Abstract
Immunoassays have made it possible to measure dozens of individual proteins and other analytes in human samples for help in establishing the diagnosis and prognosis of disease. In too many cases the results of those measurements are misleading and can lead to unnecessary treatment or missed opportunities for therapeutic interventions. These cases stem from problems inherent to immunoassays performed with human samples, which include a lack of concordance across platforms, autoantibodies, anti-reagent antibodies, and the high-dose hook effect. Tandem mass spectrometry may represent a detection method capable of alleviating many of the flaws inherent to immunoassays. We review our understanding of the problems associated with immunoassays on human specimens and describe methodologies using tandem mass spectrometry that could solve some of those problems. We also provide a critical discussion of the potential pitfalls of novel mass spectrometric approaches in the clinical laboratory.
Keywords: Immunoassays, tandem mass spectrometry, autoantibody interference, hook effect, heterophile anti-reagent antibodies, standardization
1. Immunoassays Are Imperfect
Ever since the description of the first immunoassay in the late 1950’s (Yalow and Berson, 1960), the use of antibodies to measure the concentration of proteins and small molecules in clinical samples has been changing the face of medicine. The platform for immunoassays has evolved from its initial conception as a competitive radioimmunoassay to enzyme linked immunosorbent assays on plastic surfaces to sandwich liquid phase chemiluminescent immunometric assays using paramagnetic beads on automated instruments (Bock, 2000) to microfluidic point of care testing ‘lab on chip’ immunoassays (Kartalov et al., 2008). The march of progressive technologies continues onward. Unfortunately, even after the many years we have spent optimizing antibody reagents for use in immunoassays, we frequently neglect to focus on the many limitations inherent to immunoassays as they behave with actual human samples in the clinical laboratory, which can be harmful to patients (Table 1). This review highlights how normal human biology and assay design can cause properly functioning immunoassays to harm patients and how novel approaches using mass spectrometric detection might solve those problems. It focuses on measurements of proteins rather than smaller molecules, which have been reviewed elsewhere (Chace, 2001).
Table 1.
Summary of common problems with immunoassays
| Problem | Brief Description | Effect | Examples |
|---|---|---|---|
| Lack of concordance | Assay performed with one set of reagents does not measure the same concentration as another | Patients must have measurements performed on the same platform for continuity of care | (Rawlins and Roberts, 2004; Mongia et al., 2006; Manley et al., 2007) |
| Autoantibodies | Epitopes on analyte of interest blocked from reagent antibodies by endogenous immunoglobulins | Falsely low results in sandwich immunometric assays, falsely elevated results in competitive radioimmunoassays | (Sapin et al., 1998; Spencer et al., 1998; Sapin, 2002) |
| Anti-reagent antibodies | Endogenous non-specific / multi-specific immunoglobulins bind reagent antibodies | Falsely elevated results in immunometric assays | (Willman et al., 1999; Rotmensch and Cole, 2000; Morgan and Tarter, 2001; Giovanella et al., 2007) |
| Hook effect | Reagent antibodies are saturated with analyte preventing sandwich formation | Falsely low results in the setting of extremely high analyte concentrations | (Furuya et al., 2001; Fleseriu et al., 2006; McCudden et al., 2009) |
1.1 Lack of concordance across platforms
Many of the best biomarkers for the diagnosis of human disease are proteins released at extremely low concentrations from inflamed or damaged tissues. Others are abundant in normal plasma, but vary in concentration in different disease states. Regardless of the concentration of the biomarker in plasma, the design of immunoassays always starts with antigen selection.
Antibody responses in animals can be induced against whole molecules, domains of molecules, or peptides. The antigens may contain post-translational modifications commonly found in serum, or as is often the case when made recombinantly in prokaryotes, could lack those modifications. Indeed, assays typically use different antibodies targeting different epitopes to quantify exactly the same analyte and as a result, the results of one assay can be very different from results of another assay. There are two very good examples of this in the literature. First, for thyroid stimulating hormone (TSH), comparison of 6 immunoassay platforms illustrated nicely how an analyte for which there are published guidelines and reference ranges to guide treatment, a value reported by one laboratory may indicate the need for therapy, while one from another laboratory would not (Rawlins and Roberts, 2004). Second, for CA 19-9, an important tumor marker for pancreatic and colon carcinoma, comparison of 5 different assay platforms demonstrated how a patient leaving one hospital to be treated at another (e.g. due to insurance changes), could be misdiagnosed with cancer recurrence at the new hospital when in fact the patient is still in remission (La'ulu and Roberts, 2007). The costs of unnecessary further surveillance and treatment in these two examples can be substantial financially and in terms of quality of life.
There have been efforts to standardize certain clinical analytes across assay platforms (Rodriguez-Cabaleiro et al., 2007). Unfortunately, because the purified protein standard material does not represent the molecular population of the analyte present in any one individual, assays are calibrated to closely agree for measurements of the standardized material, however differences between platforms for certain patients persist. This can be due to genetic polymorphisms that introduce changes in protein primary structure, genetic variability in glycosylation pathways, or differences in the tissue that is secreting the biomarker of interest, which could lead to variable protein processing, modification, or cross-linking. For example, it has been demonstrated that diseased or malignant tissue can secrete different proteins than those normally found in tissue or serum (Riches et al., 1991; Spencer and Lopresti, 2008). These variations in epitope structure between individuals and even within individuals lead to differences in immunoreactivity in different assays. Variability in post-translational modification has been demonstrated for human chorionic gonadotropin (hCG) (Kelly et al., 2005), prostate specific antigen (PSA) (Ward et al., 2001), TSH (Canonne et al., 1995), thyroglobulin (Spencer and Lopresti, 2008), and monoclonal paraproteins (Riches et al., 1991). Genetic variability in epitope structure has been described for mucin-1 (i.e. serum cancer antigens CA15-3, CA27-29) (Silva et al., 2001).
