced repression by simple overexpression of SMRT or N-CoR proteins (30, 31). This is presumably due to the fact that these corepressors function within much larger multiprotein complexes and that endogenous N-CoR and SMRT are already in excess to the other components of this complex. A method that has been employed to overcome this problem in the analysis of corepressor function is to overexpress just the receptor interaction domains of SMRT or NCoR. In the absence of the N-terminal repression domains, these abstracted receptor interaction domains cannot mediate repression, but instead operate as dominant-negative inhibitors of endogenous corepressor function.
To this end, we transfected CV-1 cells, which lack endogenous TRs, with TRβ1, with a luciferase reporter containing the DIV6 element from the chicken lysozyme promoter, and with increasing amounts of Myc-SMRTα(S1/S2) or Myc-SMRTτ(S1/S2). As anticipated, introduction of TRβ1 in the absence of the dominant-negative SMRT constructs repressed the luciferase reporter, presumably by recruiting the native corepressors present in these cells (Fig. 8). Co-introduction of the SMRT receptor interaction domain constructs reversed this TR-mediated repression, and the SMRTα construct was significantly more effective at doing so compared with the SMRTτ construct (Fig. 8). Immunoblotting confirmed that the stronger dominant-negative actions of the SMRTα construct were inherent properties of this protein and were not accounted for by differences in the levels of expression of the two SMRT isoform constructs (data not shown). We also performed the same dominant-negative repression assay using a synthetic construct containing the ligand-binding domain of TRβ1 fused to a Gal4 DNA-binding domain and a luciferase reporter gene with a synthetic Gal4 response element in the promoter. We obtained comparable results: SMRTα was significantly more effective at reversing Gal4-TRβ1-mediated repression than was SMRTτ (data not shown).
FIG. 8. Dominant-negative inhibition of TRβ1-mediated repression.
CV-1 cells were transfected with a TRβ1 expression vector, the lysozyme F2 element-luciferase reporter, and varying amounts of either a Myc-SMRTα(S1/S2) or Myc-SMRTτ(S1/S2) expression vector. Transfected cells were analyzed for luciferase activity. Luciferase activity for each sample is plotted versus the amount of SMRT expression vector. The unrepressed level of luciferase activity (no TRβ1) is indicated. Error bars indicate S.E. of three replicate experiments. RID, receptor interaction domain.
SMRTα and SMRTτ Are Expressed in Tissue-specific Patterns in Vivo
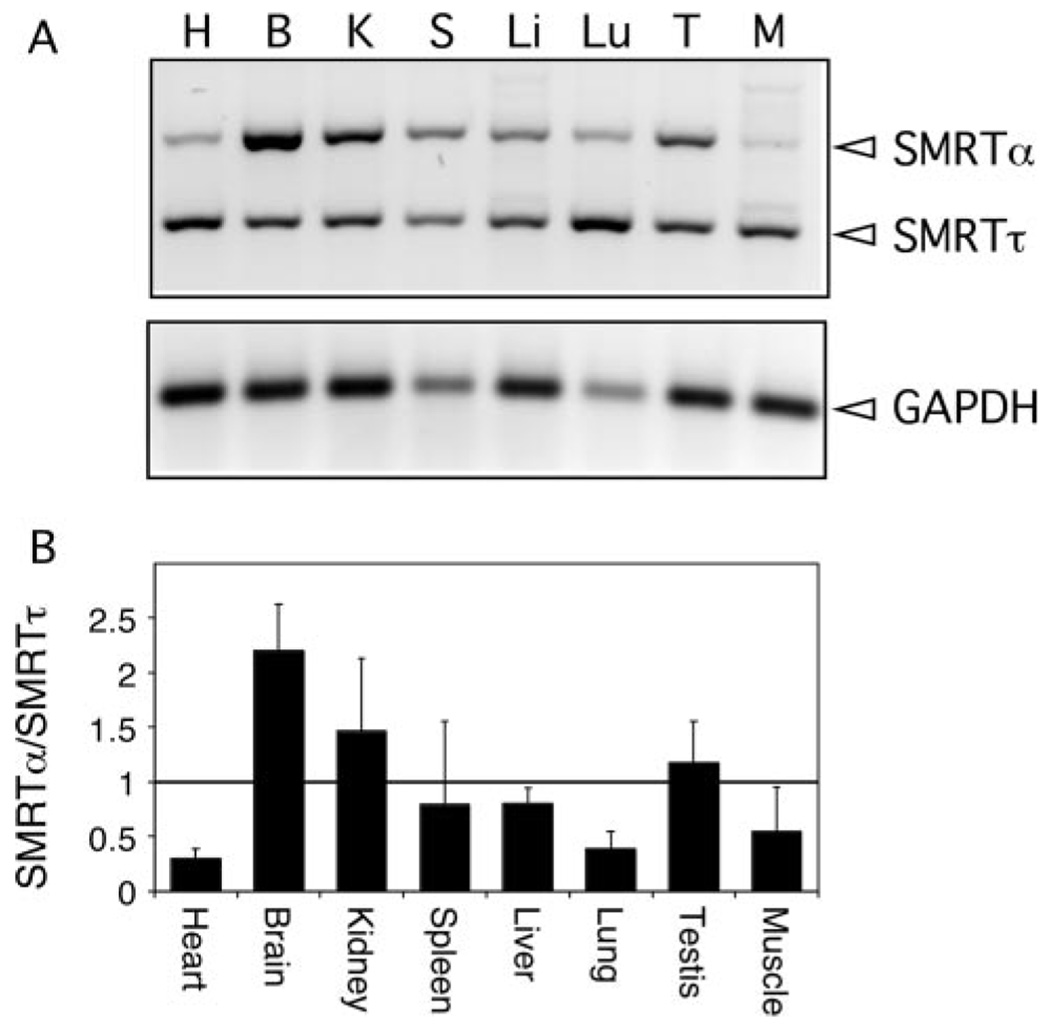
