Abstract
Transfer-messenger RNA (tmRNA) acts first as a tRNA and then as an mRNA template to rescue stalled ribosomes in eubacteria. Together with its protein partner, SmpB, tmRNA enters stalled ribosomes and transfers an Ala residue to the growing polypeptide chain. A remarkable step then occurs: the ribosome leaves the stalled mRNA and resumes translation using tmRNA as a template, adding a short peptide tag that destines the aborted protein for destruction. Exactly how the ribosome switches templates, resuming translation on tmRNA in the proper reading frame, remains unknown. Within the tmRNA sequence itself, five nucleotides (U85AGUC) immediately upstream of the first codon appear to direct frame selection. In particular, mutation of the conserved A86 results in severe loss of function both in vitro and in vivo. The A86C mutation causes translation to resume exclusively in the +1 frame. Several candidate binding partners for this upstream sequence have been identified in vitro. Using a genetic selection for tmRNA activity in E. coli, we identified mutations in the SmpB protein that restore the function of A86C tmRNA in vivo. The SmpB mutants increase tagging in the normal reading frame and reduce tagging in the +1 frame. These results demonstrate that SmpB is functionally linked with the sequence upstream of the tmRNA template; both contribute to reading frame selection on tmRNA.
Keywords: tmRNA, SmpB, trans-translation, reading frame
Stalled ribosomes in eubacteria are rescued and recycled by a highly conserved quality-control mechanism. Ribosomes stall upon reaching the 3’-end of mRNAs that lack a stop codon. With empty aminoacyl-tRNA sites (A sites), these ribosomes are trapped on the defective mRNA because they cannot efficiently bind release factors. Instead, stalled ribosomes recruit transfer-messenger RNA (tmRNA) and its binding partner, small protein B (SmpB) to their empty A sites. SmpB and aminoacylated tmRNA function first as a tRNA, adding alanine to the nascent polypeptide. The stalled ribosome then resumes translation using tmRNA as a template, translating a short open reading frame that encodes a protease-recognition sequence. Through the action of tmRNA and SmpB, known as trans-translation, the aborted polypeptides are tagged for degradation by cellular proteases and the ribosome is released at a stop codon and recycled.1,2,3
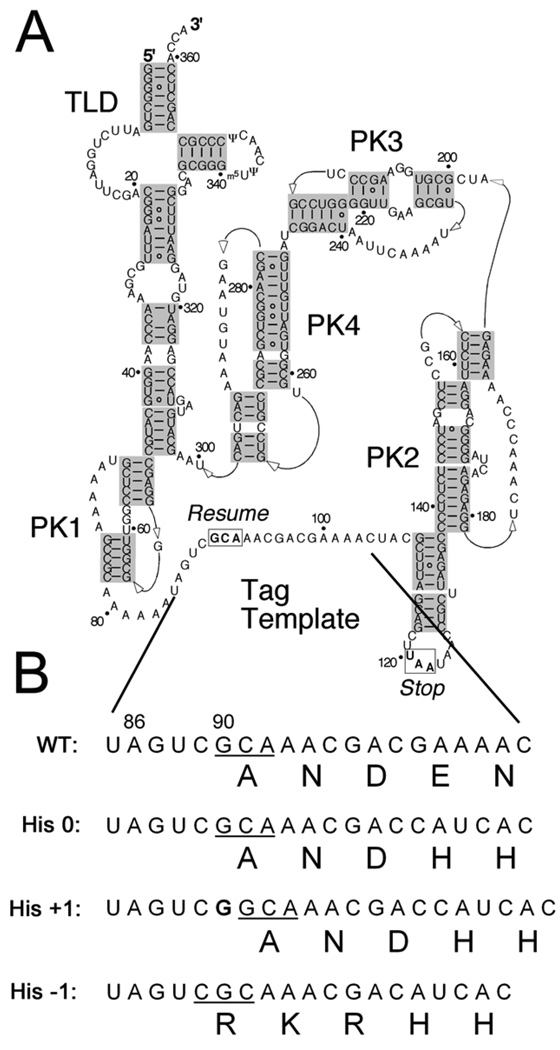
As the ribosome switches templates from the defective mRNA to tmRNA, how is the appropriate codon in tmRNA selected for translation to resume? The global structure of tmRNA plays little role in the selection of the correct frame. The four pseudoknots that dominate the tmRNA structure (Figure 1A) can be replaced with unrelated sequences with little or no loss of tmRNA activity.4,5,6 Instead, the reading frame is chosen locally, by five bases immediately upstream of the resume codon (Figure 1B).7,8,9 Mutations in this upstream sequence (U85AGUC) lead to reduced tmRNA function and errors in frame selection in vitro and in vivo. Mutation of the first two of these nucleotides is particularly deleterious: the U85A mutation, for example, partially shifts translation to the −1 frame.8,9 Mutation of the universally conserved A86 leads to severe loss of function;7,8 the A86C mutation shifts translation entirely to the +1 frame in vivo.9 From these data, we proposed that the resume codon is chosen by its placement in the ribosome as determined by the binding of an unidentified ligand to A86.9
Figure 1.
