Abstract
Background
HMGCR (3-Hydroxy-3-methylglutaryl coenzyme A reductase), the direct target of statin inhibition, undergoes alternative splicing of exon 13, which encodes part of the statin-binding domain of the enzyme. We hypothesized that HMGCR alternative splicing might be related to the interindividual variation in plasma low-density lipoprotein cholesterol response to statin treatment.
Methods and Results
We measured mRNA expression of both the full-length and the alternatively spliced HMGCR transcript lacking exon 13 (HMGCRv_1) in 170 simvastatin-incubated immortalized lymphocyte cell lines derived from participants in the Cholesterol and Pharmacogenetics (CAP) study who were treated with simvastatin 40 mg/d for 6 weeks. Greater upregulation of HMGCRv_1 in vitro was significantly correlated (P≤0.0001) with smaller in vivo reductions of plasma total cholesterol, low-density lipoprotein cholesterol, apoprotein B, and triglycerides and explained 6% to 15% of the variation in their response to treatment. In contrast, no significant relationship was found between expression of the full-length HMGCR transcript and in vivo response. By siRNA knockdown of the full-length transcript, we found that HMGCR enzyme activity measured in cells enriched in HMGCRv_1 was relatively resistant to statin inhibition, consistent with the association of increased alternative splicing with reduced statin response in the CAP study. In addition, we found that a common HMGCR single-nucleotide polymorphism (rs3846662) located within intron 13 was associated with variation in the proportion of HMGCR mRNA that is alternatively spliced.
Conclusions
Variation in the production of an HMGCR isoform with reduced statin sensitivity is a determinant of interindividual differences in low-density lipoprotein cholesterol, apolipoprotein B, and triglyceride response to statin treatment.
Keywords: cholesterol, genes, genetics, statins
Elevated total and low-density lipoprotein cholesterol (LDL-C) levels are major risk factors for cardiovascular disease, the leading cause of death in the industrialized world. HMGCR (3-Hydroxy-3-methylglutaryl coenzyme A reductase) inhibitors, or statins, inhibit endogenous cholesterol synthesis, thus stimulating uptake of LDL-C and resulting in a net reduction of plasma LDL-C. Although the efficacy of statins for lowering LDL-C and cardiovascular disease risk has been clearly demonstrated, a wide range of response exists among individuals.1–4 Known determinants of statin efficacy include age, smoking status, diet, body weight, physical activity, racial ancestry, and genetics.4–7 We recently reported that 2 common linked intronic single-nucleotide polymorphisms within HMGCR (rs17244841 and rs17238540, also designated SNPs 12 and 29) are associated with reduced simvastatin efficacy in the Cholesterol and Pharmacogenetics (CAP) study and form a haplotype (H7) with a third SNP, rs3846662 (also designated 20144), that is located in intron 13.8 This haplotype has been independently reported to be associated with attenuated LDL-C lowering with pravastatin treatment.9
The 3 SNPs that make up H7 are intronic and not linked to a common coding SNP, suggesting that they may instead influence HMGCR gene expression. mRNA alternative splicing is a mechanism of generating variation that has become recognized as an important mechanism underlying differences in drug response (reviewed by Bracco and Kearsey10). Johnson et al11 reported the existence of an alternatively spliced transcript of HMGCR, HMGCRv_1, that lacks exon 13 but retains the open reading frame. HMGCRv_1 expression has previously been identified in >80 different tissue types, including liver, fetal liver, lymph node, peripheral leukocyte, and bone marrow.12 Because exon 13 encodes a portion of the substrate-binding domain and statins act as competitive inhibitors of HMGCR, it is possible that the resulting isoform, HMGCRv_1, has altered enzyme activity and/or statin sensitivity compared with the full-length or classic isoform. Because intracellular cholesterol homeostasis is subject to precise regulation, variation in HMGCR alternative splicing also may be reflected in differences in expression of low-density lipoprotein receptor (LDLR). Thus, in this study, we sought to determine whether HMGCR alternative splicing of exon 13 is related to LDL-C response to statin treatment by measuring expression of both full-length and HMGCRv_1 transcripts in statin-incubated immortalized lymphocyte cell lines that were derived from subjects whose LDL-C response to statin had been measured previously and seeking correlations of in vitro gene expression with in vivo lipid response in the corresponding individuals. In addition, we sought evidence for functional consequences of exon 13 skipping by measuring enzyme activity in cells enriched in HMGCRv_1 via siRNA knockdown of the full-length transcript and testing for associations between HMGCR alternative splicing and LDLR gene expression. Finally, we sought to determine whether the magnitude of alternative splicing is associated with the HMGCR intron 13 SNP rs3846662 genotype.
Methods
Subjects
The CAP study included 335 black and 609 white men and women ≥30 years of age with baseline total cholesterol of 160 to 400 mg/dL.6 A 2-week run-in with placebo was conducted to exclude potential participants unable to maintain ≥90% compliance. Other exclusion criteria included use of a lipid-lowering medication, known liver disease, and uncontrolled hypertriglyceridemia, blood pressure, or diabetes mellitus.6 Baseline health, demographic, and physical examination data were obtained, and plasma total cholesterol, LDL-C, high-density lipoprotein cholesterol (HDL-C), triglycerides, and apolipoprotein (apo)B were measured on enrollment and after 4 and 6 weeks on 40 mg/d simvastatin. Compliance was assessed by pill counts every 2 weeks and averaged >95%. SNP rs3846662 was genotyped as previously described.8 The protocol for the study was approved by the Committees for the Protection of Human Subjects at Children’s Hospital and Research Center Oakland; the University of California San Francisco Medical Center; University of California, Los Angeles; University of Washington; and Cedars-Sinai Medical Center.