We next examined the expression pattern of SMRTα and SMRTτ in different mouse tissues. Organs or tissues were dissected from adult mice and pooled, and total RNA was isolated. We then analyzed these RNA samples by reverse transcription-PCR using primers that flank the alternative splice site (Fig. 9A); in this fashion, both spliced forms could be visualized simultaneously in a single analysis. The identity of both PCR products as SMRTα and SMRTτ was confirmed by isolating and sequencing the amplified DNA band from the electrophoretogram. Levels of GAPDH mRNA were also determined for each tissue to normalize for mRNA recovery and cDNA synthesis efficiency. Notably, mRNAs corresponding to SMRTα and SMRTτ were detectably expressed in all tissues tested; however, the relative amounts of each isoform varied considerably among tissues (Fig. 9B). Heart, lung, and skeletal muscle all expressed higher levels of SMRTτ, whereas brain, kidney, and testis expressed higher levels of SMRTα. Comparable results were obtained using an RNase protection assay (data not shown). We conclude that the ratio of SMRTα to SMRTτ differs significantly in different cell types and, based on these and our other results, suggest that the ability of a given nuclear receptor to recruit SMRT and to repress transcription is likely to differ correspondingly in different tissues.
FIG. 9. Analysis of expression of SMRTα and SMRTτ in various mouse tissues.
A, cDNAs from heart (H), brain (B), kidney (K), spleen (S), liver (Li), lung (Lu), testis (T), or skeletal muscle (M) were amplified with primers that span the SMRTα exon; these are expected to produce a 442-bp product for SMRTα and a 301-bp product for SMRTτ. SMRT samples were amplified for 30 cycles. The cDNA from the same mouse tissues was also amplified for 20 cycles using a primer for GAPDH, which produces a 125-bp product. B, samples from duplicate reactions from two mice were analyzed using an Alpha Innotech FluorChem 8900 densitometer. Averages of the ratio of SMRTα to SMRTτ and S.E. for each are plotted (n > 3).
DISCUSSION
The SMRT Corepressor Is Expressed as at Least Two Distinct Isoforms That Differ in Their Receptor Interaction Domains
In this study, we have provided evidence for the existence of two distinct isoforms of the corepressor SMRT that differ by the presence or absence of an in-frame 47-amino acid insert in the most C-terminal receptor interaction domain (S1 domain). This conclusion (based on alignment of published SMRT sequences) was further confirmed by analysis of data from the EST data base. Additional characterization of genomic sequence data revealed the presence of consensus splice donor sequences flanking this 141-base insert, indicating that the two SMRT isoforms arise from alternative mRNA splicing through the use of alternative 5′-splice donor sites. We refer to the longer form as SMRTα and to the shorter form as SMRTτ. When introduced ectopically, these two different SMRT isoforms accumulated in cells to comparable extents and exhibited similar stabilities. Both isoforms displayed very similar nuclear localizations when expressed as GFP fusions, observable as a punctate pattern within a more diffuse nucleoplasmic distribution; both forms of SMRT were excluded from nucleoli. This pattern is consistent with prior descriptions of the subcellular localization of both SMRT and N-CoR using GFP fusions, such as those employed here, or by immunofluorescent visualization of the endogenous corepressors (15).