1.2 Autoantibodies
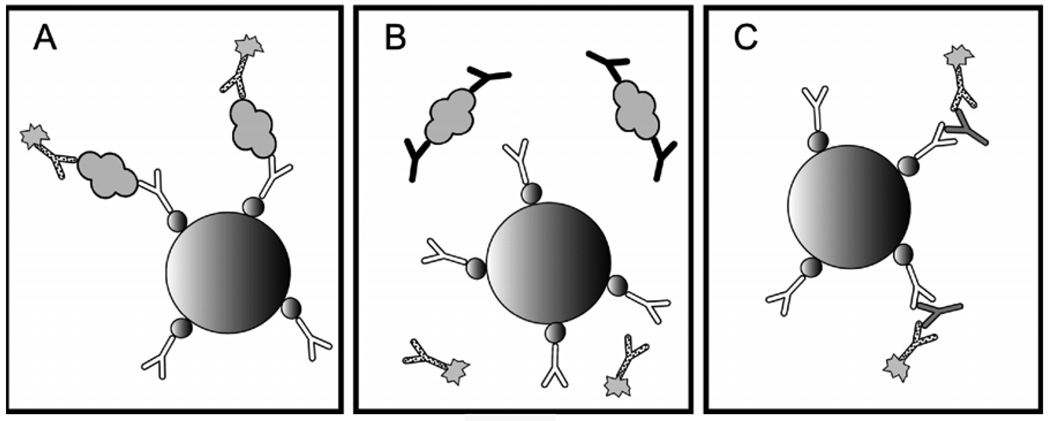
While the mechanisms for the production of self-directed antibodies remain elusive, their interference with immunoassays is an extremely important clinical problem. For thyroglobulin, this interference has been well described (Spencer et al., 1998). Up to 10% of all normal subjects are positive for anti-thyroglobulin antibodies. Importantly, this number can climb as high as 25% in patients with differentiated thyroid cancer. The reasons for such a drastic increase are unclear; however, it seems reasonable that production by the tumor of slightly abnormal thyroglobulin protein could be recognized as non-self and be the target of antibodies that cross-react with normal protein. In immunometric assays, autoantibodies bind near an epitope recognized by reagent antibodies, sterically inhibiting formation of detectable complexes (Figure 1B). This leads to decreased signal in the assay and a falsely low value. Unfortunately, autoantibody interference is unpredictable and on some platforms may lead to falsely elevated or reduced results (Spencer et al., 1998). For this reason, for patients who have detectable anti-thyroglobulin antibodies, serum thyroglobulin assay results must be interpreted with caution (Spencer and Lopresti, 2008). Other diagnostic modalities are often needed for these patients and generally include expensive imaging studies.
Figure 1. Endogenous antibody interference in sandwich immunoassays.
A modern sandwich immunoassay for thyroglobulin is pictured on the left. A streptavidin coated paramagnetic bead (large sphere) binds biotin-labeled (small spheres) capture antibody (white), which forms a sandwich with analyte (gray globule) and enzyme-labeled (sunburst) reporter antibody (dotted). Sandwiches bound to beads are separated using a magnet, unbound reporter antibody is washed away, and enzyme is detected using one of a variety of automated methodologies. Unfortunately, in a high percentage of patients, the epitopes needed for sandwich formation are sterically protected by anti-analyte antibodies (black) as pictured in the middle. In other patients, non-specific anti-reagent antibodies (gray) are able to bridge the gap between capture and reporter antibodies in the absence of analyte as picture on the right.
While thyroglobulin is the assay for which autoantibody interference has been best characterized, autoantibodies to other proteins are well documented. For example, the heavily glycosylated tumor antigen mucin-1, contains the epitopes detected in the CA15-3 and CA27.29 tumor marker immunoassays, which are used as markers for recurrent breast cancer. Autoantbodies to mucin-1 are not uncommon in patients with breast cancer and are a good prognostic sign (von Mensdorff-Pouilly et al., 1996). Even though patients are known to make autoantibodies to this tumor antigen, there have been no published studies demonstrating the performance of either CA15-3 or CA27.29 in the population of patients with autoantibodies to mucin-1. The likelihood of interference is substantial and would lead to missed or inappropriate treatment, depending on the platform. Antibodies to other antigens have also been demonstrated and include PSA (Taylor et al., 2008), TSH (Sapin et al., 1998), C-reactive protein (Wettero et al., 2009), and insulin (Sapin, 2002). Studies into the influence of autoantibodies on the performance of these assays are warranted.
1.3 Anti-reagent antibodies
Human anti-reagent antibodies have always plagued immunoassays. They include non-specific antibodies, often called heterophile antibodies, and other miscellaneous antibodies directed at reagent immunoglobulins (Kricka, 1999), biotin (Dale et al., 1994), or other epitopes (Levinson and Miller, 2002; Sapin et al., 2007). Other proteins in plasma that interfere with immunoassays and cause non-specific aggregation could also be included in this category simply due to our lack of biochemical understanding of these interferences.
Anti-reagent antibodies can have drastic implications in patient care. In sandwich assays, they are able to bridge the gap between capture and reporter antibodies and lead to inaccurate results (Figure 1C). False elevations in patients screened for choriocarcinoma have resulted in unwarranted therapy including surgery and chemotherapy (Rotmensch and Cole, 2000). In at least one case, a false positive prostate specific antigen result led to unnecessary hormonal ablation and external beam radiation (Morgan and Tarter, 2001). Other markers that have been affected by anti-reagent antibodies include chromogranin A (Giovanella et al., 2007), human immunodeficiency virus (Willman et al., 1999), thyroglobulin (Preissner et al., 2003), and many others.
For decades, endogenous antibodies that bind reagent proteins in immunoassays have been called heterophile or heterophilic antibodies, a term which implies multiple non-specific targets. The term ‘heterophile antibodies’ in this context is often confused by clinicians with an assay for heterophile antibodies to animal red blood cells. The assay, commonly performed as the infectious mononucleosis spot test, or monospot, is used in the diagnosis of acute Ebstein-Barr virus infection and is in no way predictive of interference with immunoassays. Although clinicians and laboratory scientists both use the term ‘heterophile antibody,’ they typically mean very different things.
Some investigators have used the term anti-animal antibodies to denote antibodies that develop after extensive exposure to animals, or after working with or receiving animal-derived therapeutics (Kricka, 1999), and assays have been designed to test for antibodies specific to mouse immunoglobulins, also called human anti-mouse antibodies (HAMAs). These acquired anti-animal antibodies are uncommon, but generally have a high titer and cause significant analytical interference. Most anti-reagent antibodies are naturally occurring, low titer, and polyspecific, i.e. they bind immunoglobulins from several different species (Hennig et al., 2000). Rheumatoid factors (auto-antihuman IgG) may also react with IgG of other species (e.g. rabbit) and interfere with many immunoassays (Knoblock et al., 2002). The targets of interfering antibodies are not limited to the antibody portion of the assay (Willman et al., 1999). Indeed, the endogenous antibodies, which appear to have multiple targets (Levinson and Miller, 2002) and likely some non-specific affinity for one another, could be directed toward any combination of label, linker, solid phase, and immunoglobulins involved in the assay to give a falsely elevated result. In vitro diagnostic companies have worked hard to overcome the problem by including decoy antibodies and other proprietary reagents to reduce the incidence of anti-reagent interference. There are also available several stand-alone anti-reagent antibody blocking products with variable performance (Reinsberg, 1996).