Sequence and structure of tmRNA. A) Secondary structure of E. coli tmRNA, including the tRNA-like domain (TLD), four pseudoknots (PK1-4), and the resume and stop codons boxed at either end of the tag template sequence.32 B) Fragment of the tmRNA sequence including the first five codons in the tag template as well as five nucleotides of the upstream sequence known to play a role in frame selection. The resume codon is underlined. The His 0 template encodes ANDH6D in the natural frame. Addition of a G before G90 leads to the synthesis of this tag only when the +1 frame is read in the His +1 reporter. Likewise, deletion of C98 leads to synthesis of RKRH6D only upon translation in the −1 frame in the His −1 reporter.
A variety of candidates have been reported to bind the sequence upstream of the resume codon. One suggestion is that the last of these upstream nucleotides, the so called −1 triplet (G87UC), is recognized directly by rRNA in the ribosomal decoding center,10 although this hypothesis does not withstand analysis of tmRNA activity in vivo.9 Another candidate is ribosomal protein S1, previously shown to crosslink to U85.11 Cryo-electron microscopy structures of tmRNA bound inside 70S ribosomes reveal that S1 affects the structure of the tmRNA template sequence.12 Though S1 cannot interact directly with tmRNA on the ribosome, it has been proposed that free S1 binds tmRNA and stabilizes a functional, open complex that is then passed to stalled ribosomes.12 In support of this model, one study presents evidence that S1 is required for tmRNA to serve as a template in vitro.13 There are also reports that refute this proposed role for S1, however, using reconstituted translation systems14,15 as well as in vivo functional analysis.16
Another promising candidate is SmpB, a protein that plays a role in the stability and aminoacylation of tmRNA and is required for its entry into the ribosome. SmpB binds the tRNA-like domain (TLD) of tmRNA in a well-characterized interaction.17 Interest in SmpB has focused on its ability to license tmRNA entry into the ribosomal A site through interacting with the decoding center.18,19,20 Recent reports suggest that more than one binding site for SmpB exists in tmRNA, raising the possibility of additional functions for this protein. Felden and co-workers showed that SmpB binding reduces the accessibility of the upstream sequence to nucleases in probing assays and proposed that SmpB plays a role in resume codon selection.21 After further characterization with surface plasmon resonance and filter binding assays, they report that this interaction is higher in affinity than SmpB binding to the TLD.22 An interaction between SmpB and the upstream region was likewise reported by Himeno and co-workers using chemical probing assays. Intriguingly, the site of SmpB binding shifted in tmRNA mutants known to alter the frame in which translation resumes.23 On the other hand, several crosslinking, chemical probing, and hydroxyl-radical cleavage assays have failed to detect an interaction between SmpB and the tmRNA upstream sequence.24,25,26,27
The role of S1 and SmpB in frame selection remains controversial because binding has only been detected in some assays in vitro and these RNA-protein binding events have not been shown to affect tmRNA activity either in vitro or in vivo. To test for a functional interaction between SmpB and the upstream region of tmRNA, we identified mutations in SmpB that restore the function of an A86C mutant tmRNA. The A86C mutation strongly reduces tmRNA activity in several assays and causes tmRNA to be translated exclusively in the +1 frame.9 Several SmpB mutations were identified which rescue tmRNA function and alter frame selection on A86C tmRNA. These results demonstrate definitively that SmpB plays a biologically relevant role in setting the frame on tmRNA.
SmpB mutations restore A86C tmRNA function
We used a genetic selection to identify mutations in SmpB that suppress the defect (improper frame selection) in A86C tmRNA. The selection relies on the tmRNA tagging process to complete the synthesis of the kanamycin resistance protein (KanR).4,9 Ribosomes are programmed to stall at the end of a truncated KanR protein that lacks the C-terminal 15 amino acids. tmRNA rescues these stalled ribosomes and tags the nascent polypeptide with the missing 15 amino acids; these are encoded by an altered tmRNA template sequence. In this way, tmRNA function completes the KanR protein and makes the cells kanamycin resistant (Figure 2). While cells with wild-type (A86) tmRNA survive equally well with or without kanamycin, only about 1 in 106 cells expressing the A86C tmRNA mutant form colonies on selective media, even at the lowest stringency conditions (15 µg/mL kanamycin at 25 °C). This low background survival rate allowed us to select for SmpB mutants that suppress the A86C defect and restore high levels of tmRNA function and kanamycin resistance.