SMRTα and SMRTτ Differ in Their Interactions with Different Nuclear Receptors
Notably, the alternative spliced sequences in SMRTα and SMRTτ map within the S1 receptor interaction domain of the corepressor, only 5 amino acids down-stream from a CoRNR box that serves as a key binding surface between corepressor and its nuclear receptor partners. The CoRNR box itself appears to form an α-helical domain that fits into a docking surface composed of helices 3, 5, and 6 of the receptor hormone-binding domain. Corepressor sequences immediately flanking the CoRNR box are known to play a role in defining the specificity of the corepressor for different nuclear receptors; and indeed, SMRTα and SMRTτ differ substantially in their affinity for TRs bound to DNA response elements. The most dramatic differences were seen in the relative affinity of the two SMRT isoforms for TRβ1 homodimers bound to the lysozyme F2 element, with SMRTα displaying a 48-fold greater interaction compared with SMRTτ. SMRTα also exhibited a strong preference for TRα. The preferential ability of the SMRTα isoform to interact with TRs in vitro was also manifested in vivo as the greater ability of a dominant-negative SMRTα construct to inhibit TR-mediated repression relative to a comparable SMRTα construct.
In contrast to TRα and TRβ, the relative affinities of SMRTα and SMRTτ (assayed as the combined S1 and S2 domains) for RARα are virtually indistinguishable. Previous work has shown that, whereas TRs interact with both the S1 and S2 domains of SMRT, RARs interact primarily with the S2 domain (15). The S2 domain of SMRT is unaltered by the alternative splicing event that generates the SMRTα and SMRTτ isoforms; and therefore, it is not unexpected that both SMRT isoforms exhibit comparable interactions with RARα homodimers. The RXR molecule in an RAR/RXR heterodimer is thought to interact with the corepressor S1 domain, leaving the S2 domain available to interact with the RAR moiety; nonetheless RARα/RXRα heterodimers also displayed nearly equal apparent affinities for SMRTα and SMRTτ in our assays.
The Nature of the DNA-binding Site Can Influence the Apparent Affinity of SMRTα and SMRTτ for the Nuclear Receptor Partner
Previous work has demonstrated that binding of a nuclear receptor to DNA can influence its ability to interact with corepressor (48). Although RARs and TRs exhibit nearly equal affinities for SMRT and N-CoR in the absence of a DNA response element (i.e. in a GST pull-down or two-hybrid assay), these receptors display preferential interaction with SMRT or N-CoR when assayed as receptor dimers bound to DNA in a supershift EMSA protocol (42). A similar phenomenon was observed here in our studies of the two different SMRT isoforms. When assayed in a GST pull-down assay in the absence of DNA, SMRTα displayed a somewhat lower ability compared with SMRTτ to bind to TRs (data not shown), yet SMRTα displayed a much higher apparent affinity for TRs when bound to DNA in the EMSA protocol. Consistent with the importance of the DNA-binding site in determining corepressor specificity, we also determined that the preference of both the TRα1 and TRβ1 homodimers for SMRTα over SMRTτ was highest for a DIV6 DNA element (i.e. as found in the lysozyme F2 promoter) and reproducibly less for the same receptors bound to the DR4 DNA element. This suggests that the nature of the DNA response element has the potential to influence the identity of the corepressor recruited to that element. This DNA modulation of corepressor recruitment may operate through an allosteric mechanism by which DNA binding alters the conformation or accessibility of the corepressor docking surface on the receptor, or it may arise from the alterations in the topology of the receptor dimer when arrayed on response elements bearing different half-site orientations and/or spacings.