Simvastatin Incubation of Immortalized Lymphocytes
Lymphocytes were isolated from each CAP subject with IsoPrep (Robbins Scientific Corp, Sunnyvale, Calif) and immortalized by Epstein-Barr virus transformation.13–15 Simvastatin, kindly provided by Merck Inc (Whitehouse Station, NJ), was 98% converted to the active form as previously described.16 A subset of 170 cell lines were grown at 37°C and 5% CO2 in RPMI Medium 1640 (Invitrogen, Carlsbad, Calif) supplemented with 10% FBS (HyClone, Logan, Utah), 500 U/mL penicillin/streptomycin, and 2 nmol/L GlutaMAX (Invitrogen) and exposed in replicate to 1.8 µmol/L (n=119) or 14.5 µmol/L (n=51) activated simvastatin or a control solution for 24 hours.
HMGCR mRNA Quantification
RNA from replicate experiments was pooled, and HMGCR gene expression was measured with 3 TaqMan assays spanning exons 6 to 7 (H5), exons 12 to 13 (H13), and exons 12 to 14 (H14) to determine the total, full-length, and HMGCRv_1 transcripts, respectively, via prequantified serially diluted standards. The H14 assay consisted of primers (CTCCAGTACCTACCTTACAGGGATT and GCTGCTG GCACCTCCA) and probe (5′-FAM-CAAGCAAGGAGTAATTAT-nonfluorescent quencher-3′), and the H5 and H13 assays were purchased from Applied Biosystems (Foster City, Calif). LDLR gene expression also was measured via TaqMan Assay (Applied Biosystems). Each real-time polymerase chain reaction (PCR) was performed in triplicate on an ABI PRISM 7900 Sequence Detection System with standard reagents and 125 ng cDNA (Applied Biosystems).
siRNA Transfection
FreeStyle 293 human kidney-derived cells (Invitrogen) were transfected with 2 siRNA duplexes (Dharmacon, Lafayette, Colo) targeting exon 13 (UAUCCAAUAACAUUCUCACUU and UUGCUCUGCAGCCUCUAUUU) or a nontargeting control siRNA, siCONTROL pool 2 (Dharmacon), at a final concentration of 100 nmol/L with 293fectin (Invitrogen). HMGCR gene expression and enzyme activity were measured in siRNA-transfected and control cells 16 hours after transfection.
HMGCR Enzymatic Activity
HMGCR enzyme activity was measured in the presence or absence of simvastatin (0.15, 1.5, and 6.0 nmol/L) as previously described.17 Activity was expressed as picomole of mevalonate formed per minute per 500 000 homogenized live cells. The traditional units of per 1 mg protein may be inappropriate because protein may be quantified from both living and dead cells, whereas only living homogenized cells contribute to activity. Cell count was determined on the Guava Personal Cell Analysis fluorescent-activated cell sorting (Guava Technologies Inc, Hayward, Calif).
Statistical Analyses
The real-time assay replicates were normalized against SLC7A and CLPTM1,18 the expression of which was validated to be unchanged by simvastatin treatment (data not shown). Fold changes were calculated as the mRNA quantity of the statin-treated sample divided by the buffer-exposed sample. The ratio of fold changes, HMGCRv_1 to total HMGCR, was used to assess splicing independently of the effects on overall transcription. Multivariate regression models were created with the absolute and percent changes of in vivo lipid parameters as dependent variables and in vitro HMGCR expression as independent variables. Relationships between gene expression and plasma lipid and lipoprotein changes were examined separately for each in vitro statin dose (1.8 µmol/L [n=119] and 14.5 µmol/L [n=51]); the results were combined because similar associations were seen at both doses and because the magnitude of induction of either HMGCR transcript did not differ significantly between the 2 doses. All models included adjustment for age, race, and smoking status because these variables were associated with LDL-C response to simvastatin in CAP,6 as well as sex and body mass index. Plasma triglyceride concentrations were log transformed. Bonferroni adjustment was used to estimate significance after adjustment for multiple comparisons. All statistical analyses were performed with JMP version 6.0.3 (SAS Institute, Inc, Cary, NC).
The authors had full access to and take full responsibility for the integrity of the data. All authors have read and agree to the manuscript as written.
Results
Identification of HMGCRv_1 Expression in Immortalized Lymphocytes
Reverse-transcription PCR with primers spanning exon 12 to 14 revealed variation in the magnitude of HMGCRv_1 expression across cell lines derived from CAP subjects (Figure 1). Real-time PCR analysis of 170 immortalized lymphocyte cell lines showed that basal levels of HMGCRv_1 transcripts varied widely, ranging from 15 372 to 492 992 molecules of HMGCRv_1/125 ng cDNA and representing between 7.5% and 95.3% of the total HMGCR transcripts. On average, HMGCRv_1 transcripts represented 41.5% of the total HMGCR transcripts quantified. Reverse-transcription PCR of various sets of all HMGCR exon-exon junctions did not identify any other alternatively spliced HMGCR transcripts (data not shown).
Figure 1.
A, HMGCR exon 13 alternative splicing patterns. B, Full-length HMGCR and HMGCRv_1 expression in immortalized lymphocyte cell lines. Reverse-transcription PCR with primers spanning exon 12 to 14 was used to amplify cDNA from 10 different immortalized lymphocyte cell lines from CAP subjects.