Figure 2.
Genetic selection for SmpB mutants that restore A86C tmRNA activity. Translation of a truncated KanR gene is stalled during termination at the sequence Glu-Pro-Stop. The resulting KanR protein lacks the C-terminal 15 amino acids (red) and is inactive unless these stalled ribosomes are rescued by tmRNA that has been altered to encode the last 14 amino acids, ANKLQFHLMLDEFF.4 Roughly 108 SmpB mutants were screened to identify those that restore tagging levels sufficient to synthesize KanR and confer cellular survival on kanamycin plates. SmpB library construction – EagI and EcoRV cloning sites in the selection plasmid9 were used to insert the SmpB gene mutagenized by error-prone PCR33 with the following primers: 392, GGTATCAACAGGGACACCAGG and 470, CCAGTCACGTAGCGAAGATC. The SmpB library was introduced into MegaX DH10B competent cells (Invitrogen) by electroporation and was amplified, purified, and then introduced into the ΔssrA-smpB strain (a gift from Brice Felden)34 for selection in the KanR assay. KanR assay for tmRNA activity – ΔssrA-smpB cells expressing A86C tmRNA and SmpB from the selection plasmid were grown overnight in 2xYT with ampicillin. Saturated cultures were diluted to an OD600 of approximately 0.3 in fresh media containing 2% arabinose to induce KanR expression and grown for 4 hours. The cells were plated onto selective media: 2xYT, ampicillin, chloramphenicol, 2% arabinose, and 15 µg/mL kanamycin. Growth comparisons (selective vs. non-selective plates) were made after incubation for 48 h at 25 °C. Mutant smpB genes from selected clones were amplified by PCR and cloned into fresh selection vector and re-introduced into the selection strain to verify their phenotype.
We generated a library of ~108 SmpB mutants using error-prone PCR and subjected it to the KanR selection with A86C tmRNA, obtaining survival levels 100-fold higher than background. The SmpB genes were recloned from this enriched pool and reselected, with nearly all the cells surviving on selective plates. Sequencing revealed three related but distinct SmpB clones that were designated A1, A2 and A5 (Table 1). The A1 clone has two changes, Tyr24Cys and Val129Ala. A2 has these and the additional Glu107Val mutation. A5 shares the same Tyr24Cys mutation as A1 and A2 but coupled instead with Ala130Gly. The tagging activity of these three SmpB clones was measured by plating cells on media containing 15 µg/mL kanamycin at 25 °C. All three clones showed at least 50% survival, several orders of magnitude higher than the background level of 1 in 106. These findings show that mutations in SmpB can compensate for deficiencies in the function of the upstream region of tmRNA and the critical nucleotide A86 in particular.
Table 1.
SmpB clones that restore A86C tmRNA activity
| Clone | SmpB mutations | |||
|---|---|---|---|---|
| A1 | Tyr24Cys | Val129Ala | ||
| A2 | Tyr24Cys | Glu107Val | Val129Ala | |
| A5 | Tyr24Cys | Ala130Gly | ||
Changes in the SmpB sequence are shown for three clones that survived the KanR selection for tmRNA activity. The codons for the Tyr24Cys and Val129Ala mutations are the same at the DNA level in each clone that they appear.
SmpB mutations affect frame choice on A86C tmRNA
To verify that the selected SmpB mutants restore tmRNA activity, we directly measured the tmRNA tagging levels with immunoblots. In the immunoblot assay, tmRNA directs the addition of a His6 tag to the end of the full-length GST protein. The sequence Glu-Pro-Opal was used at the C-terminus of GST to induce stalling during termination.28 The tmRNA template sequence was altered to encode ANDH6D. Tagging in the natural (or 0) frame was detected by immunoblot using an anti-His6 antibody. We also developed additional tools to detect translation of the tmRNA template in other frames.9 We created a +1 frame construct by inserting a single G before G90, the first nucleotide in the resume codon. This tmRNA encodes a His6-tag only if tmRNA is translated in the +1 frame (Figure 1B). Likewise, tmRNA in which C98 is deleted only encodes the His6-tag when read in the −1 frame. These three tmRNA constructs allow tagging in all three frames (−1, 0, +1) to be visualized on an immunoblot for any given tmRNA and SmpB mutant pair.
