Our global eyes were opened once again by the unexpected outbreak of swine influenza in April 2009. The continued testing of the world's pandemic preparedness should be considered beneficial and analogous to a societal vaccination in that it primes surveillance and research communities and public health and government officials for the statistical likelihood of a pandemic, but it should not promote complacency. A large pandemic may never occur, but as we travel more often and further, outbreaks similar to that seen for swine influenza should be expected. From a phenotypic stance, the differences between outbreak and pandemic classification lie in the virulence and communicability of the outbreak strain, and the selective pressure that promotes viral genetic drift and shift. Social factors that affect whether an outbreak becomes a pandemic include the timing of the outbreak and the recognition of its severity by health organizations. The emergence of this novel influenza virus in North America highlights the continuing need for basic and clinical research to develop new vaccines and chemotherapeutic agents, as well as the need to establish and maintain effective lines of communication among the scientific community and civil authorities. Continued reevaluation of these factors will assist in the assessment of our preparedness in light of the current outbreak and provide a gap analysis for improvement.
INFLUENZA: THE STATISTICS
Influenza types A and B virus infections in humans results in an estimated 150,000–200,000 hospitalizations and 30,000–50,000 deaths in the United States annually (1, 2). Annual epidemics have a large economic impact, costing more than $11 billion in direct medical costs, $16 billion in indirect loss of earnings and life, and $88 billion per year total (hospitalization costs and lost productivity) in the United States alone (3). In the event of a highly pathogenic pandemic, the Centers for Disease Control and Prevention (CDC) has predicted a 3- to 7-fold increase in hospitalization and mortality rates and at least a 20-fold increase in economic impact in the United States alone. Influenza pandemics are likely to be many times more devastating in regions of the world where health resources are lacking (4, 5). Finally, the potential decimation of the domestic fowl (6, 7) or swine (8) populations by pathogenic influenza would have a tremendous economic impact on worldwide markets. The potential social and economic disruption ensuing from a human pandemic combined with the economic disaster resulting from the destruction of domesticated animal populations has prompted the NIAID to classify the influenza virus as a Category C priority pathogen and the CDC to classify highly pathogenic avian influenza as a select agent (9).
Vaccination remains the principal means for controlling influenza. The selection process for the strains incorporated into the trivalent vaccine (generally composed of an H1N1, an H3N2, and a B-type strain) requires year-round surveillance. Every year in late January, the FDA's Vaccines and Related Biological Products Advisory Committee (VRBPAC) reviews global surveillance data, and recommends one or more of the three strains to be included in the vaccine for the subsequent influenza season. The World Health Organization (WHO) completes its review and makes recommendations for the Northern Hemisphere vaccine by mid-February and for the Southern hemisphere vaccine by September. In March, VRBPAC meets to finalize the recommendations for the U.S. influenza vaccine. The effectiveness of the resultant trivalent vaccine depends upon how well the chosen strains match the predicted circulating strains. Although there is no guarantee that the strains picked for the vaccine will be the circulating strains during the subsequent flu season, there is an estimated 90% match between vaccine strains and circulating strains each season (10). Vaccination rates are also crucial to outbreak management. For the 2005–2006 season, estimated influenza vaccination coverage for persons with high risk conditions for the age groups 18–49 and 50–64 were 30.5% and 48.4%, respectively; for normal individuals the corresponding fractions were 18.3% and 32.2%. In addition, 69.3% of all persons older than 65 years of age were vaccinated (11). Clearly, increasing the number of vaccinated people is one strategy for mitigating seasonal influenza outbreaks and remains a high priority of the national Pandemic Preparedness Plan. The recent outbreak justifies continued and aggressive community education programs that encourage people to become vaccinated.
THE INFLUENZA REPLICATION CYCLE, SHIFT, AND DRIFT
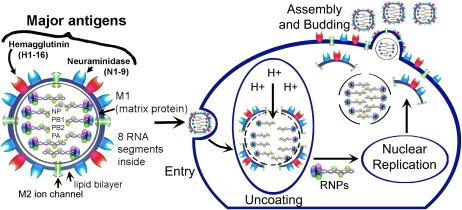
Influenza particles are spherical (∼ 100 nm diameter), with a lipid bilayer derived from the host plasma membrane (12). The virion contains a matrix protein (M1) and three transmembrane proteins (hemagglutinin [HA] and neuraminidase [NA]), which are the major antigenic determinants, and a small membrane-bound protein (M2), which forms a homo-tetrameric proton channel that is sparsely distributed throughout the viral lipid envelope (Figure 1). Beneath the matrix coat is the helical ribonucleocapsid, which includes the vRNA genome, nucleoprotein (NP), nuclear export protein (NEP), and the three viral polymerase subunits (PB1, PB2, PA) that form the polymerase holoenzyme. There are 16 different classifications of HA, and nine different NAs. The types of HA and NA together describe the virus characterization and constitute part of the virus nomenclature (e.g., H3N2, H1N1, or H5N1).