The Identification of Two Isoforms of SMRT Supports a Role for SMRT, as Well as N-CoR, in TR-mediated Repression
The role of SMRT in TR-mediated repression has been of some debate. SMRT was first isolated by a yeast two-hybrid screen using a TR allele as “bait,” and both SMRT and N-CoR interact strongly with TRs in two-hybrid and GST pull-down procedures (29–31). It was also noted, however, that TRs preferentially interact with N-CoR over SMRT when bound to DNA response elements, leading to the suggestion that N-CoR, and not SMRT, is the primary effector of TR-mediated repression in cells (42). Paradoxically, both SMRT and N-CoR were found associated with TRs at physiologically relevant, T3-regulated promoters during Xenopus laevis metamorphosis using chromosome immunoprecipitation (35). Differences in the isoform of SMRT employed in these analyses could account for at least some of these differences in the reported ability of SMRT to interact with TR. In our own experiments, SMRTτ interacted with TRs quite weakly compared with N-CoR, whereas SMRTα and N-CoR both interacted with TRs strongly. Notably, a recent publication confirmed that the alternative mRNA splicing event that gives rise to the SMRTα and SMRTτ isoforms is conserved in Xenopus as well as in mammals and that SMRTα predominates at least at some stages of Xenopus development (86). We suggest that SMRTα is the isoform most likely associated with TR in the previously published studies of Xenopus metamorphosis (35, 86).
Alternative mRNA Splicing Is a General Means of Diversifying Corepressor Expression
In common with SMRT, there are also alternatively spliced isoforms of N-CoR that differ in their interaction with nuclear receptors (87–89). Intriguingly, this alternative mRNA splicing of N-CoR operates through a very distinct mode from that described here for SMRT. There is no evidence in the EST data base for a form of N-CoR bearing a splice junction equivalent to that of SMRTτ (i.e. virtually all identified N-CoR clones contain the SMRTα-like 47-amino acid region within the N1 interaction domain). Instead, N-CoR is expressed as two isoforms (denoted N-CoR and RIP13Δ1) that differ by the inclusion or exclusion of a third receptor interaction domain (N3) that is absent from all known SMRT isolates (32). The longer N-CoR isoform interacts strongly with TRs, primarily through N3 and N2 contacts, whereas the alternatively spliced RIP13Δ1 isoform lacks the N3 receptor interaction domain and interacts only weakly with TRs (32, 89). Conversely, RIP13Δ1 interacts more strongly than does N-CoR with the orphan nuclear receptors COUP-TF (chicken ovalbumin upstream promoter transcription factor) and Rev-ErbA (90, 91). Thus, SMRT receptor specificity is regulated by altering the amino acid sequences flanking the S1 CoRNR box; N-CoR receptor specificity is regulated by insertion or removal of a supernumery third CoRNR box.
Although the focus of this study has been on splicing events that alter the receptor specificity of SMRT (and N-CoR), additional alternative splicing events that map outside the receptor interaction domains are also known for these two corepressors. TRAC-1 and SMRTβ variants that lack various portions of the N-terminal repression domains found in SMRTα and N-CoR have been reported; the physiological significance of these putative isoforms has not been fully established (31, 75, 76). Similarly, a RIP13a variant of N-CoR that has an altered RD-3 has been reported (32). Inspection of the EST data base provides evidence for still additional variants of both N-CoR and SMRT,3 and a recent report using both bioinformatic and reverse transcription-PCR approaches has confirmed that a multiplicity of alternatively spliced SMRT mRNAs are expressed in vertebrates (86).
In conclusion, two distinct genetic loci in vertebrates (SMRT and N-CoR) give rise to two distinct corepressor paralogs. Alternative splicing of each of these loci gives rise to a still broader repertoire of corepressor proteins that differ in their receptor specificities and (potentially) transcriptional regulatory properties. These diverse corepressor variants are expressed at different levels in different cell types. We suggest that, by regulating the expression of corepressor isoforms in a tissue-dependent manner, the cellular response to different hormones can be modulated. This may contribute to the known tissue selectivity of hormone response and the tissue-selective effects of nuclear receptor-mediated repression during development.
Footnotes
This work was supported in part by United States Public Health Service Grant DK53538 from NIDDK. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The abbreviations used are: TRs, thyroid hormone receptors; RARs, retinoic acid receptors; RXRs, retinoid × receptors; GST, glutathione S-transferase; EST, expressed sequence tag; GFP, green fluorescent protein; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; MEKK1, mitogen-activated protein kinase/extracellular signal-regulated kinase kinase kinase-1; EMSA, electrophoretic mobility shift assay; T3, triiodothyronine.
Jonas, B. A., and Privalsky, M. L. (2004) J. Biol. Chem. 279, 54676–54686.