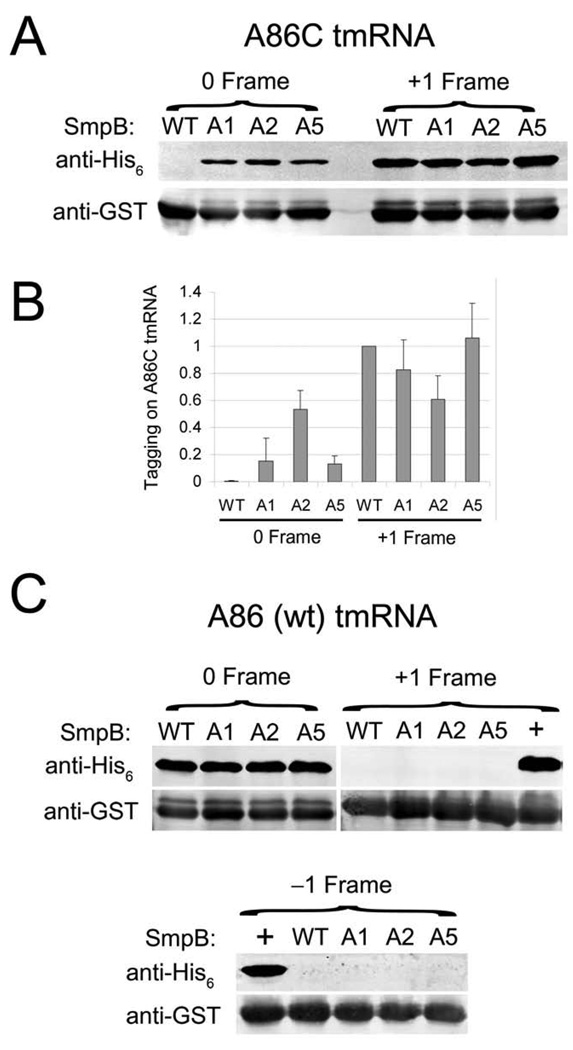
Wild-type SmpB yields no detectable activity in the 0 frame and high levels of +1 frame tagging with A86C tmRNA (Figure 3A). In contrast, the selected A1, A2, and A5 SmpB clones all restore significant levels of tagging in the 0 frame; the A2 mutant is the most active. The A2 mutant also reduces the +1 frame tagging seen with wild-type SmpB (t-test P < 0.01). It appears that the total level of tagging remains constant in the A2 mutant—tagging in the +1 frame decreases at about the same level as tagging in the 0 frame increases (Figure 3B). No significant reduction of +1 frame tagging was detected with A1 or A5, but this is expected since these mutants restore 0 frame tagging at lower levels. No tagging in the −1 frame was detected with the wild-type or mutant SmpB clones (data not shown). The restoration of tagging in the natural frame and reduction in the +1 frame suggests that these SmpB mutants rescue tmRNA function in the KanR selection by restoring the proper reading frame on A86C tmRNA. These data corroborate the genetic findings that SmpB is functionally tied to the upstream region of tmRNA. They also provide evidence that SmpB plays a role in selection of the reading frame.
Figure 3.
SmpB mutants alter frame selection on A86C tmRNA but not wild-type tmRNA. tmRNA was altered to express an ANDH6D tag in the −1, 0, or +1 frame (see Fig. 1B). The complete GST protein with the known stalling signal Glu-Pro-Stop at the C-terminus served as a substrate for tagging. Addition of the tag was visualized with an anti-His6 antibody as described.9 An anti-GST control was visualized on the same blot. Note that the higher band recognized by the anti-GST antibody corresponds to GST containing the tmRNA tag. A) 0 and +1 frame tagging by A86C tmRNA with the selected SmpB clones. B) Quantification of the level of A86C tmRNA tagging divided by the level of GST expression, normalized to the level of +1 tagging with wild-type SmpB. Error bars report the standard deviation of three independent experiments. C) Tagging by wild-type tmRNA (A86) in all three frames in combination with various SmpB clones. A86C and U85A were used as positive controls for the detection of +1 and −1 frame tagging, respectively, with wild-type SmpB (the “+” lanes).
Analysis of the role of the selected mutations in restoring tagging
The selected SmpB mutants alter the reading frame on A86C tmRNA, but what effect do they have on wild-type tmRNA (A86)? We analyzed reading frame selection with SmpB clones A1, A2, and A5 with wild-type (A86) tmRNA. Immunoblot analysis revealed no evidence of increased translation in either the +1 or −1 frame (Figure 3C). A86C and U85A mutants were used as positive controls to verify that tagging in the +1 or −1 frames, respectively, could be visualized. This result shows that our SmpB mutants are not sufficient to alter frame recognition alone—they only do so in the context of the A86C mutation.