Despite these preventive measures, in the rare patient for whom these naturally occurring anti-reagent antibodies are present in high titer and high avidity, the impact on immunoassays can be clinically very significant. This will likely remain a problem as long as immunometric assays are used to measure proteins in human serum and plasma. Clinicians should always keep the possibility of interference in mind when the clinical picture and the laboratory result do not agree. Many patients have been—and in the future will continue to be—harmed by anti-reagent antibodies and their effects in immunoassays.
1.4 Hook effect
Ideally, as concentrations of analyte in plasma or serum increase, the response from sandwich immunoassays increases as well. The increase in signal should be monotonic with concentration. However, as the concentration of analyte increases above a certain point, the system gets saturated and the signal begins to decline, the plot of which resembles a fish-hook. As a result, the phenomenon earned the name “high-dose hook effect.” Theoretically, this issue is only applicable to sandwich immunometric assays without a wash step between reagent additions. But in all sandwich assays, the signal begins to plateau with high concentrations of analyte due to limiting amounts of reagent immunoglobulins and rare samples with extremely high concentrations of analyte can even lead to the hook effect in assays with a wash step.
To avoid reporting falsely low results, some laboratories make a pool of patient specimens and compare the pool to the expected average of the batch of samples. If the pooled specimen is higher than expected, one of the samples in the pool was inaccurately quantified due to the hook effect. Other laboratories dilute every specimen looking for diluted samples that after correction for dilution measure higher than neat samples. These are suspicious for the hook effect. There are still other laboratories that inadequately control for the possibility of the hook effect in their assays. Many assays have been reported to suffer from the hook effect in patients with high analyte values and include assays for prolactin (Fleseriu et al., 2006), serum free light chains (McCudden et al., 2009), and PSA (Furuya et al., 2001).
2. Could mass spectrometry be the answer?
In the 1980’s, scientists at Yale described robust methods for ionizing intact molecules and introducing them into mass analyzers (Whitehouse et al., 1985). These ionization strategies combined with gas-phase fragmentation permitted the de novo identification of peptides and proteins from complex mixtures, thus birthing the field of proteomics. Later advances in electronics and architecture have made modern mass analyzers capable of directly detecting attomoles of analyte. With this level of sensitivity and properly designed assays, it is possible that mass spectrometry could replace elements of immunoassays in the future. We will highlight recent developments in the targeted quantification of known biomarker proteins in clinical specimens rather than approaches for the discovery of novel biomarkers, which has been reviewed elsewhere (Resing and Ahn, 2005; Han et al., 2008).
2.1 The mass spectrometric experiment
The overall experiment to quantify a known biomarker peptide starts with the digestion of all proteins in a sample using a protease and the subsequent separation of the resulting peptides using liquid chromatography (HPLC). Then, using the mass spectrometer, one can select the mass-to-charge ratio (m/z) of a peptide from the protein of interest, break the peptide into fragments, and then quantify the fragment of interest (Figure 2). The specificity of the approach thus lies in the three-step separation of peptides based on (1) retention time (HPLC), (2) precursor m/z (peptide of interest), and (3) fragment m/z (peptide fragment of interest).
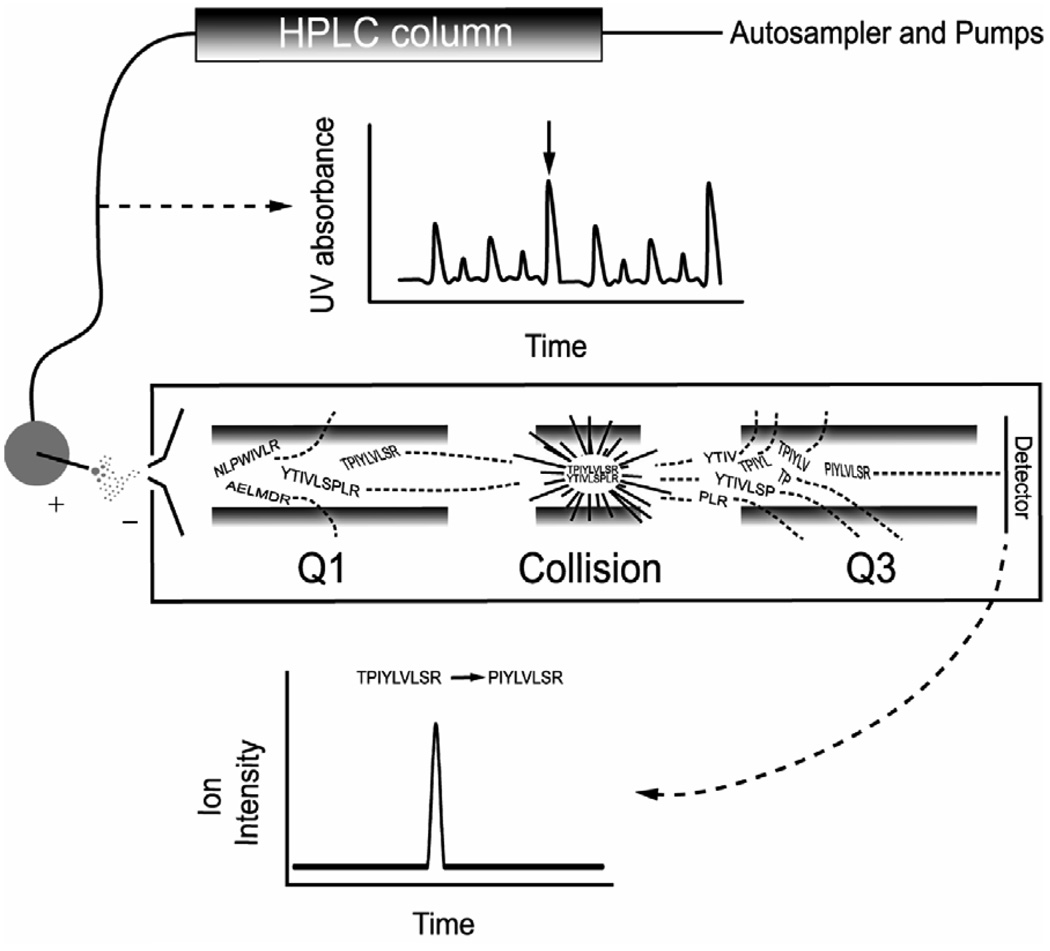
Figure 2. HPLC-tandem mass spectrometry.