In Vivo Response to Simvastatin Treatment
The demographic characteristics and lipid and apoprotein responses to simvastatin treatment in the subset of CAP subjects from whom the cell lines in the present analysis were derived are shown in Table 1 and Table 2. The subset was enriched in blacks and had a slightly greater proportion of female subjects than did the entire cohort. In addition, the population tested had somewhat greater baseline triglycerides, LDL-C, and HDL-C, as well as lower total cholesterol, LDL-C, and HDL-C changes, with statin treatment.
Table 1.
Clinical Characteristics of Study Participants
| Tested Subset | Entire CAP Cohort | |
|---|---|---|
| N | 170 | 944 |
| Women, % | 52.0* | 48.6 |
| BMI | 29.31 ± 0.46 | 28.56 ± 0.19 |
| Black, % | 64.7* | 35.5 |
| Age, y | 56.25 ± 1.01* | 54.40 ± 0.41 |
| Smoking >1 pack/d, % | 20.0 | 19.9 |
BMI indicates body mass index.
The mean was significantly greater than the entire CAP cohort (P<0.01).
Table 2.
Lipid Measurements in Study Participants
| Tested Subset Only |
Baseline, mg/dL | Absolute Difference, mg/dL |
Difference, % |
|---|---|---|---|
| Total cholesterol |
212.92 ± 2.51 | −59.49 ± 1.92† | −27.43 ± 0.75† |
| LDL-C | 135.24 ± 2.36* | −56.87 ± 1.75† | −41.59 ± 0.97 |
| HDL-C | 55.97 ± 1.21* | 1.42 ± 0.40† | 3.05 ± 0.77 |
| ApoB | 94.01 ± 1.54 | −29.08 ± 1.12 | −30.05 ± 1.10 |
| Triglycerides | 109.03 ± 4.09† | −19.84 ± 2.87 | −15.58 ± 1.93 |
The mean was significantly greater than the entire CAP cohort (P<0.01).
The mean was significantly lower than the entire CAP cohort (P<0.05).
Upregulation of HMGCR mRNA Expression in Statin-Treated Lymphocytes
To determine whether HMGCRv_1 expression is upregulated with statin treatment, 170 immortalized lymphocyte cell lines were incubated with activated simvastatin (1.8 µmol/L [n=119] or 14.5 µmol/L [n=51]) or sham buffer. Full-length HMGCR and HMGCRv_1 transcripts were induced to a similar degree, 1.53 ± 0.03-fold and 1.45 ± 0.04-fold (average ± SE), respectively. Induced levels of both transcripts were slightly higher in men than women, with full-length HMGCR upregulated 1.56 ± 0.04-fold in men compared with 1.44 ± 0.04-fold in women (P<0.01) and HMGCRv_1 upregulated 1.57 ± 0.07-fold in men versus 1.38 ± 0.04-fold in women (P<0.01). On the other hand, no significant differences were found in the magnitude of induction of either transcript between cell lines derived from blacks and those derived from whites. In a subset of cell lines (n=5), each incubated with 7 simvastatin concentrations ranging from 0.6 to 14.5 µmol/L, it was determined that 0.83 µmol/L was required for half-maximal induction of both HMGCR transcripts (data not shown).
HMGCR mRNA Expression in Statin-Treated Lymphocytes Is Correlated With In Vivo Lipid Response
We next tested whether the magnitude of transcriptional upregulation of either the full-length or HMGCRv_1 transcripts in response to in vitro simvastatin incubation was related to interindividual differences in statin efficacy as assessed by lipid and lipoprotein changes in vivo (Figure 2). No significant relationships were found between in vitro expression of the full-length HMGCR transcript and in vivo response (data not shown). In contrast, greater in vitro induction of HMGCRv_1 was significantly (P<0.001) associated with smaller in vivo absolute and percent reductions of total cholesterol, LDL-C, apoB, and triglycerides (Figure 2). HMGCRv_1 expression was not significantly associated with the absolute or percentage changes in HDL-C.
Figure 2.
Associations between in vitro HMGCR alternative splicing after statin treatment and in vivo percentage response of total cholesterol, LDL-C, apoB, triglycerides, and HDL-C. Results are for the combined analysis of 170 subjects whose cells were incubated with 1.8 µmol/L (n=119) or 14.5 µmol/L (n=51) (see Methods). P values are for multiple regression models including age, race, smoking status, sex, and body mass index.
In regression models, HMGCRv_1 expression explained 9% of the variance in changes of both total and LDL-C and 15% of the variance in change of apoB. The magnitude of LDL-C change associated with variation in HMGCRv_1 expression ranged from −68% to −3% of baseline. In vitro HMGCRv_1 expression in the absence of statin was not associated with in vivo lipid or lipoprotein measurements at baseline (data not shown)
The relationship between expression of HMGCRv_1 and statin efficacy was significant in both races (n=110 blacks, n=60 whites; both P<0.01), and no significant difference was found in HMGCRv_1 expression after statin treatment between the 2 races. However, the ratio of HMGCRv_1 to total HMGCR was slightly higher in blacks than whites, 0.99 ± 0.02 and 0.96 ± 0.06, respectively (P=0.29).