Are the selected SmpB clones specific for C86 or do they support tmRNA activity with the wild-type tmRNA? Immunoblot analysis with A86 tmRNA revealed that the mutants show no loss of function in the normal frame (Figure 3C). We also tested the A1, A2, and A5 clones in the KanR assay against A86 tmRNA and found that the SmpB mutants did not lower the level of survival. These findings indicate that specificity for the tmRNA upstream sequence was relaxed, rather than altered to be specific for the cytidine nucleotide. Relaxed specificity is the simpler solution in most cases; in directed evolution experiments a negative selection against the wild-type sequence must be used to obtain truly altered specificity.29
Which SmpB mutations are necessary to restore function and how do they work together? We tested all four mutations found in A1, A2, and A5 both individually and in pairs. The following mutants were assayed by immunoblot with A86C tmRNA: the single point mutations Y24C, E107V, V129A, A130G, and double mutants Y24C/E107V, E107V/V129A, E107V/A130G, and V129A/A130G. None of these yielded a detectable amount of tagging in the 0 frame (data not shown), in contrast to the substantial signals seen with the original pairings. We conclude that at least two mutations are required to rescue A86C tmRNA: Tyr24Cys and either Val129Ala or Ala130Gly.
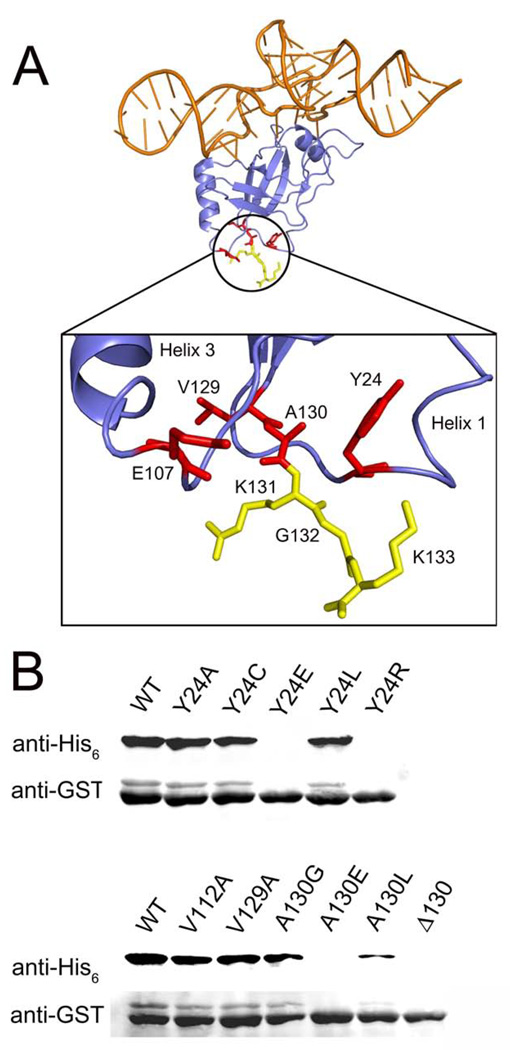
When mapped onto the atomic structure of Thermus thermophilus SmpB,17 these three residues cluster in a junction at the bottom of the protein, away from the well-characterized tmRNA binding site (Figure 4A). This junction is formed from the beginning of the C-terminal tail and loops linked to helices 1 and 3. The fourth point mutation, Glu107Val, is found nearby in the loop following helix 3. The clustering of these mutations at this single site implicates this junction at the bottom of SmpB in the frame selection process.
Figure 4.
Structure and mutagenesis of SmpB. A) The structure of the SmpB-tmRNA complex from T. thermophilus is shown with the tRNA-like domain of tmRNA in orange and SmpB in blue.17 Residues mutated in the selected SmpB clones are shown in red. The first three amino acids in the C-terminal tail (K131GK) are shown in yellow. Residue labels depict the E. coli numbering and sequence. The corresponding T. thermophilus residues are Y14, K96, and L119ARGK. Created with Pymol. B) Tagging of stalled GST was measured by immunoblot as in Fig. 3 but with wild-type (A86) tmRNA encoding ANDH6D in the natural (0) frame. This series of SmpB mutants was designed to test the structural role of residues at the junction where the C-terminal tail begins.