Complex mixtures of peptides are first separated by HPLC based on hydrophobicity. If a UV detector were placed in-line after the HPLC column, peptides would be detected as they elute from the column. In this example, four peptides elute simultaneously (e.g. at the black arrow in the UV trace). In the first quadrupole (Q1), the mass-to-charge ratio (m/z) of the precursor peptide is selected. Here, two peptides of identical m/z make it to the second quadrupole (Collision), where they are fragmented by collision induced dissociation. A single fragment m/z is selected in the third quadrupole (Q3) and any ions that reach the detector have the correct precursor m/z and fragment ion m/z. Specificity in the HPLC-tandem mass spectrometric experiment thus arises from the retention time on the HPLC column and the precursor-fragment ion pair, which is also called a transition. In a multiple reaction monitoring experiment, multiple transitions are monitored.
In the first step using HPLC, the peptides partition between the solid phase of the column and the liquid phase. In reverse phase chromatography, which is the most commonly used approach for the quantification of peptides, the solid phase is apolar and strongly binds hydrophobic molecules. This permits extensive washing of the peptides before their successive elution from the column using a gradient of organic solvent. Thus in most mass spectrometric analyses of proteins, the peptides are separated based on hydrophobicity.
The second step of the separation of peptides uses the mass spectrometer to distinguish peptides based on their m/z. When placed in an electric field, ions are deflected by a constant force and the acceleration of each ion is inversely proportional to its m/z. Mass analyzers take advantage of this fact and are capable of selecting only the m/z of the peptide of interest (Figure 2). We use the triple quadrupole tandem mass spectrometer as our example instrument because it is the most commonly used instrument in clinical laboratories; however, there are many other types of mass spectrometers that are becoming clinically useful. The quadrupole is named for the four parallel rods across which voltages are placed to select for the specific m/z of interest. If a peptide has the correct m/z, it will pass through the first quadrupole and continue to the next quadrupole in the instrument.
In complex mixtures, the m/z of interest selected in step 2 may include the peptide of interest and several other peptides of equal m/z as they elute from the HPLC column (e.g. two peptides with the same m/z are shown in Figure 2). In order to differentiate irrelevant molecules from the peptide of interest, a second quadrupole filled with an inert collision gas is used to fragment the molecules from the first quadrupole. The peptide fragments generated in the second quadrupole are then analyzed in the third quadrupole, and if a fragment has the correct m/z, it passes through the third quadrupole, strikes the detector, and registers a signal.
When a specific precursor m/z ratio is selected in the first quadrupole and a specific fragment m/z is selected in the third quadrupole, the combination is called a transition (e.g. the TPIYLVLSR → PIYLVLSR transition depicted in Figure 2 would detect only the peptide TPIYLVLSR because that transition would not be possible from peptide YTIVLSPLR, which has the same precursor mass). When multiple transitions are simultaneously analyzed during an HPLC elution program, it is called multiple reaction monitoring (MRM). Methods utilizing MRM approaches can simultaneously quantify dozens of analytes in one specimen, making mass spectrometry an exciting method for not only solving the problems inherent to immunoassays, but for multiplexing protein measurements in the clinical laboratory as well (Anderson and Hunter, 2006).
2.2 Proteolytic digestion and direct quantification using isotope dilution
The simplest method to provide relative quantification of proteins using mass spectrometry is to proteolytically digest them with enzyme and determine the peak area of one or more analyte-specific peptides in the digest. While HPLC effectively separates the analyte of interest from many contaminants, co-eluting molecules that may vary in concentration from patient to patient can suppress the ionization of the molecule of interest (Annesley, 2003). This combined with the fact that mass spectrometers perform differently over the course of a day of analysis, makes it imperative to normalize the peak area from the analyte of interest to an internal standard to control for ion suppression and variable instrument performance. The most common approach to this normalization is to incorporate stable-isotopes (i.e. deuterium, 13C, or 15N) into the peptide. These peptides behave chemically identically to unlabeled peptide, yet they can be distinguished by their mass difference in the mass spectrometer. Proteolytic digestion and direct quantification with isotope dilution has been used to quantify Zn-α2-glycoportein with a limit of detection of 90 nmol/L (Bondar et al., 2007). It has even been used to quantify ceruloplasmin in dried blood spots in newborn screening for Wilson’s disease (deWilde et al., 2008).
2.3 Fractionation of serum or plasma prior to proteolytic digestion
Methods to improve the detection of low abundance serum proteins invariably include some form of fractionation of serum or plasma to enrich for the analyte of interest. One group has successfully used fractionation of proteins to quantify C-reactive protein in human serum with a limit of detection of 7 nmol/L (Kuhn et al., 2004). Enrichment for specific post-translational modifications has also been carried out using solid phase strategies after protein digestion (Rush et al., 2005). The most specific method to enrich for the analyte of interest from serum is immunoaffinity enrichment (Berna et al., 2007).
2.4 Immunoaffinity peptide enrichment-tandem mass spectrometry
The most important approach for detecting very low concentrations of serum proteins is one using antibodies to enrich for specific peptides of interest from a proteolytic digest of serum that are then quantified using tandem mass spectrometry. It is possible to control for the variability of the immunoaffinity enrichment step by including stable-isotope labeled peptide at that point. This methodological approach has been termed by Anderson et al. “Stable Isotope Standards with Capture by Anti-Peptide Antibodies” or SISCAPA (Anderson et al., 2004).
2.5 Thyroglobulin case study
As described above, thyroglobulin is one of the best characterized tumor markers in clinical medicine. Performance issues with the assay have been well described and have actually taught us much about analogous problems with other immunoassays. With this ground, our laboratory attempted to solve many of these problems using tandem mass spectrometry (Hoofnagle et al., 2008). A clinically useful serum thyroglobulin assay using mass spectrometry would have a functional limit of quantification of 1 ng/mL (1.5 fM) or lower. Our efforts using proteolytic digestion and isotope dilution tandem mass spectrometry supported the findings of previous experiments with a limit of detection of 30 µg/mL. We improved the limit of detection for thyroglobulin approximately 10-fold by taking advantage of the large size of the molecule (660 kDa) and partially separating it from other serum proteins using size exclusion chromatography. Because this was still three orders of magnitude away from a clinically useful assay, we turned to immunoaffinity peptide enrichment using polyclonal antibodies. Using this methodology, we achieved a limit of detection of 2.6 ng/mL, nearly good enough for clinical utility in tumor marker monitoring in patients treated for differentiated thyroid carcinoma.