Cellular Enrichment of HMGCRv_1 Results in Resistance of HMGCR Activity to Simvastatin
The association between greater alternative splicing and lower statin response raised the possibility that exon 13 skipping results in an HMGCR isoform resistant to statin inhibition compared with the full-length isoform. To test this hypothesis, we produced 293 human kidney cells enriched in the HMGCRv_1 transcript by knocking down the expression of the full-length transcript using 2 siRNA duplexes targeted to exon 13. siRNA transfection resulted in ≈68% knockdown of the full-length transcript with minimal effects on HMGCRv_1 (Figure 3A). In addition, siRNA-transfected cells had greater residual enzymatic activity when treated with concentrations of simvastatin ranging from 150 pmol/L to 6.0 nmol/L (Figure 3B). Transfection with a nontargeting control siRNA duplex did not alter the expression of either HMGCR transcript, nor did it affect statin sensitivity of the enzyme (data not shown), demonstrating that the change in statin sensitivity was due specifically to HMGCRv_1 enrichment.
Figure 3.
HMGCR gene expression (A) and enzyme activity (B) after knockdown of the full-length HMGCR transcript via siRNA. A, HMGCR gene expression measured by real-time PCR in 293 kidney cells (n=8) after 16 hours of transfection with 2 siRNA duplexes targeting exon 13. B, Comparison of HMGCR enzyme activity measured in the presence of 3 concentrations of simvastatin from siRNA-transfected vs control cells (n=8). No changes in either HMGCR transcript levels or enzyme activity were seen after transfection with a nontargeting control siRNA duplex (data not shown).
Relationship Between HMGCR Alternative Splicing and LDLR Gene Expression in Response to Statin
As predicted by their coregulation by sterol regulatory element-binding proteins, the magnitude of LDLR with statin incubation upregulation was positively correlated with total HMGCR induction (r=0.30, P=0.0001). However, greater alternative splicing of HMGCR with statin treatment was associated with reduced upregulation of LDLR (r=−0.10, P=0.0342). Because statins induce LDL-C uptake as a consequence of LDLR-mediated LDL-C clearance, smaller statin-induced upregulation of LDLR is consistent with our finding that HMGCR exon 13 alternative splicing influences the ability of statin to reduce plasma LDL-C levels.
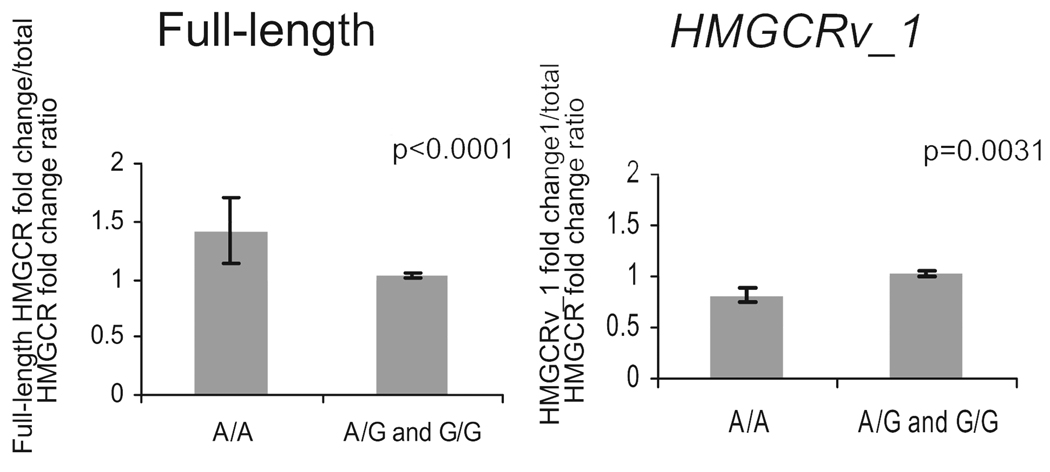
Relationship Between HMGCR SNP rs3846662 Genotype and Exon 13 Alternative Splicing
Figure 4 shows that incubation of cells with 1.8 µmol/L simvastatin resulted in a 40% greater induction of the full-length HMGCR transcript in SNP rs3846662 A/A homozygotes than in either A/G or G/G subjects (P<0.0001). Conversely, expression of HMGCRv_1 in the A/A individuals was 20% lower than in carriers of 1 copy of the G allele (P=0.0031). No statistically significant differences were found between induction of either the full-length HMGCR or HMGCRv_1 transcripts between the A/G and G/G subjects.
Figure 4.
Association of the HMGCR SNP rs3846662 genotype with statin-induced expression of the unspliced and alternatively spliced HMGCR transcripts. HMGCR gene expression was measured in immortalized lymphocytes derived from CAP subjects with 3 TaqMan assays specific for the total HMGCR transcripts, the full-length HMGCR transcript (including exon 13), and HMGCRv_1 (lacking exon 13) after 24 hours of incubation with 1.8 µmol/L simvastatin (n=119). HMGCR SNP rs3846662 was genotyped with Ampliflor primers, and associations between baseline and statin-treated HMGCR gene expression with SNP rs3846662 genotype were tested with a dominant model in JMP 6.0. Ratios of the fold changes in both the HMGCRv_1 and full-length HMGCR transcripts to the fold change in total HMGCR transcripts were used to adjust the measurements of exon 13 alternative splicing for overall changes in HMGCR gene expression.