Figure 1.
The components of the influenza virus, and the steps of viral replication Viral attachment and entry into the cell occur via the endosomal pathway. Acidification of the endosome promotes viral membrane fusion and activates the M2 ion channel, which pumps protons (H+) into the interior of the viral core to initiate uncoating of the M1 protein. Nuclear replication occurs, and viral gene products are transported to the plasma membrane for assembly. Nascent virions are assembled at the apical domain of the plasma membrane, and complete virus particles bud and are released from the cell (27).
The viral replication cycle begins with infection of the host cell by HA-mediated binding to cell surface sialic acids (which are ubiquitously present on membrane-associated glycoproteins and glycolipids), followed by internalization by receptor-mediated endocytosis. Viruses with proteolytically activated HA (HA with a cleaved fusion peptide) fuse with the endosomal membrane. Acidification of the endosome promotes viral membrane fusion and activates the M2 ion channel, which pumps protons (H+) into the interior of the viral core to initiate uncoating of the M1 protein, promoting viral membrane fusion and release of the viral ribonucleoprotein (vRNP) into the cytoplasm. Nuclear-localization signals on the NP facilitate transport of the vRNP to the nucleus, where viral mRNA transcription (vRNA→vmRNA) and genomic replication (vRNA→cRNA→vRNA) occurs. Once translated, one of the virally encoded nonstructural proteins, NS1, binds double-stranded RNA and host mRNA processing factors to inhibit the cellular interferon-induced antiviral response (reviewed in Ref. 13). Another recently discovered nonstructural protein, PB1-F2, is thought to promote host cell apoptosis by insertion into mitochondrial and cell membranes (14). The polymerase holoenzyme performs host mRNA cap-recognition and -snatching to provide capped mRNA primers for initiation of viral transcription (15). Replicated vRNPs are exported from the nucleus, assisted by NEP, and transported to the plasma membrane for assembly and with the envelope proteins (NA, M2, HA, M1). Virus budding and release by NA-facilitated cleavage of terminal sialic acid residues on the cell surface glycoligands completes the cycle (12) (Figure 1).
Modern research attributes the seasonal recurrence of influenza both to its nature as a segmented negative-strand RNA virus, and to selective pressure to coexist in a dual host system consisting of the primary avian reservoir and a secondary mammalian host such as humans, pigs, horses, dogs, cats, and ferrets, all of which can contract the influenza virus (16). The high rate of annual influenza infection is attributed partially to the ability of the virus to acquire random mutations in its surface proteins (antigenic drift) that allow it to evade immunogenic recognition, and partially to its ability to shuffle its genomic constellation (antigenic shift), resulting in a genetic reassortment that may combine separate strain idiosyncrasies into a single virus with increased virulence. Any RNA virus is subject to random mutation (genetic drift) due to the lack of replicative fidelity in RNA polymerases. Humans form antibodies against hemagglutinin and neuraminidase; unfortunately, these two surface proteins are also the primary determinants of influenza antigenic drift. This strategy allows seasonal influenza to evade humoral immunity established in previous seasons by subtly and unpredictably altering the sequence of the HA and NA to make them unrecognizable by existing antibodies. In contrast, antigenic shift is a drastic change in the sequence of one or more of the eight virus genomic segments (genetic constellation) that results in a novel strain with uncharacterized properties. This “shifting” can occur through inter-reservoir crossover of strains (i.e., from avian to human), or through an intermediate reservoir (e.g., swine) that can be infected by separate strains that are normally specific for either birds or humans. Genetic reassortment (shift) of more than one viral strain in an intermediate reservoir can create a new human influenza A subtype virus. This is probably what happened in the case of the recent swine flu emergence, although the involvement of swine has not been confirmed. Antigenic shift and drift demand a new vaccine every year and rationalize the need for continued virological surveillance against the rise of particularly virulent influenza strains. This is the reasoning supporting the statistical certainty of another pandemic, a prospect that is continually emphasized by the recent emergence of novel human strains in North America, and the last decade's highly pathogenic avian influenza strains in Asia and their spread over a large portion of the world (17). Most important, the novel swine influenza strain has characteristics of a pandemic candidate, namely that the virus easily spreads from person to person in a sustained pattern, and resulted in a life-threatening illness in Mexico.