This novel approach to detect low abundance proteins in complex mixtures has been developed over the last five years and has the potential to eliminate interference by endogenous immunoglobulins, eliminate the hook effect, and provide a well standardized method across laboratories. Interfering immunoglobulins are removed by digestion with trypsin. The hook effect is no longer a problem because the detectors used in mass spectrometry are not saturated at the amount of peptide that can be purified with the antibodies; however, limiting amounts of reagent antibody will lead to signal plateauing at high analyte levels. Perhaps most importantly, the method specifically detects peptides derived from the protein, not epitopes. Provided one selects analyte-derived peptides that are free from post-translational modification and polymorphisms, the peptides will be representative of the amount of protein in the sample and will enable calibration of assays across laboratories. Recent advances in this approach have led to more automated sample preparation (Anderson et al., 2009) and to multiplexing by enriching for peptides from more than one protein at once (Kuhn et al., 2009).
3. Potential Pitfalls of LC-MS/MS in the clinical quantification of proteins
However exciting mass spectrometric assays for protein quantification might seem, they will undoubtedly have significant problems of their own. In this section we will highlight recognized issues with mass spectrometric assays, but we must keep in mind that unanticipated problems that will also likely surface over time. Indeed, the abovementioned problems with immunoassays were only elucidated after years of clinical use.
3.1 Complexity of serum and plasma
The complexity of the human plasma proteome is an immense problem for the measurement of proteins by mass spectrometry. Protein concentrations span 10 orders of magnitude (Hortin et al., 2008) and as an example, for every one thyroglobulin peptide in plasma, there are 40 million peptides from albumin. The problem could be likened to finding a needle in a needlestack. For this reason, a lot of investigation has centered on simplifying plasma using immunoaffinity depletion of abundant proteins. Unfortunately, proteins of interest can bind to immunoglobulins (e.g. thyroglobulin may bind to autoantibodies) and to albumin (Gundry et al., 2007), two abundant plasma proteins. Therefore, depletion of these proteins would accidentally remove the protein of interest in the specimens most in need of a better assay.
Another problem associated with the complexity of plasma is the amount of starting material required to generate enough peptide from low abundance proteins to be detected reliably by the mass spectrometer (typically femtomoles). Thus for many analytes, this amounts to at least 0.02–0.1 mL plasma, or more than 1 mg of protein—a challenging amount of protein to digest to completion by any protocol. Incomplete digestion leads to variability in digestion from sample to sample, but despite this variability, multiple reports employing serum digestion and quantification by mass spectrometry have yielded promising results (Kuhn et al., 2004; Bondar et al., 2007; deWilde et al., 2008; Hoofnagle et al., 2008; Kuhn et al., 2009). The ideal method to control for variable digestion would be a stable isotope labeled version of the protein of interest that is folded similarly to endogenous protein. Variability in the digestion of endogenous protein would then be normalized by the internal standard protein. This approach has yet to be demonstrated in clinical specimens.
A third problem caused by the complexity of plasma is the unanticipated interference of homologous peptides in immunoaffinity peptide enrichment. Although a completed sequence of the human genome is available, it is impossible to predict the extent to which similar peptides will compete for antibody binding sites. In fact, the plasma proteome is still largely undefined and the potential for peptides identical to the peptide of interest to be present at low concentrations within protein fragments or protein chimera must be recognized.
3.2 Problems shared by immunoassay and mass spectrometry
While 99% of each human genome is identical, there are more than 3 million differences between individuals. Polymorphisms that lie in exons and result in amino acid substitutions likely play a role in the poor interlaboratory concordance seen for many protein immunoassays. They are likely to influence mass spectrometric assays as well. Any amino acid changes in peptides of interest will lead to a loss of detection by the mass spectrometer. It is likely that quantifying more than two peptides for each protein will be helpful in identifying specimens in which a peptide is underrepresented compared with the other peptides from the same protein, but this has not been demonstrated. More exciting is the possibility that the 1,000 Genomes Project (Siva, 2008) will identify non-polymorphic peptides within analytes of interest that will help the community select peptides to be used for mass spectrometric assays.
In the same way that polymorphisms will affect both immunoassays and mass spectrometric assays, post-translational modifications will complicate analysis by both as well. Many post-translational modifications have already been determined for analytes of interest. For example, thyroglobulin has extensive post-translational modifications that include glycosylation, iodination, phosphorylation, oxidation, and sulfation that total 10% of the normal protein mass of thyroglobulin (Hoofnagle et al., 2008). These events must be considered when selecting peptides for mass spectrometric assays, but it is impossible to fathom all the post-translational modifications that take place in diseased or transformed tissue and therefore unimaginable what peptides will be affected by the problem. As for genetic polymorphisms, measurement of multiple peptides for each protein will help reduce the chance of missing modifications. An important initiative funded by the National Cancer Institute called the Clinical Proteomic Technology Assessment for Cancer (CPTAC) is developing tools for use in SISCAPA assays. For each protein analyte of interest, the team is developing monoclonal antibodies to five peptides in each protein. This will allow five independent measurements of protein concentration in each assay. Therefore, loss of peptides due to polymorphism or post-translational modification will be easier to detect, and in fact, such variations could be informative if they are disease related. Current targets for development by CPTAC include thyroglobulin, hCG, and PSA, three of the most difficult and complicated protein analytes in human serum (N. Leigh Anderson, personal communication).
A third complication shared by immunoassays and mass spectrometric assays is embodied by the multiple isoforms of many proteins present in plasma. These may arise from splicing events at the transcriptional level or from post-translational proteolytic cleavages, which can accumulate in the plasma due to kidney or liver dysfunction. Importantly, detecting a peptide does not mean detecting intact protein. The impact of this on clinical analytes in clinical specimens could be substantial.
3.3 Mass spectrometry as a new methodology
While mass spectrometry has been used in medical laboratories for years, the workforce to run the assays is not universally well-trained to do so. This is problematic since the quality control procedures for traditional small molecule mass spectrometric assays will need to be rigorously applied to protein quantification as well. This includes careful monitoring for ion suppression, which varies from specimen to specimen, use of ion ratios to detect isobaric interference, and evaluation of carryover which can vary from 0.01% to 1% depending on the autosampler and analyte. The latter issue is of great concern for tumor antigens like CA19-9 and hCG that can vary in concentration over more than 8 orders of magnitude across the population. Ideal curve-fitting algorithms and standardized approaches for calibrating assays also need to be developed. While detection of peptides in mass spectrometry generally produces a linear response, linearity is not consistently observed and cannot be assumed to be present in clinical specimens.