Discussion
mRNA alternative splicing is a mechanism of generating variation that has been implicated as a mechanism regulating cardiovascular disease risk.19–24 Recently, alternative splicing has become recognized as an important mechanism underlying differences in drug response.25 For example, alternative splicing of the sodium-potassium-chloride transporter 2 (NKCC2) has been shown to influence furosemide treatment for hypertension because of differential transport kinetics of the resulting isoforms.26
Here, we provide evidence that interindividual variation in the magnitude of expression of an alternatively spliced HMGCR mRNA lacking exon 13, HMGCRv_1, contributes to variability in LDL-C, apoB, and triglyceride reduction with simvastatin therapy. Greater in vitro upregulation of HMGCRv_1 after statin treatment was significantly associated with reduced in vivo statin-induced change in total cholesterol, LDL-C, apoB, and triglyceride. This relationship is attributable specifically to HMGCR alternative splicing because no association between full-length HMGCR mRNA expression and statin response was found. Additionally, greater induction of HMGCRv_1 with statin treatment was associated with reduced upregulation of LDLR, one of the major mechanisms by which statins lower levels of plasma LDL-C, apoB, and triglyceride-rich lipoproteins. Thus, in vitro HMGCRv_1 expression is a molecular marker for the magnitude of LDL-C and apoB reductions with statin therapy.
On the basis of our current data, HMGCRv_1 expression explains only a fraction of the variation in LDL-C, apoB, and triglyceride response to statins (9%, 15%, and 6%, respectively); however, this proportion probably is highly underestimated because of the nature of the experiment. A direct measurement of hepatic HMGCRv_1 in a subject currently undergoing statin treatment would likely be much more closely related to his or her lipid and lipoprotein response than our in vitro measurement in a statin-incubated immortalized lymphocyte cell line. In addition, when age, race, smoking status, and body mass index are added to the model, the combination of these parameters accounts for 24% of the variation in LDL-C response, 29% of the variation in apoB response, and 8% of the variation in triglyceride, with HMGCRv_1 gene expression the most significant predictor of response in all 3. Because previous studies of genetic variation associated with statin response have explained <2% to 7.5% of the variation in LDL-C and HDL-C,8,27 our findings point to a pathway that has much more substantial impact than previously identified SNPs. This finding is the first to establish a substantial contribution of a biological process to variation in statin efficacy. Furthermore, because HMGCR is directly involved in cholesterol homeostasis, these results demonstrate that variation in pharmacodynamics can have relevance to drug efficacy in addition to traditional pharmacokinetic mechanisms.
In the original description of HMGCRv_1, Johnson et al11 assumed that the resulting HMGCR isoform would be catalytically inactive because exon 13 (residues 522 to 574) encodes a portion of the catalytic domain, generally described as exons 11 through 20 (residues 426 to 888).11,28–30 However, the key residues for substrate binding, known as the cis loop (residues 684 to 692), are encoded by exon 16 and thus may not be affected by exon 13 skipping. Although catalytic function may be preserved, the deletion may diminish enzymatic activity owing to the loss of Glu559, a residue involved in catalysis, and the truncation of the L domain (residues 528 to 590 and 694 to 872), which makes multiple contacts with HMG-CoA.28,31 Verification of this hypothesis requires identification and characterization of a protein isoform or expressed construct specifically lacking exon 13.
Statins competitively inhibit HMGCR activity by forming polar interactions between the HMG moiety of the statin and the cis loop of HMGCR. In addition, statins form multiple van der Waals interactions with HMGCR residues such as Leu562 (encoded by exon 13); thus, its loss may contribute to the apparent reduction in simvastatin sensitivity that we observed. Other statins such as rosuvastatin, atorvastatin, and fluvastatin form additional interactions with Arg590, Arg568, and Ser565.32 Further investigation is required to determine whether HMGCRv_1 has additional resistance to these statins. The change in primary structure with the loss of exon 13 also could influence the secondary, tertiary, or quaternary structure in a manner that could alter either substrate or inhibitor binding.28
An earlier example of an HMGCR mRNA splicing aberration was described in the UT2* cell line, a mutagenized Chinese hamster ovary cell line containing 2 HMGCR splice donor site mutations. These mutations interrupt the open reading frame, and the predicted truncated protein would lack the entire catalytic domain.33 Despite these mutations, the UT2* cell line expresses a smaller but enzymatically active isoform of HMGCR that has lower specific activity (52 versus 275 pmol·min−1·mg−1) and statin sensitivity compared with the normal isoform.34 The authors were unable to identify an HMGCR transcript that could account for this novel isoform; however, this finding suggests that splicing mutations around exon 13 can result in an active, and partially statin-resistant, isoform.
Our finding of a correlation between greater statin-induced levels of HMGCRv_1 and reduced LDLR upregulation suggests that exon 13 skipping has a functional effect on HMGCR statin sensitivity. In further support of the functional effect of HMGCR alternative splicing, we found that greater alternative splicing at baseline is correlated with greater expression of LDLR (r=0.26, P=0.0001; data not shown). This relationship could account for the lack of a correlation between HMGCR alternative splicing and LDL-C levels at baseline because concordant regulation would be expected to maintain cholesterol homeostasis.
Our studies have demonstrated that SNP rs3846662, a common genetic polymorphism of human HMGCR, is related to the proportion of HMGCR mRNA that is alternatively spliced. The G allele was reciprocally associated with an increase in HMGCRv_1 expression and a decrease in the expression of the full-length transcript, suggesting that the allele containing this SNP specifically modulates exon 13 alternative splicing and not overall HMGCR gene expression. The SNP is located within intron 13 but is not found in either the splice donor or acceptor sites of exon 13 or 14. However, its location proximal to the site of alternative splicing suggests that it may lie in a regulatory motif responsible for binding factors that modulate exon 13 skipping.