The Tyr24 side chain makes hydrophobic interactions with several other residues in this junction region. We introduced mutations to test the importance of these hydrophobic interactions for overall SmpB function (paired with wild-type tmRNA). Replacement of Tyr24 with charged residues Glu or Arg results in a complete loss of SmpB function (Figure 4B), while mutation to Leu is well tolerated. Mutation to the smaller side chains Cys or Ala also has little or no effect on SmpB activity. Since Ala is tolerated at residue 24, we wondered it could substitute for Cys in our selected SmpB clones, all of which contain the Tyr24Cys mutation (Table 1). Perhaps the loss of the aromatic side chain of Tyr24 introduces structural plasticity in this junction region; alternatively, the Cys side chain itself may play a role in the new activity of these clones. To distinguish between these possibilities, we replaced the Tyr24Cys mutation with Tyr24Ala in the A2 mutant. The resulting SmpB protein showed survival levels similar to the original A2 mutant in the KanR assay with A86C tmRNA. Together these data show that Tyr24 is in a hydrophobic environment and loss of its side chain is a key component in allowing our SmpB mutants to function on A86C tmRNA.
Analysis of the atomic structure suggests that one residue that interacts with Tyr24 is Ala130. Mutation of Ala130 to the charged Glu or the bulky Leu dramatically reduce SmpB function (Figure 4B). Deletion of Ala130, effectively replacing it with Lys131, likewise destroys SmpB function. These mutagenesis results support the conclusion that Tyr24 and Ala130 form hydrophobic interactions that are essential for SmpB activity. The mutations identified in our screen, taken individually, do not inhibit SmpB activity (Figure 4B), but likely introduce subtle structural changes while retaining the basic structure of this region. We propose that the Tyr24Cys, Val129Ala, and Ala130Gly mutations identified in our selection introduce structural plasticity required to restore A86C tmRNA function and proper frame selection.
Conclusions
Our findings demonstrate that SmpB and the region upstream of the resume codon on tmRNA are functionally linked; both play a role in establishing the reading frame on tmRNA. The A86C mutation in tmRNA leads to the total loss of 0 frame tagging and high levels of +1 frame tagging. We identified several SmpB mutants that restore the function of A86C tmRNA both in a genetic selection and in a direct assay for the addition of the tmRNA tag. The A2 mutant, for example, restores 0 frame tagging at about the same level that it reduces +1 frame tagging (Figure 3B). This suggests that the overall tagging efficiency is the same and that A2 increases tmRNA activity by simply restoring the proper frame in tmRNA translation.
How might SmpB set the frame during trans-translation? SmpB binds to the tRNA-like domain of tmRNA to form a structure that mimics normal tRNAs.17 Positioned as an anticodon stem mimic, SmpB licenses the entry of the SmpB-tmRNA complex into the ribosomal A site. Following accommodation and peptidyl transfer, this complex moves to the P site.27,30 SmpB binds the 30S P site with high affinity even without tmRNA.25,31 We envision a model in which interactions with the P site position SmpB precisely such that it draws tmRNA into the A site. With the first codon (GCA) lying in the mRNA channel in the decoding center, translation begins with tmRNA as a template. Intriguingly, the SmpB residues identified in our selection as affecting frame selection (Tyr24, Val129, and Ala130) cluster together in a hydrophobic pocket located on what would be the A-site face of SmpB. We propose that the selected mutations act together to induce structural plasticity at this site. The changes in SmpB structure induced by these mutations may alter the positioning of the conserved K131GKK sequence found at the beginning of the C-terminal tail and immediately downstream of the Val129Ala and Ala130Gly mutations. Interestingly, these residues were recently implicated in SmpB′s interaction with the ribosomal decoding center.20 These conserved amino acids (Tyr24 and V129AKGKK) may therefore play a critical role in decoding and a role in frame selection.
How might SmpB draw the tmRNA template sequence into the A site and position the resume codon for translation to resume? Several studies have reported a direct interaction between SmpB and the region upstream of the tag template in tmRNA (especially U85). Our results demonstrate a genetic and functional link between these molecules but cannot determine whether their interaction is direct or indirect. Experiments are underway to further characterize biochemically the binding interface between SmpB and the upstream sequence of tmRNA.
Acknowledgments
The authors thank Brice Felden for the ΔssrA-smpB strain. This work was supported by grant GM77633 to A.B. from the National Institutes of Health.