4. Conclusions
Over the past five decades, we have learned that immunoassays can be used to help diagnose human disease. Sadly, we have also learned that immunoassays can hurt patients. It is important for clinicians to remember how immunoassays can give inaccurate results in many clinical situations. It is also important for the community as a whole to look for solutions, which may reside in novel detection technologies like tandem mass spectrometry. And as with all novel technologies, we must be prepared for unexpected complications from the infinitely complex reservoir of human biology.
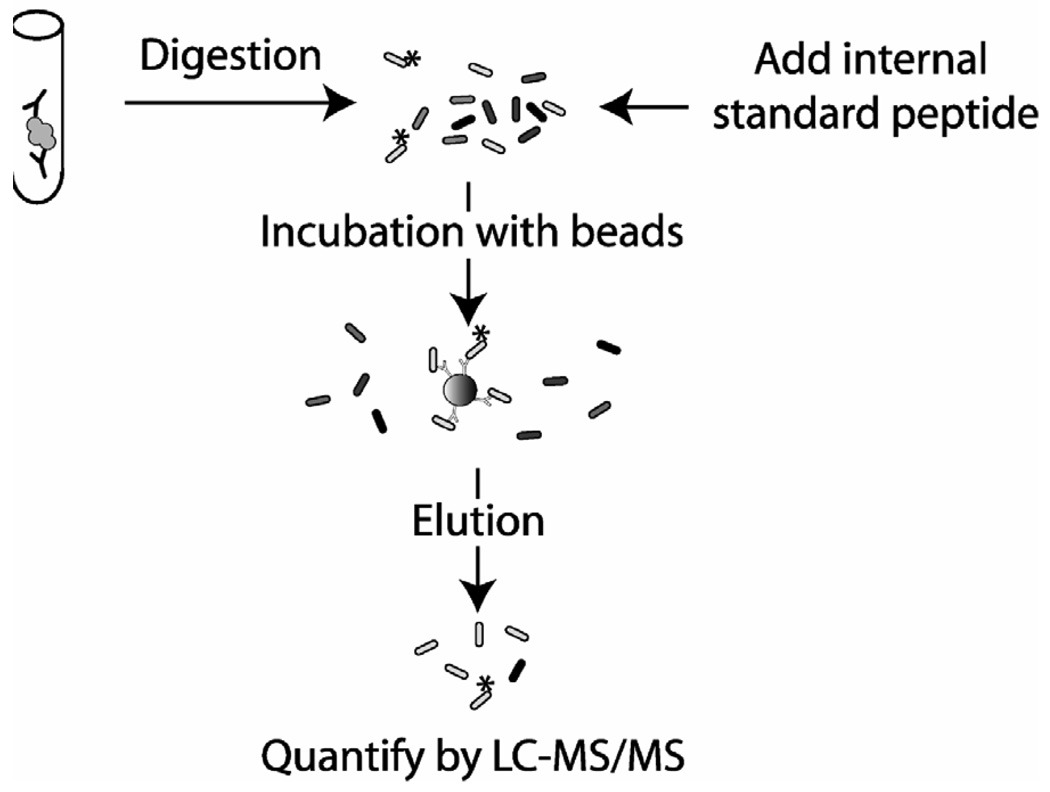
Figure 3. Immunoaffinity peptide enrichment and isotope dilution tandem mass spectrometry.
All proteins in the sample are digested to peptides with trypsin, which digests analyte (gray globule) and any interfering endogenous immunoglobulins (black). Stable isotope labeled peptide is added after digestion and the peptides are incubated with beads coated in polyclonal anti-peptide antibody. Unbound peptides are washed away and bound peptides are eluted and then analyzed using HPLC-tandem mass spectrometry.
Abbreviations
- HPLC
high performance liquid chromatography
- TSH
thyroid stimulating hormone
- CEA
carcinoembryonic antigen
- hCG
human chorionic gonadotropin
- MRM
multiple reaction monitoring
- PSA
prostate specific antigen
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Anderson L, Hunter CL. Quantitative mass spectrometric multiple reaction monitoring assays for major plasma proteins. Mol Cell Proteomics. 2006;5:573–588. doi: 10.1074/mcp.M500331-MCP200. [DOI] [PubMed] [Google Scholar]
- Anderson NL, Anderson NG, Haines LR, Hardie DB, Olafson RW, Pearson TW. Mass spectrometric quantitation of peptides and proteins using Stable Isotope Standards and Capture by Anti-Peptide Antibodies (SISCAPA) J Proteome Res. 2004;3:235–244. doi: 10.1021/pr034086h. [DOI] [PubMed] [Google Scholar]
- Anderson NL, Jackson A, Smith D, Hardie D, Borchers C, Pearson TW. SISCAPA peptide enrichment on magnetic beads using an inline beadtrap device. Mol Cell Proteomics. 2009 doi: 10.1074/mcp.M800446-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Annesley TM. Ion suppression in mass spectrometry. Clin Chem. 2003;49:1041–1044. doi: 10.1373/49.7.1041. [DOI] [PubMed] [Google Scholar]
- Berna MJ, Zhen Y, Watson DE, Hale JE, Ackermann BL. Strategic use of immunoprecipitation and LC/MS/MS for trace-level protein quantification: myosin light chain 1, a biomarker of cardiac necrosis. Anal Chem. 2007;79:4199–4205. doi: 10.1021/ac070051f. [DOI] [PubMed] [Google Scholar]
- Bock JL. The new era of automated immunoassay. Am J Clin Pathol. 2000;113:628–646. doi: 10.1309/DUDM-3Y6L-3R1L-QP15. [DOI] [PubMed] [Google Scholar]
- Bondar OP, Barnidge DR, Klee EW, Davis BJ, Klee GG. LC-MS/MS quantification of Zn-alpha2 glycoprotein: a potential serum biomarker for prostate cancer. Clin Chem. 2007;53:673–678. doi: 10.1373/clinchem.2006.079681. [DOI] [PubMed] [Google Scholar]
- Canonne C, Papandreou MJ, Medri G, Verrier B, Ronin C. Biological and immunochemical characterization of recombinant human thyrotrophin. Glycobiology. 1995;5:473–481. doi: 10.1093/glycob/5.5.473. [DOI] [PubMed] [Google Scholar]
- Chace DH. Mass spectrometry in the clinical laboratory. Chem Rev. 2001;101:445–477. doi: 10.1021/cr990077+. [DOI] [PubMed] [Google Scholar]
- Dale GL, Gaddy P, Pikul FJ. Antibodies against biotinylated proteins are present in normal human serum. J Lab Clin Med. 1994;123:365–371. [PubMed] [Google Scholar]
- deWilde A, Sadilkova K, Sadilek M, Vasta V, Hahn SH. Tryptic peptide analysis of ceruloplasmin in dried blood spots using liquid chromatography-tandem mass spectrometry: application to newborn screening. Clin Chem. 2008;54:1961–1968. doi: 10.1373/clinchem.2008.111989. [DOI] [PubMed] [Google Scholar]
- Fleseriu M, Lee M, Pineyro MM, Skugor M, Reddy SK, Siraj ES, Hamrahian AH. Giant invasive pituitary prolactinoma with falsely low serum prolactin: the significance of 'hook effect'. J Neurooncol. 2006;79:41–43. doi: 10.1007/s11060-005-9108-7. [DOI] [PubMed] [Google Scholar]
- Furuya Y, Cho S, Ohta S, Sato N, Kotake T, Masai M. High dose hook effect in serum total and free prostate specific antigen in a patient with metastatic prostate cancer. J Urol. 2001;166:213. [PubMed] [Google Scholar]
- Giovanella L, Ceriani L, Lumastro C, Ghelfo A. False-positive serum chromogranin A assay due to heterophile antibody interference. Clin Chim Acta. 2007;379:171–172. doi: 10.1016/j.cca.2006.12.001. [DOI] [PubMed] [Google Scholar]