Although the SNP rs3846662 genotype is related to statin-induced expression of HMGCRv_1, the fact that we identified both the full-length and HMGCRv_1 transcripts in all subjects tested regardless of genotype demonstrates that SNP rs3846662 is not the sole determinant of exon 13 alternative splicing. This may account for our finding that in the full CAP population, the SNP rs3846662 genotype by itself was not significantly associated with statin response.8 Further studies are required to determine whether rs3846662 has a direct effect on HMGCR alternative splicing and what other factors may modulate the extent of splicing.
Previously, we reported that association of statin resistance with H7, a haplotype that contains SNP rs3846662, was observed in blacks but not in whites in the CAP study.8 In contrast, the HMGCRv_1 relationship to reduced statin response was found in both the black and white subpopulations tested in the present report. This discrepancy may be due to the fact that a large racial difference exists in the allele frequency of SNP rs3846662, the G allele being almost twice as prevalent in blacks (allele frequency, 87.9%) compared with whites (allele frequency, 47.4%).8 This is consistent with the possibility that the CAP trial may have been underpowered to detect the H7 statin-resistant phenotype in whites.
One limitation of the present study is the use of immortalized lymphocyte cell lines as the model system compared with measurements of gene expression in vivo or in fresh lymphocytes. Moreover, statins act primarily in the liver, and it is not known whether the HMGCR alternative splicing that we have observed in lymphocytes occurs in parallel in the liver. However, HMGCRv_1 has been previously identified in the liver, and peripheral mononuclear cell gene expression has been shown to be a marker for hepatic cholesterol metabolism with concordant HMGCR regulation between the 2 tissues.11,35,36
Our findings point to a major role of HMGCR alternative splicing in influencing cholesterol response to statin treatment and thus exemplify how alternative splicing can act as a modifier of drug response. Although measurement of HMGCRv_1 expression on its own does not yet have clinical utility, its importance lies in highlighting new pathways and effects mediated by alternative splicing and its impacts on mechanisms related to cholesterol metabolism. This information also may lead to improved prediction of individuals who would be most likely to benefit from statin treatment and to the identification of new drug targets for improving statin efficacy.
CLINICAL PERSPECTIVE.
Statins reduce low-density lipoprotein cholesterol by inhibiting 3-hydroxy-3-methylglutaryl-coenzyme A reductase (HMGCR), a rate-limiting enzyme for cholesterol synthesis. Although statins are generally efficacious, a wide range of low-density lipoprotein cholesterol–lowering response exists among individuals. We have identified an alternatively spliced transcript of HMGCR that lacks exon 13, HMGCRv_1, and measured its expression in 170 simvastatin-incubated immortalized lymphocyte cell lines derived from participants in the Cholesterol and Pharmacogenetics (CAP) study who were treated with simvastatin 40 mg/d for 6 weeks. We found that greater upregulation of HMGCRv_1 in vitro was significantly correlated (P≤0.0001) with smaller in vivo reductions of plasma total and low-density lipoprotein cholesterol, triglycerides, and apolipoprotein B and explained 6% to 15% of the variation in the statin response of these measurements. Artificial enrichment of HMGCRv_1 via siRNA produced cells relatively resistant to statin inhibition, consistent with the association of increased alternative splicing with reduced statin response in the CAP study. Our findings point to a major role of HMGCR alternative splicing in influencing cholesterol response to statin treatment and exemplify how alternative splicing can act as a modifier of drug response. Although measurement of HMGCRv_1 expression on its own does not yet have clinical utility, its importance lies in highlighting new pathways and effects mediated by alternative splicing and its impacts on mechanisms related to cholesterol metabolism. This information also may lead to improved prediction of individuals who would be most likely to benefit from statin treatment and to the identification of new drug targets for improving statin efficacy.
Acknowledgments
We thank Dr Lara Mangravite for developing the cell culture protocols, Sheila Pressman for supervising the establishment of the lymphoblastoid cell lines, Dr Sandra Erickson for consultation on the assay of HMGCR, and the Children’s Hospital Oakland Research Institute Core Lipoprotein Laboratory for lipid and apoprotein measurements.
Sources of Funding
This work was funded by National Institutes of Health grant U01 HL69757, with additional support from Pfizer Inc, and the Cedars-Sinai Board of Governor’s Chair in Medical Genetics.
Footnotes
Disclosures
Dr Krauss has received grant support from Merck, Merck-Schering Plough, and Pfizer and has consulted for Merck and Merck-Schering Plough. The other authors report no conflicts.