The abbreviations used are
- SmpB
small protein B
- tmRNA
transfer-messenger RNA
- A site
aminoacyl-tRNA site
- P site
peptidyl-tRNA site
- TLD
tRNA-like domain of tmRNA
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Moore SD, Sauer RT. The tmRNA system for translational surveillance and ribosome rescue. Annu Rev Biochem. 2007;76:101–124. doi: 10.1146/annurev.biochem.75.103004.142733. [DOI] [PubMed] [Google Scholar]
- 2.Withey JH, Friedman DI. A SALVAGE PATHWAY FOR PROTEIN SYNTHESIS: tmRNA and Trans-Translation. Annu Rev Microbiol. 2003;57:101–123. doi: 10.1146/annurev.micro.57.030502.090945. [DOI] [PubMed] [Google Scholar]
- 3.Dulebohn D, Choy J, Sundermeier T, Okan N, Karzai AW. Trans-translation: the tmRNA-mediated surveillance mechanism for ribosome rescue, directed protein degradation, and nonstop mRNA decay. Biochemistry. 2007;46:4681–4693. doi: 10.1021/bi6026055. [DOI] [PubMed] [Google Scholar]
- 4.Tanner DR, Dewey JD, Miller MR, Buskirk AR. Genetic analysis of the structure and function of transfer messenger RNA pseudoknot 1. J Biol Chem. 2006;281:10561–10566. doi: 10.1074/jbc.M600167200. [DOI] [PubMed] [Google Scholar]
- 5.Nameki N, Tadaki T, Himeno H, Muto A. Three of four pseudoknots in tmRNA are interchangeable and are substitutable with single-stranded RNAs. FEBS Lett. 2000;470:345–349. doi: 10.1016/s0014-5793(00)01349-1. [DOI] [PubMed] [Google Scholar]
- 6.Wower IK, Zwieb C, Wower J. Escherichia coli tmRNA lacking pseudoknot 1 tags truncated proteins in vivo and in vitro. Rna. 2009;15:128–137. doi: 10.1261/rna.1192409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Williams KP, Martindale KA, Bartel DP. Resuming translation on tmRNA: a unique mode of determining a reading frame. Embo J. 1999;18:5423–5433. doi: 10.1093/emboj/18.19.5423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lee S, Ishii M, Tadaki T, Muto A, Himeno H. Determinants on tmRNA for initiating efficient and precise trans-translation: some mutations upstream of the tag-encoding sequence of Escherichia coli tmRNA shift the initiation point of trans-translation in vitro. Rna. 2001;7:999–1012. doi: 10.1017/s1355838201010342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Miller MR, Healey DW, Robison SG, Dewey JD, Buskirk AR. The role of upstream sequences in selecting the reading frame on tmRNA. BMC Biol. 2008;6:29. doi: 10.1186/1741-7007-6-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lim VI, Garber MB. Analysis of recognition of transfer-messenger RNA by the ribosomal decoding center. J Mol Biol. 2005;346:395–398. doi: 10.1016/j.jmb.2004.11.061. [DOI] [PubMed] [Google Scholar]
- 11.Wower IK, Zwieb CW, Guven SA, Wower J. Binding and cross-linking of tmRNA to ribosomal protein S1, on and off the Escherichia coli ribosome. Embo J. 2000;19:6612–6621. doi: 10.1093/emboj/19.23.6612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gillet R, Kaur S, Li W, Hallier M, Felden B, Frank J. Scaffolding as an organizing principle in trans-translation. The roles of small protein B and ribosomal protein S1. J Biol Chem. 2007;282:6356–6363. doi: 10.1074/jbc.M609658200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Saguy M, Gillet R, Skorski P, Hermann-Le Denmat S, Felden B. Ribosomal protein S1 influences trans-translation in vitro and in vivo. Nucleic Acids Res. 2007;35:2368–2376. doi: 10.1093/nar/gkm100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Qi H, Shimizu Y, Ueda T. Ribosomal protein S1 is not essential for the trans-translation machinery. J Mol Biol. 2007;368:845–852. doi: 10.1016/j.jmb.2007.02.068. [DOI] [PubMed] [Google Scholar]
- 15.Takada K, Takemoto C, Kawazoe M, Konno T, Hanawa-Suetsugu K, Lee S, Shirouzu M, Yokoyama S, Muto A, Himeno H. In vitro trans-translation of Thermus thermophilus: ribosomal protein S1 is not required for the early stage of trans-translation. Rna. 2007;13:503–510. doi: 10.1261/rna.363207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McGinness KE, Sauer RT. Ribosomal protein S1 binds mRNA and tmRNA similarly but plays distinct roles in translation of these molecules. Proc Natl Acad Sci U S A. 2004;101:13454–13459. doi: 10.1073/pnas.0405521101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bessho Y, Shibata R, Sekine