- Gundry RL, Fu Q, Jelinek CA, Van Eyk JE, Cotter RJ. Investigation of an albumin-enriched fraction of human serum and its albuminome. Proteomics: Clin Appl. 2007;1:73–88. doi: 10.1002/prca.200600276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han X, Aslanian A, Yates JR., 3rd Mass spectrometry for proteomics. Curr Opin Chem Biol. 2008;12:483–490. doi: 10.1016/j.cbpa.2008.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hennig C, Rink L, Fagin U, Jabs WJ, Kirchner H. The influence of naturally occurring heterophilic anti-immunoglobulin antibodies on direct measurement of serum proteins using sandwich ELISAs. J Immunol Methods. 2000;235:71–80. doi: 10.1016/s0022-1759(99)00206-9. [DOI] [PubMed] [Google Scholar]
- Hoofnagle AN, Becker JO, Wener MH, Heinecke JW. Quantification of thyroglobulin, a low-abundance serum protein, by immunoaffinity peptide enrichment and tandem mass spectrometry. Clin Chem. 2008;54:1796–1804. doi: 10.1373/clinchem.2008.109652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hortin GL, Sviridov D, Anderson NL. High-abundance polypeptides of the human plasma proteome comprising the top 4 logs of polypeptide abundance. Clin Chem. 2008;54:1608–1616. doi: 10.1373/clinchem.2008.108175. [DOI] [PubMed] [Google Scholar]
- Kartalov EP, Lin DH, Lee DT, Anderson WF, Taylor CR, Scherer A. Internally calibrated quantification of protein analytes in human serum by fluorescence immunoassays in disposable elastomeric microfluidic devices. Electrophoresis. 2008;29:5010–5016. doi: 10.1002/elps.200800297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly LS, Kozak M, Walker T, Pierce M, Puett D. Lectin immunoassays using antibody fragments to detect glycoforms of human chorionic gonadotropin secreted by choriocarcinoma cells. Anal Biochem. 2005;338:253–262. doi: 10.1016/j.ab.2004.12.011. [DOI] [PubMed] [Google Scholar]
- Knoblock RJ, Lehman CM, Smith RA, Apple FS, Roberts WL. False-positive AxSYM cardiac troponin I results in a 53-year-old woman. Arch Pathol Lab Med. 2002;126:606–609. doi: 10.5858/2002-126-0606-FPACTI. [DOI] [PubMed] [Google Scholar]
- Kricka LJ. Human anti-animal antibody interferences in immunological assays. Clin Chem. 1999;45:942–956. [PubMed] [Google Scholar]
- Kuhn E, Addona T, Keshishian H, Burgess M, Mani DR, Lee RT, Sabatine MS, Gerszten RE, Carr SA. Developing Multiplexed Assays for Troponin I and Interleukin-33 in Plasma by Peptide Immunoaffinity Enrichment and Targeted Mass Spectrometry. Clin Chem. 2009;55 doi: 10.1373/clinchem.2009.123935. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn E, Wu J, Karl J, Liao H, Zolg W, Guild B. Quantification of C-reactive protein in the serum of patients with rheumatoid arthritis using multiple reaction monitoring mass spectrometry and 13C-labeled peptide standards. Proteomics. 2004;4:1175–1186. doi: 10.1002/pmic.200300670. [DOI] [PubMed] [Google Scholar]
- La'ulu SL, Roberts WL. Performance characteristics of five automated CA 19-9 assays. Am J Clin Pathol. 2007;127:436–440. doi: 10.1309/H52VET3M6P7GYWG1. [DOI] [PubMed] [Google Scholar]
- Levinson SS, Miller JJ. Towards a better understanding of heterophile (and the like) antibody interference with modern immunoassays. Clin Chim Acta. 2002;325:1–15. doi: 10.1016/s0009-8981(02)00275-9. [DOI] [PubMed] [Google Scholar]
- Manley SE, Stratton IM, Clark PM, Luzio SD. Comparison of 11 human insulin assays: implications for clinical investigation and research. Clin Chem. 2007;53:922–932. doi: 10.1373/clinchem.2006.077784. [DOI] [PubMed] [Google Scholar]
- McCudden CR, Voorhees PM, Hammett-Stabler CA. A case of hook effect in the serum free light chain assay using the Olympus AU400e. Clin Biochem. 2009;42:121–124. doi: 10.1016/j.clinbiochem.2008.10.005. [DOI] [PubMed] [Google Scholar]
- Mongia SK, Rawlins ML, Owen WE, Roberts WL. Performance characteristics of seven automated CA 125 assays. Am J Clin Pathol. 2006;125:921–927. doi: 10.1309/NBA3-12W0-LANR-XYH9. [DOI] [PubMed] [Google Scholar]
- Morgan BR, Tarter TH. Serum heterophile antibodies interfere with prostate specific antigen test and result in over treatment in a patient with prostate cancer. J Urol. 2001;166:2311–2312. [PubMed] [Google Scholar]
- Preissner CM, O'Kane DJ, Singh RJ, Morris JC, Grebe SK. Phantoms in the assay tube: heterophile antibody interferences in serum thyroglobulin assays. J Clin Endocrinol Metab. 2003;88:3069–3074. doi: 10.1210/jc.2003-030122. [DOI] [PubMed] [Google Scholar]
- Rawlins ML, Roberts WL. Performance characteristics of six third-generation assays for thyroid-stimulating hormone. Clin Chem. 2004;50:2338–2344. doi: 10.1373/clinchem.2004.039156. [DOI] [PubMed] [Google Scholar]
- Reinsberg J. Different efficacy of various blocking reagents to eliminate interferences by human antimouse antibodies with a two-site immunoassay. Clin Biochem. 1996;29:145–148. doi: 10.1016/0009-9120(95)02044-6. [DOI] [PubMed] [Google Scholar]