References
- 1.Brousseau ME. Statins, super-statins and cholesterol absorption inhibitors. IDrugs. 2003;6:458–463. [PubMed] [Google Scholar]
- 2.LaRosa JC. Statins and risk of coronary heart disease. JAMA. 2000;283:2935–2936. [PubMed] [Google Scholar]
- 3.Sacks FM, Moye LA, Davis BR, Cole TG, Rouleau JL, Nash DT, Pfeffer MA, Braunwald E. Relationship between plasma LDL concentrations during treatment with pravastatin and recurrent coronary events in the Cholesterol and Recurrent Events trial. Circulation. 1998;97:1446–1452. doi: 10.1161/01.cir.97.15.1446. [DOI] [PubMed] [Google Scholar]
- 4.Shear CL, Franklin FA, Stinnett S, Hurley DP, Bradford RH, Chremos AN, Nash DT, Langendorfer A. Expanded Clinical Evaluation of Lovastatin (EXCEL) study results: effect of patient characteristics on lovastatin-induced changes in plasma concentrations of lipids and lipoproteins. Circulation. 1992;85:1293–1303. doi: 10.1161/01.cir.85.4.1293. [DOI] [PubMed] [Google Scholar]
- 5.Hunt D, Young P, Simes J, Hague W, Mann S, Owensby D, Lane G, Tonkin A. Benefits of pravastatin on cardiovascular events and mortality in older patients with coronary heart disease are equal to or exceed those seen in younger patients: results from the LIPID trial. Ann Intern Med. 2001;134:931–940. doi: 10.7326/0003-4819-134-10-200105150-00007. [DOI] [PubMed] [Google Scholar]
- 6.Simon JA, Lin F, Hulley SB, Blanche PJ, Waters D, Shiboski S, Rotter JI, Nickerson DA, Yang H, Saad M, Krauss RM. Phenotypic predictors of response to simvastatin therapy among African-Americans and Caucasians: the Cholesterol and Pharmacogenetics (CAP) Study. Am J Cardiol. 2006;97:843–850. doi: 10.1016/j.amjcard.2005.09.134. [DOI] [PubMed] [Google Scholar]
- 7.Streja L, Packard CJ, Shepherd J, Cobbe S, Ford I. Factors affecting low-density lipoprotein and high-density lipoprotein cholesterol response to pravastatin in the West of Scotland Coronary Prevention Study (WOSCOPS) Am J Cardiol. 2002;90:731–736. doi: 10.1016/s0002-9149(02)02599-7. [DOI] [PubMed] [Google Scholar]
- 8.Krauss RM, Mangravite LM, Smith JD, Medina MW, Wang D, Guo X, Rieder MJ, Simon JA, Hulley SB, Waters D, Saad M, Williams PT, Taylor KD, Yang H, Nickerson DA, Rotter JI. Variation in the 3-hydroxyl-3-methylglutaryl coenzyme a reductase gene is associated with racial differences in low-density lipoprotein cholesterol response to simvastatin treatment. Circulation. 2008;117:1537–1544. doi: 10.1161/CIRCULATIONAHA.107.708388. [DOI] [PubMed] [Google Scholar]
- 9.Chasman DI, Posada D, Subrahmanyan L, Cook NR, Stanton VP, Jr, Ridker PM. Pharmacogenetic study of statin therapy and cholesterol reduction. JAMA. 2004;291:2821–2827. doi: 10.1001/jama.291.23.2821. [DOI] [PubMed] [Google Scholar]
- 10.Bracco L, Kearsey J. The relevance of alternative RNA splicing to pharmacogenomics. Trends Biotechnol. 2003;21:346–353. doi: 10.1016/S0167-7799(03)00146-X. [DOI] [PubMed] [Google Scholar]
- 11.Johnson JM, Castle J, Garrett-Engele P, Kan Z, Loerch PM, Armour CD, Santos R, Schadt EE, Stoughton R, Shoemaker DD. Genome-wide survey of human alternative pre-mRNA splicing with exon junction microarrays. Science. 2003;302:2141–2144. doi: 10.1126/science.1090100. [DOI] [PubMed] [Google Scholar]
- 12.Johnson J, Armour C, Castle J, Garrett-Engele P, inventors. Rosetta Inpharmatics LLC, assignee. Alternatively spliced isoforms of human HMG-CoA reductase. WO 03/102209 US patent. 2003 November 12;
- 13.Anderson MA, Gusella JF. Use of cyclosporin A in establishing Epstein-Barr virus-transformed human lymphoblastoid cell lines. In Vitro. 1984;20:856–858. doi: 10.1007/BF02619631. [DOI] [PubMed] [Google Scholar]
- 14.Boyum A. Separation of white blood cells. Nature. 1964;204:793–794. doi: 10.1038/204793a0. [DOI] [PubMed] [Google Scholar]
- 15.Pressman S, Rotter JI. Epstein-Barr virus transformation of cryopreserved lymphocytes: prolonged experience with technique. Am J Hum Genet. 1991;49:467. [PMC free article] [PubMed] [Google Scholar]
- 16.Brown MS, Faust JR, Goldstein JL, Kaneko I, Endo A. Induction of 3-hydroxy-3-methylglutaryl coenzyme A reductase activity in human fibroblasts incubated with compactin (ML-236B), a competitive inhibitor of the reductase. J Biol Chem. 1978;253:1121–1128. [PubMed] [Google Scholar]
- 17.Erickson SK, Lear SR, Barker ME, Musliner TA. Regulation of cholesterol metabolism in the ethionine-induced premalignant rat liver. J Lipid Res. 1990;31:933–945. [PubMed] [Google Scholar]
- 18.Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, Speleman F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002;3:34.31–0034.0011. doi: 10.1186/gb-2002-3-7-research0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mango R, Biocca S, del Vecchio F, Clementi F, Sangiuolo F, Amati F, Filareto A, Grelli S, Spitalieri P, Filesi I, Favalli C, Lauro R, Mehta JL, Romeo F, Novelli G. In vivo and in vitro studies support that a new splicing isoform of OLR1 gene is protective against acute myocardial infarction. Circ Res. 2005;97:152–158. doi: 10.1161/01.RES.0000174563.62625.8e. [DOI] [PubMed] [Google Scholar]