M. L. Goodson and M. L. Privalsky, unpublished data.
REFERENCES
- 1.Sucov HM, Evans RM. Mol. Neurobiol. 1995;10:169–184. doi: 10.1007/BF02740674. [DOI] [PubMed] [Google Scholar]
- 2.Yen PM. Physiol. Rev. 2001;81:1097–1142. doi: 10.1152/physrev.2001.81.3.1097. [DOI] [PubMed] [Google Scholar]
- 3.Tsai MJ, O’Malley BW. Annu. Rev. Biochem. 1994;63:451–486. doi: 10.1146/annurev.bi.63.070194.002315. [DOI] [PubMed] [Google Scholar]
- 4.Zhang J, Lazar MA. Annu. Rev. Physiol. 2000;62:439–466. doi: 10.1146/annurev.physiol.62.1.439. [DOI] [PubMed] [Google Scholar]
- 5.Kastner P, Mark M, Chambon P. Cell. 1995;83:859–869. doi: 10.1016/0092-8674(95)90202-3. [DOI] [PubMed] [Google Scholar]
- 6.Gronemeyer H, Miturski R. Cell. Mol. Biol. Lett. 2001;6:3–52. [PubMed] [Google Scholar]
- 7.Chambon P. FASEB J. 1996;10:940–954. [PubMed] [Google Scholar]
- 8.Mangelsdorf DJ, Thummel C, Beato M, Herrlich P, Schutz G, Umesono K, Blumberg B, Kastner P, Mark M, Chambon P, Evans RM. Cell. 1995;83:835–839. doi: 10.1016/0092-8674(95)90199-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Glass CK. J. Endocrinol. 1996;150:349–357. doi: 10.1677/joe.0.1500349. [DOI] [PubMed] [Google Scholar]
- 10.Mangelsdorf DJ, Evans RM. Cell. 1995;83:841–850. doi: 10.1016/0092-8674(95)90200-7. [DOI] [PubMed] [Google Scholar]
- 11.Whitfield GK, Jurutka PW, Haussler CA, Haussler MR. J. Cell. Biochem. 1999;532:110–122. doi: 10.1002/(sici)1097-4644(1999)75:32+<110::aid-jcb14>3.0.co;2-t. [DOI] [PubMed] [Google Scholar]
- 12.Aranda A, Pascual A. Physiol. Rev. 2001;81:1269–1304. doi: 10.1152/physrev.2001.81.3.1269. [DOI] [PubMed] [Google Scholar]
- 13.Zhang XK, Wills KN, Graupner G, Tzukerman M, Hermann T, Pfahl M. New Biol. 1991;3:169–181. [PubMed] [Google Scholar]
- 14.Glass CK, Rosenfeld MG. Genes Dev. 2000;14:121–141. [PubMed] [Google Scholar]
- 15.Privalsky ML. Annu. Rev. Physiol. 2004;66:315–360. doi: 10.1146/annurev.physiol.66.032802.155556. [DOI] [PubMed] [Google Scholar]
- 16.Koide T, Downes M, Chandraratna RA, Blumberg B, Umesono K. Genes Dev. 2001;15:2111–2121. doi: 10.1101/gad.908801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sachs LM, Jones PL, Havis E, Rouse N, Demeneix BA, Shi YB. Mol. Cell. Biol. 2000;22:8527–8538. doi: 10.1128/MCB.22.24.8527-8538.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Matsushita A, Misawa H, Andoh S, Natsume H, Nishiyama K, Sasaki S, Nakamura H. J. Endocrinol. 2000;167:493–503. doi: 10.1677/joe.0.1670493. [DOI] [PubMed] [Google Scholar]
- 19.Yoh SM, Privalsky ML. Mol. Cell. Endocrinol. 2000;159:109–124. doi: 10.1016/s0303-7207(99)00201-4. [DOI] [PubMed] [Google Scholar]
- 20.Yen PM. Trends Endocrinol. Metab. 2003;14:327–333. doi: 10.1016/s1043-2760(03)00114-0. [DOI] [PubMed] [Google Scholar]
- 21.Yoh SM, Chatterjee VK, Privalsky ML. Mol. Endocrinol. 1997;11:470–480. doi: 10.1210/mend.11.4.9914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chatterjee VK. Biochem. Soc. Trans. 2001;29:227–231. doi: 10.1042/bst0290227. [DOI] [PubMed] [Google Scholar]
- 23.Huber BR, Desclozeaux M, West BL, Cunha-Lima ST, Nguyen HT, Baxter JD, Ingraham HA, Fletterick RJ. Mol. Endocrinol. 2003;17:107–116. doi: 10.1210/me.2002-0097. [DOI] [PubMed] [Google Scholar]