OFF SEASON OUTBREAKS: PRECURSOR TO PANDEMIC?
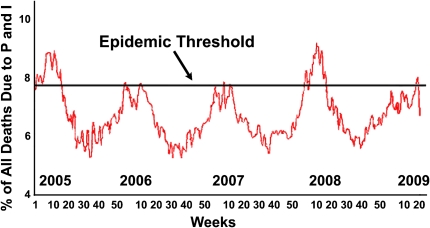
Peak influenza cases are typically reported during December through March in North America, although the influenza season officially ends May 17. Discounting the recent H1N1 swine influenza activity, the 2008–2009 influenza season was mild when compared with the 2007–2008 season (Figure 2). In most cases the vaccine promoted sufficient immunity to ward off significant outbreaks. In addition, antiviral drugs (such as oseltamivir and amantadine), are widely available and are effective against influenza, if prescribed either prophylactically or shortly after influenza infection. However, their overwhelming use coupled with the selective pressure for survival on the influenza genome has resulted in virus strains with almost global resistance to some of these antivirals, raising questions about their continued use (18). During the most recent influenza season, an increasing resistance in H1N1 isolates to the neuraminidase inhibitor oseltamivir was observed. As of December 2008, of 50 H1N1 viruses from 12 states, 98% tested by the CDC were resistant to oseltamivir, and 2% of these were resistant to zanamivir. One hundred percent of H3N2 viruses were resistant to amantadine, but none of these were resistant to either neuraminidase inhibitor (19). The compelling epidemiology suggests what we can expect in the coming season when considering the efficacy of existing antivirals and their intervening role in the control of seasonal influenza. The development and outbreak of a novel influenza strain in Mexico complicates the prediction of dominant circulating virus strains in the upcoming season because it occurred (and at the time of this writing, is still occurring) outside the expected window of seasonal influenza and after the vaccine strains for the upcoming season have already been finalized. Occurrence after the standard influenza season is an uncommon event and is in sharp contrast to other outbreaks, such as the 1976 outbreak, which began in February in New Jersey (20). For a more recent comparison, during the 2007−2008 influenza season, a total of 86 influenza-associated deaths were reported, but only one of these deaths occurred after May 18, 2008. No human cases of influenza A were reported to the National Notifiable Diseases Surveillance System during the summer months (21). Because influenza is notably less communicable in the warmer and more humid conditions present in the northern hemisphere during summer (22) the communicability of the virus in Mexico in April (one of the three hottest months, on average, for Mexico City, with temperatures around 80°F for most of the country) is an interesting puzzle that will not be immediately solved.
Figure 2.
Pneumonia and influenza mortality for 122 U.S. cities, week ending April 18th, 2009. Adapted from Ref. 28.
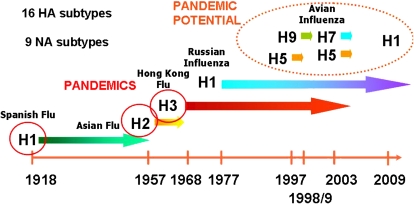
Notwithstanding, the novel swine influenza virus has several characteristics that make it a candidate for a pandemic, similar to other pandemic viral strains isolated within the last century (Figure 3). First, the late-season outbreak likely excludes it from inclusion in the next season's trivalent vaccine. Considering the possibility that the same H1N1 virus could be in prominent circulation at the beginning of the next influenza season, this suggests that next year's vaccine may be less effective than expected. This complication will likely be moderated by contingencies set in place during the previous decade's pandemic planning. The global vaccine production capability is well enough established that retooling of vaccine production to include the novel swine influenza strain is possible and expected. Second, the combination of low apparent common immunity coupled with the remarkable human-to-human communicability in warm weather conditions makes it a candidate for persistence in the global population. Third, the elevated apparent mortality in regions without modern supportive medical care suggests an increased pathogenicity, but this must be confirmed statistically. Finally, the observed likelihood of otherwise healthy, non–risk group individuals (generally age 18–49) to become infected with the virus is reminiscent of the recorded behavior of previous pandemics, most notably of the 1918 H1N1 influenza virus (23). This again emphasizes the importance of early detection, intervention, and modern supportive care on human mortality.