- 20.Komamura K, Iwai N, Kokame K, Yasumura Y, Kim J, Yamagishi M, Morisaki T, Kimura A, Tomoike H, Kitakaze M, Miyatake K. The role of a common TNNT2 polymorphism in cardiac hypertrophy. J Hum Genet. 2004;49:129–133. doi: 10.1007/s10038-003-0121-4. [DOI] [PubMed] [Google Scholar]
- 21.Dunn DM, Ishigami T, Pankow J, von Niederhausern A, Alder J, Hunt SC, Leppert MF, Lalouel JM, Weiss RB. Common variant of human NEDD4L activates a cryptic splice site to form a frameshifted transcript. J Hum Genet. 2002;47:665–676. doi: 10.1007/s100380200102. [DOI] [PubMed] [Google Scholar]
- 22.Fava C, von Wowern F, Berglund G, Carlson J, Hedblad B, Rosberg L, Burri P, Almgren P, Melander O. 24-h ambulatory blood pressure is linked to chromosome 18q21–22 and genetic variation of NEDD4L associates with cross-sectional and longitudinal blood pressure in Swedes. Kidney Int. 2006;70:562–569. doi: 10.1038/sj.ki.5001590. [DOI] [PubMed] [Google Scholar]
- 23.Nikitin AG, Chudakova DA, Spitsina EV, Minushkina LO, Zateishchikov DA, Nosikov VV, Debabov VG. Association of GNB3 gene C825T polymorphism with coronary heart disease [in Russian] Genetika. 2007;43:1129–1133. [PubMed] [Google Scholar]
- 24.Zhu H, Tucker HM, Grear KE, Simpson JF, Manning AK, Cupples LA, Estus S. A common polymorphism decreases low-density lipoprotein receptor exon 12 splicing efficiency and associates with increased cholesterol. Hum Mol Genet. 2007;16:1765–1772. doi: 10.1093/hmg/ddm124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Heinzen EL, Yoon W, Tate SK, Sen A, Wood NW, Sisodiya SM, Goldstein DB. Nova2 interacts with a cis-acting polymorphism to influence the proportions of drug-responsive splice variants of SCN1A. Am J Hum Genet. 2007;80:876–883. doi: 10.1086/516650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gimenez I, Isenring P, Forbush B. Spatially distributed alternative splice variants of the renal Na-K-Cl cotransporter exhibit dramatically different affinities for the transported ions. J Biol Chem. 2002;277:8767–8770. doi: 10.1074/jbc.C200021200. [DOI] [PubMed] [Google Scholar]
- 27.Winkelmann BR, Hoffmann MM, Nauck M, Kumar AM, Nandabalan K, Judson RS, Boehm BO, Tall AR, Ruano G, Marz W. Haplotypes of the cholesteryl ester transfer protein gene predict lipid-modifying response to statin therapy. Pharmacogenomics J. 2003;3:284–296. doi: 10.1038/sj.tpj.6500195. [DOI] [PubMed] [Google Scholar]
- 28.Istvan ES, Palnitkar M, Buchanan SK, Deisenhofer J. Crystal structure of the catalytic portion of human HMG-CoA reductase: insights into regulation of activity and catalysis. EMBO J. 2000;19:819–830. doi: 10.1093/emboj/19.5.819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liscum L, Finer-Moore J, Stroud RM, Luskey KL, Brown MS, Goldstein JL. Domain structure of 3-hydroxy-3-methylglutaryl coenzyme A reductase, a glycoprotein of the endoplasmic reticulum. J Biol Chem. 1985;260:522–530. [PubMed] [Google Scholar]
- 30.Nakajima T, Iwaki K, Hamakubo T, Kodama T, Emi M. Genomic structure of the gene encoding human 3-hydroxy-3-methyl-glutaryl coenzyme A reductase: comparison of exon/intron organization of sterolsensing domains among four related genes. J Hum Genet. 2000;45:284–289. doi: 10.1007/s100380070017. [DOI] [PubMed] [Google Scholar]
- 31.Friesen JA, Rodwell VW. The 3-hydroxy-3-methylglutaryl coenzyme-A (HMG-CoA) reductases. Genome Biol. 2004;5:248.241–248.247. doi: 10.1186/gb-2004-5-11-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Istvan ES, Deisenhofer J. Structural mechanism for statin inhibition of HMG-CoA reductase. Science. 2001;292:1160–1164. doi: 10.1126/science.1059344. [DOI] [PubMed] [Google Scholar]
- 33.Engfelt WH, Masuda KR, Paton VG, Krisans SK. Splice donor site mutations in the 3-hydroxy-3-methylglutaryl coenzyme A reductase gene cause a deficiency of the endoplasmic reticulum 3-hydroxy-3-methylglutaryl coenzyme A reductase protein in UT2 cells. J Lipid Res. 1998;39:2182–2191. [PubMed] [Google Scholar]
- 34.Aboushadi N, Shackelford JE, Jessani N, Gentile A, Krisans SK. Characterization of peroxisomal 3-hydroxy-3-methylglutaryl coenzyme A reductase in UT2 cells: sterol biosynthesis, phosphorylation, degradation, and statin inhibition. Biochemistry. 2000;39:237–247. doi: 10.1021/bi9916325. [DOI] [PubMed] [Google Scholar]
- 35.Aggarwal D, Freake HC, Soliman GA, Dutta A, Fernandez ML. Validation of using gene expression in mononuclear cells as a marker for hepatic cholesterol metabolism. Lipids Health Dis. 2006;5:22.21–22.24. doi: 10.1186/1476-511X-5-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Powell EE, Kroon PA. Low density lipoprotein receptor and 3-hydroxy-3-methylglutaryl coenzyme A reductase gene expression in human mononuclear leukocytes is regulated coordinately and parallels gene expression in human liver. J Clin Invest. 1994;93:2168–2174. doi: 10.1172/JCI117213. [DOI] [PMC free article] [PubMed] [Google Scholar]