- 24.Tomita A, Buchholz DR, Obata K, Shi YB. J. Biol. Chem. 2003;278:30788–30795. doi: 10.1074/jbc.M303309200. [DOI] [PubMed] [Google Scholar]
- 25.Segalla S, Rinaldi L, Kilstrup-Nielsen C, Badaracco G, Minucci S, Pelicci PG, Landsberger N. Mol. Cell. Biol. 2003;23:8795–8808. doi: 10.1128/MCB.23.23.8795-8808.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lin RJ, Egan DA, Evans RM. Trends Genet. 1999;15:179–184. doi: 10.1016/s0168-9525(99)01710-2. [DOI] [PubMed] [Google Scholar]
- 27.Lin RJ, Sternsdorf T, Tini M, Evans RM. Oncogene. 2001;20:7204–7215. doi: 10.1038/sj.onc.1204853. [DOI] [PubMed] [Google Scholar]
- 28.Hong SH, David G, Wong CW, Dejean A, Privalsky ML. Proc. Natl. Acad. Sci. U. S. A. 1997;94:9028–9033. doi: 10.1073/pnas.94.17.9028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Horlein AJ, Naar AM, Heinzel T, Torchia J, Gloss B, Kurokawa R, Ryan A, Kamei Y, Soderstrom M, Glass CK, Rosenfeld MG. Nature. 1995;377:397–404. doi: 10.1038/377397a0. [DOI] [PubMed] [Google Scholar]
- 30.Chen JD, Evans RM. Nature. 1995;377:454–457. doi: 10.1038/377454a0. [DOI] [PubMed] [Google Scholar]
- 31.Sande S, Privalsky ML. Mol. Endocrinol. 1996;10:813–825. doi: 10.1210/mend.10.7.8813722. [DOI] [PubMed] [Google Scholar]
- 32.Seol W, Mahon MJ, Lee YK, Moore DD. Mol. Endocrinol. 1996;10:1646–1655. doi: 10.1210/mend.10.12.8961273. [DOI] [PubMed] [Google Scholar]
- 33.Li J, Wang J, Nawaz Z, Liu JM, Qin J, Wong J. EMBO J. 2000;19:4342–4350. doi: 10.1093/emboj/19.16.4342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Guenther MG, Lane WS, Fischle W, Verdin E, Lazar MA, Shiekhattar R. Genes Dev. 2000;14:1048–1057. [PMC free article] [PubMed] [Google Scholar]
- 35.Tomita A, Buchholz DR, Shi YB. Mol. Cell. Biol. 2004;24:3337–3346. doi: 10.1128/MCB.24.8.3337-3346.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yoon HG, Chan DW, Huang ZQ, Li J, Fondell JD, Qin J, Wong J. EMBO J. 2003;22:1336–1346. doi: 10.1093/emboj/cdg120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang J, Kalkum M, Chait BT, Roeder RG. Mol. Cell. 2002;9:611–623. doi: 10.1016/s1097-2765(02)00468-9. [DOI] [PubMed] [Google Scholar]
- 38.Nagy L, Kao HY, Love JD, Li C, Banayo E, Gooch JT, Krishna V, Chatterjee K, Evans RM, Schwabe JW. Genes Dev. 1999;13:3209–3216. doi: 10.1101/gad.13.24.3209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Perissi V, Staszewski LM, McInerney EM, Kurokawa R, Krones A, Rose DW, Lambert MH, Milburn MV, Glass CK, Rosenfeld MG. Genes Dev. 1999;13:3198–3208. doi: 10.1101/gad.13.24.3198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Webb P, Anderson CM, Valentine C, Nguyen P, Marimuthu A, West BL, Baxter JD, Kushner PJ. Mol. Endocrinol. 2000;14:1976–1985. doi: 10.1210/mend.14.12.0566. [DOI] [PubMed] [Google Scholar]
- 41.Cohen RN, Brzostek S, Kim B, Chorev M, Wondisford FE, Hollenberg AN. Mol. Endocrinol. 2001;15:1049–1061. doi: 10.1210/mend.15.7.0669. [DOI] [PubMed] [Google Scholar]
- 42.Hu X, Lazar MA. Nature. 1999;402:93–96. doi: 10.1038/47069. [DOI] [PubMed] [Google Scholar]