- Resing KA, Ahn NG. Proteomics strategies for protein identification. FEBS Lett. 2005;579:885–889. doi: 10.1016/j.febslet.2004.12.001. [DOI] [PubMed] [Google Scholar]
- Riches PG, Sheldon J, Smith AM, Hobbs JR. Overestimation of monoclonal immunoglobulin by immunochemical methods. Ann Clin Biochem. 1991;28(Pt 3):253–259. doi: 10.1177/000456329102800310. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Cabaleiro D, Van Uytfanghe K, Stove V, Fiers T, Thienpont LM. Pilot study for the standardization of insulin immunoassays with isotope dilution liquid chromatography/tandem mass spectrometry. Clin Chem. 2007;53:1462–1469. doi: 10.1373/clinchem.2007.088393. [DOI] [PubMed] [Google Scholar]
- Rotmensch S, Cole LA. False diagnosis and needless therapy of presumed malignant disease in women with false-positive human chorionic gonadotropin concentrations. Lancet. 2000;355:712–715. doi: 10.1016/S0140-6736(00)01324-6. [DOI] [PubMed] [Google Scholar]
- Rush J, Moritz A, Lee KA, Guo A, Goss VL, Spek EJ, Zhang H, Zha XM, Polakiewicz RD, Comb MJ. Immunoaffinity profiling of tyrosine phosphorylation in cancer cells. Nat Biotechnol. 2005;23:94–101. doi: 10.1038/nbt1046. [DOI] [PubMed] [Google Scholar]
- Sapin R. The interference of insulin antibodies in insulin immunometric assays. Clin Chem Lab Med. 2002;40:705–708. doi: 10.1515/CCLM.2002.121. [DOI] [PubMed] [Google Scholar]
- Sapin R, Agin A, Gasser F. Efficacy of a new blocker against anti-ruthenium antibody interference in the Elecsys free triiodothyronine assay. Clin Chem Lab Med. 2007;45:416–418. doi: 10.1515/CCLM.2007.064. [DOI] [PubMed] [Google Scholar]
- Sapin R, d'Herbomez M, Schlienger JL, Wemeau JL. Anti-thyrotropin antibody interference in thyrotropin assays. Clin Chem. 1998;44:2557–2559. [PubMed] [Google Scholar]
- Silva F, Carvalho F, Peixoto A, Seixas M, Almeida R, Carneiro F, Mesquita P, Figueiredo C, Nogueira C, Swallow DM, Amorim A, David L. MUC1 gene polymorphism in the gastric carcinogenesis pathway. Eur J Hum Genet. 2001;9:548–552. doi: 10.1038/sj.ejhg.5200677. [DOI] [PubMed] [Google Scholar]
- Siva N. 1000 Genomes project. Nat Biotechnol. 2008;26:256. doi: 10.1038/nbt0308-256b. [DOI] [PubMed] [Google Scholar]
- Spencer CA, Lopresti JS. Measuring thyroglobulin and thyroglobulin autoantibody in patients with differentiated thyroid cancer. Nat Clin Pract Endocrinol Metab. 2008;4:223–233. doi: 10.1038/ncpendmet0757. [DOI] [PubMed] [Google Scholar]
- Spencer CA, Takeuchi M, Kazarosyan M, Wang CC, Guttler RB, Singer PA, Fatemi S, LoPresti JS, Nicoloff JT. Serum thyroglobulin autoantibodies: prevalence, influence on serum thyroglobulin measurement, and prognostic significance in patients with differentiated thyroid carcinoma. J Clin Endocrinol Metab. 1998;83:1121–1127. doi: 10.1210/jcem.83.4.4683. [DOI] [PubMed] [Google Scholar]
- Taylor BS, Pal M, Yu J, Laxman B, Kalyana-Sundaram S, Zhao R, Menon A, Wei JT, Nesvizhskii AI, Ghosh D, Omenn GS, Lubman DM, Chinnaiyan AM, Sreekumar A. Humoral response profiling reveals pathways to prostate cancer progression. Mol Cell Proteomics. 2008;7:600–611. doi: 10.1074/mcp.M700263-MCP200. [DOI] [PubMed] [Google Scholar]
- von Mensdorff-Pouilly S, Gourevitch MM, Kenemans P, Verstraeten AA, Litvinov SV, van Kamp GJ, Meijer S, Vermorken J, Hilgers J. Humoral immune response to polymorphic epithelial mucin (MUC-1) in patients with benign and malignant breast tumours. Eur J Cancer. 1996;32A:1325–1331. doi: 10.1016/0959-8049(96)00048-2. [DOI] [PubMed] [Google Scholar]
- Ward AM, Catto JW, Hamdy FC. Prostate specific antigen: biology, biochemistry and available commercial assays. Ann Clin Biochem. 2001;38:633–651. doi: 10.1258/0004563011901055. [DOI] [PubMed] [Google Scholar]
- Wettero J, Nilsson L, Jonasson L, Sjowall C. Reduced serum levels of autoantibodies against monomeric C-reactive protein (CRP) in patients with acute coronary syndrome. Clin Chim Acta. 2009;400:128–131. doi: 10.1016/j.cca.2008.10.002. [DOI] [PubMed] [Google Scholar]
- Whitehouse CM, Dreyer RN, Yamashita M, Fenn JB. Electrospray interface for liquid chromatographs and mass spectrometers. Anal Chem. 1985;57:675–679. doi: 10.1021/ac00280a023. [DOI] [PubMed] [Google Scholar]
- Willman JH, Martins TB, Jaskowski TD, Hill HR, Litwin CM. Heterophile antibodies to bovine and caprine proteins causing false-positive human immunodeficiency virus type 1 and other enzyme-linked immunosorbent assay results. Clin Diagn Lab Immunol. 1999;6:615–616. doi: 10.1128/cdli.6.4.615-616.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yalow RS, Berson SA. Immunoassay of endogenous plasma insulin in man. J Clin Invest. 1960;39:1157–1175. doi: 10.1172/JCI104130. [DOI] [PMC free article] [PubMed] [Google Scholar]