S, Murayama K, Higashijima K, Hori-Takemoto C, Shirouzu M, Kuramitsu S, Yokoyama S. Structural basis for functional mimicry of long-variable-arm tRNA by transfer-messenger RNA. Proc Natl Acad Sci U S A. 2007;104:8293–8298. doi: 10.1073/pnas.0700402104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sundermeier TR, Dulebohn DP, Cho HJ, Karzai AW. A previously uncharacterized role for small protein B (SmpB) in transfer messenger RNA-mediated trans-translation. Proc Natl Acad Sci U S A. 2005;102:2316–2321. doi: 10.1073/pnas.0409694102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shimizu Y, Ueda T. SmpB triggers GTP hydrolysis of elongation factor Tu on ribosomes by compensating for the lack of codon-anticodon interaction during trans-translation initiation. J Biol Chem. 2006;281:15987–15996. doi: 10.1074/jbc.M512165200. [DOI] [PubMed] [Google Scholar]
- 20.Nonin-Lecomte S, Germain-Amiot N, Gillet R, Hallier M, Ponchon L, Dardel F, Felden B. Ribosome hijacking: a role for small protein B during trans-translation. EMBO Rep. 2009;10:160–165. doi: 10.1038/embor.2008.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Metzinger L, Hallier M, Felden B. Independent binding sites of small protein B onto transfer-messenger RNA during trans-translation. Nucleic Acids Res. 2005;33:2384–2394. doi: 10.1093/nar/gki534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Metzinger L, Hallier M, Felden B. The highest affinity binding site of small protein B on transfer messenger RNA is outside the tRNA domain. Rna. 2008;14:1761–1772. doi: 10.1261/rna.1185808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Konno T, Kurita D, Takada K, Muto A, Himeno H. A functional interaction of SmpB with tmRNA for determination of the resuming point of trans-translation. Rna. 2007 doi: 10.1261/rna.604907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wower J, Zwieb CW, Hoffman DW, Wower IK. SmpB: a protein that binds to double-stranded segments in tmRNA and tRNA. Biochemistry. 2002;41:8826–8836. doi: 10.1021/bi0201365. [DOI] [PubMed] [Google Scholar]
- 25.Kurita D, Sasaki R, Muto A, Himeno H. Interaction of SmpB with ribosome from directed hydroxyl radical probing. Nucleic Acids Res. 2007 doi: 10.1093/nar/gkm677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Barends S, Karzai AW, Sauer RT, Wower J, Kraal B. Simultaneous and functional binding of SmpB and EF-Tu-TP to the alanyl acceptor arm of tmRNA. J Mol Biol. 2001;314:9–21. doi: 10.1006/jmbi.2001.5114. [DOI] [PubMed] [Google Scholar]
- 27.Ivanova N, Lindell M, Pavlov M, Holmberg Schiavone L, Wagner EG, Ehrenberg M. Structure probing of tmRNA in distinct stages of trans-translation. Rna. 2007;13:713–722. doi: 10.1261/rna.451507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hayes CS, Bose B, Sauer RT. Proline residues at the C terminus of nascent chains induce SsrA tagging during translation termination. J Biol Chem. 2002;277:33825–33832. doi: 10.1074/jbc.M205405200. [DOI] [PubMed] [Google Scholar]
- 29.Wang L, Brock A, Herberich B, Schultz PG. Expanding the genetic code of Escherichia coli. Science. 2001;292:498–500. doi: 10.1126/science.1060077. [DOI] [PubMed] [Google Scholar]
- 30.Bugaeva EY, Shpanchenko OV, Felden B, Isaksson LA, Dontsova OA. One SmpB molecule accompanies tmRNA during its passage through the ribosomes. FEBS Lett. 2008;582:1532–1536. doi: 10.1016/j.febslet.2008.03.049. [DOI] [PubMed] [Google Scholar]
- 31.Ivanova N, Pavlov MY, Bouakaz E, Ehrenberg M, Schiavone LH. Mapping the interaction of SmpB with ribosomes by footprinting of ribosomal RNA. Nucleic Acids Res. 2005;33:3529–3539. doi: 10.1093/nar/gki666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Burks J, Zwieb C, Muller F, Wower I, Wower J. Comparative 3-D modeling of tmRNA. BMC Mol Biol. 2005;6:14. doi: 10.1186/1471-2199-6-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cadwell RC, Joyce GF. Mutagenic PCR. PCR Methods Appl. 1994;3:S136–S140. doi: 10.1101/gr.3.6.s136. [DOI] [PubMed] [Google Scholar]
- 34.Hallier M, Ivanova N, Rametti A, Pavlov M, Ehrenberg M, Felden B. Pre-binding of small protein B to a stalled ribosome triggers trans-translation. J Biol Chem. 2004;279:25978–25985. doi: 10.1074/jbc.M314086200. [DOI] [PubMed] [Google Scholar]