- 43.Farboud B, Hauksdottir H, Wu Y, Privalsky ML. Mol. Cell. Biol. 2003;23:2844–2858. doi: 10.1128/MCB.23.8.2844-2858.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang J, Hu X, Lazar MA. Mol. Cell. Biol. 1999;19:6448–6457. doi: 10.1128/mcb.19.9.6448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Marimuthu A, Feng W, Tagami T, Nguyen H, Jameson JL, Fletterick RJ, Baxter JD, West BL. Mol. Endocrinol. 2002;16:271–286. doi: 10.1210/mend.16.2.0777. [DOI] [PubMed] [Google Scholar]
- 46.Carlberg C. J. Steroid Biochem. Mol. Biol. 2004;89–90:227–232. doi: 10.1016/j.jsbmb.2004.03.112. [DOI] [PubMed] [Google Scholar]
- 47.Pissios P, Tzameli I, Kushner P, Moore DD. Mol. Cell. 2000;6:245–253. doi: 10.1016/s1097-2765(00)00026-5. [DOI] [PubMed] [Google Scholar]
- 48.Hu X, Li Y, Lazar MA. Mol. Cell. Biol. 2001;21:1747–1758. doi: 10.1128/MCB.21.5.1747-1758.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Makowski A, Brzostek S, Cohen RN, Hollenberg AN. Mol. Endocrinol. 2003;17:273–286. doi: 10.1210/me.2002-0310. [DOI] [PubMed] [Google Scholar]
- 50.Wong CW, Privalsky ML. Mol. Cell. Biol. 1998;18:5724–5733. doi: 10.1128/mcb.18.10.5724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Evans RM. Science. 1988;240:889–895. doi: 10.1126/science.3283939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Forrest D, Vennstrom B. Thyroid. 2000;10:41–52. doi: 10.1089/thy.2000.10.41. [DOI] [PubMed] [Google Scholar]
- 53.Keightley MC. Mol. Cell. Endocrinol. 1998;137:1–5. doi: 10.1016/s0303-7207(97)00236-0. [DOI] [PubMed] [Google Scholar]
- 54.Chatterjee VK, Tata JR. Cancer Surv. 1992;14:147–167. [PubMed] [Google Scholar]
- 55.Chambon P. Semin. Cell Biol. 1994;5:115–125. doi: 10.1006/scel.1994.1015. [DOI] [PubMed] [Google Scholar]
- 56.Zelent A, Mendelsohn C, Kastner P, Krust A, Garnier JM, Ruffenach F, Leroy P, Chambon P. EMBO J. 1991;10:71–81. doi: 10.1002/j.1460-2075.1991.tb07922.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Leroy P, Krust A, Zelent A, Mendelsohn C, Garnier JM, Kastner P, Dierich A, Chambon P. EMBO J. 1991;10:59–69. doi: 10.1002/j.1460-2075.1991.tb07921.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Koenig RJ, Lazar MA, Hodin RA, Brent GA, Larsen PR, Chin WW, Moore DD. Nature. 1989;337:659–661. doi: 10.1038/337659a0. [DOI] [PubMed] [Google Scholar]
- 59.Izumo S, Mahdavi V. Nature. 1988;334:539–542. doi: 10.1038/334539a0. [DOI] [PubMed] [Google Scholar]
- 60.Lazar MA. Endocr. Rev. 1993;14:184–193. doi: 10.1210/edrv-14-2-184. [DOI] [PubMed] [Google Scholar]
- 61.Li H, Leo C, Schroen DJ, Chen JD. Mol. Endocrinol. 1997;11:2025–2037. doi: 10.1210/mend.11.13.0028. [DOI] [PubMed] [Google Scholar]
- 62.Yoh SM, Privalsky ML. J. Biol. Chem. 2001;276:16857–16867. doi: 10.1074/jbc.M010022200. [DOI] [PubMed] [Google Scholar]
- 63.Cohen RN, Putney A, Wondisford FE, Hollenberg AN. Mol. Endocrinol. 2000;14:900–914. doi: 10.1210/mend.14.6.0474. [DOI] [PubMed] [Google Scholar]
- 64.Guan KL, Dixon JE. Anal. Biochem. 1991;192:262–267. doi: 10.1016/0003-2697(91)90534-z. [DOI] [PubMed] [Google Scholar]
- 65.Yang Z, Privalsky ML. Mol. Endocrinol. 2001;15:1170–1185. doi: 10.1210/mend.15.7.0656. [DOI] [PubMed] [Google Scholar]
- 66.Hauksdottir H, Farboud B, Privalsky ML. Mol. Endocrinol. 2003;17:373–385. doi: 10.1210/me.2002-0340. [DOI] [PubMed] [Google Scholar]
- 67.Laemmli UK. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 68.Wong C, Sridhara S, Bardwell JC, Jakob U. BioTechniques. 2000;28:426–432. doi: 10.2144/00283bm07. [DOI] [PubMed] [Google Scholar]
- 69.Chen HW, Privalsky ML. Mol. Cell. Biol. 1993;13:5970–5980. doi: 10.1128/mcb.13.10.5970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 71.Myers EW, Miller W. Comput. Appl. Biosci. 1988;4:11–17. doi: 10.1093/bioinformatics/4.1.11. [DOI] [PubMed] [Google Scholar]
- 72.Pearson WR, Lipman DJ. Proc. Natl. Acad. Sci. U. S. A. 1988;85:2444–2448. doi: 10.1073/pnas.85.8.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Pearson WR. Methods Enzymol. 1990;183:63–98. doi: 10.1016/0076-6879(90)83007-v. [DOI] [PubMed] [Google Scholar]
- 74.Sarge KD, Park-Sarge OK, Kirby JD, Mayo KE, Morimoto RI. Biol. Reprod. 1994;50:1334–1343. doi: 10.1095/biolreprod50.6.1334. [DOI] [PubMed] [Google Scholar]
- 75.Park EJ, Schroen DJ, Yang M, Li H, Li L, Chen JD. Proc. Natl. Acad. Sci. U. S. A. 1999;96:3519–3524. doi: 10.1073/pnas.96.7.3519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ordentlich P, Downes M, Xie W, Genin A, Spinner NB, Evans RM. Proc. Natl. Acad. Sci. U. S. A. 1999;96:2639–2644. doi: 10.1073/pnas.96.6.2639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Hong SH, Privalsky ML. Mol. Cell. Biol. 2000;20:6612–6625. doi: 10.1128/mcb.20.17.6612-6625.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zamir I, Zhang J, Lazar MA. Genes Dev. 1997;11:835–846. doi: 10.1101/gad.11.7.835. [DOI] [PubMed] [Google Scholar]
- 79.Miyamoto T, Suzuki S, DeGroot LJ. Mol. Endocrinol. 1993;7:224–231. doi: 10.1210/mend.7.2.8469235. [DOI] [PubMed] [Google Scholar]
- 80.Yen PM, Darling DS, Carter RL, Forgione M, Umeda PK, Chin WW. J. Biol. Chem. 1992;267:3565–3568. [PubMed] [Google Scholar]
- 81.Carey J. Proc. Natl. Acad. Sci. U. S. A. 1988;85:975–979. doi: 10.1073/pnas.85.4.975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Fried MG. Electrophoresis. 1989;10:366–376. doi: 10.1002/elps.1150100515. [DOI] [PubMed] [Google Scholar]
- 83.Senear DF, Brenowitz M. J. Biol. Chem. 1991;266:13661–13671. [PubMed] [Google Scholar]
- 84.Fried M, Crothers DM. Nucleic Acids Res. 1981;9:6505–6525. doi: 10.1093/nar/9.23.6505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Letovsky J, Dynan WS. Nucleic Acids Res. 1989;17:2639–2653. doi: 10.1093/nar/17.7.2639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Malartre M, Short S, Sharpe C. Nucleic Acids Res. 2004;32:4676–4686. doi: 10.1093/nar/gkh786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Burke LJ, Downes M, Laudet V, Muscat GE. Mol. Endocrinol. 1998;12:248–262. doi: 10.1210/mend.12.2.0061. [DOI] [PubMed] [Google Scholar]
- 88.Bailey PJ, Dowhan DH, Franke K, Burke LJ, Downes M, Muscat GE. J. Steroid Biochem. Mol. Biol. 1997;63:165–174. doi: 10.1016/s0960-0760(97)00079-4. [DOI] [PubMed] [Google Scholar]
- 89.Downes M, Burke LJ, Bailey PJ, Muscat GE. Nucleic Acids Res. 1996;24:4379–4386. doi: 10.1093/nar/24.22.4379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Muscat GE, Burke LJ, Downes M. Nucleic Acids Res. 1998;26:2899–2907. doi: 10.1093/nar/26.12.2899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Bailey P, Sartorelli V, Hamamori Y, Muscat GE. Nucleic Acids Res. 1998;26:5501–5510. doi: 10.1093/nar/26.23.5501. [DOI] [PMC free article] [PubMed] [Google Scholar]