Figure 3.
Timeline of emergence of Influenza A viruses in humans. The viruses isolated from pandemics in the last century are indicated by time of origin and subtype. Virus subtypes with pandemic potential are circled.
CHALLENGES FOR EXPANDED RESEARCH AND SURVEILLANCE
The influenza A subtype represents a great threat to civic welfare. It is the only strain to exhibit antigenic shift (24) and has accounted for all known high-mortality epidemics and pandemics (25). Although a typical influenza season poses only a moderate health risk to the general population, large portions of the community are routinely endangered due to age-related susceptibility or compromised immunity, and influenza remains one of the 10 most common causes of death in the United States (26). However, these typical assumptions must be reconsidered when planning for a pandemic because historical and recent events have shown that it is quite possible for novel influenza strains to infect and spread prevalently through the general population of healthy adults. The issuance by the U.S. Department of Health and Human Services of a nationwide public health emergency declaration and the raising of the Pandemic Alert to Level 5 by the WHO in response to swine influenza was precautionary but prudent; in contrast, the mild hysteria propagated by the media serves only to confuse the facts and obscure the threat about influenza outbreaks. It is incumbent on the medical and academic professions to promote increasing community education and encourage understanding of the factors involved in influenza evolution, predicting seasonal and pandemic statistics, and strategies to reduce communicability. In the coming months, critiques by government and world health organizations will outline gap analyses and suggestions for improvement to our surveillance, outbreak, and pandemic response plans. Forensic epidemiology has already allowed the rapid identification of the earliest swine influenza patient and measured the time from the first known infection to outbreak across several continents at 3 to 4 weeks. Public notification of containment efforts combined with increased public alertness continues to promote the reporting of patients with any flu-like symptoms, although this may serve in the short term to obscure the number of genuine swine influenza cases until confirmation is available. It is predicted that the nation's response to this recent outbreak will receive high marks, primarily for maintaining the perceptions of control during a growing outbreak without unduly alarming public awareness.
A more critical eye might be turned toward world influenza surveillance efforts. As many of us were looking east for the pandemic to emerge in Asia and possibly from avian populations, a novel virus erupted in a neighboring country and then spread to our own backyard. As of yet there is no direct evidence that swine influenza originated in swine and was subsequently transmitted to humans, making the designation of “swine flu” something of a misnomer. Although the virus has not been reported to cause illness in pigs in the United States, a new strain of the swine influenza virus, different from any other ever reported in U.S. swine herds, serves as a reminder of the need for strict and enforceable surveillance and biosecurity for pork production operations.
Several avenues of research are suggested by the observed disparate mortality rates between infected patients in Mexico and the United States. Of primary concern is the level and quality of supportive care available to patients in the two countries and the environmental differences (e.g., pollution, health of the general population, and diet) that may contribute to higher mortality when respiratory diseases are involved. The emergence of novel influenza strains during the past decade have conventionally led to expectations that the next pandemic would emerge from the Asian part of the world, given the converging population density of both humans and poultry found in that region. The unexpected surfacing of a potentially pandemic strain from Mexico clearly emphasizes that multiple crucibles for pandemic influenza exist, and none should be downplayed. Because of modern travel, the outbreak mirrored the pattern of emergence seen for the 2003 SARS outbreak, with near simultaneous occurrences of infection disseminating from individuals in proximity to and traveling from patient zero, and these examples suggest that containment in the modern world may not be a practical goal. The recent and uncharacteristically late-season outbreak of a novel influenza virus in North America highlights the continuing need for research into the factors influencing evolution of influenza. Also, the levels of vaccination, avenues of treatment, the judicious application of antiviral stocks, and the directions of continued surveillance efforts should all be considered carefully to promote early intervention and to facilitate rapid diagnosis, as well as appropriate infection control measures.
This work was supported in part by National Institutes of Health grants 1R01AI071393 to J.W.N. and 2R01HL031197 to S.M., J.W.N., and D.L.N.
Conflict of Interest Statement: None of the authors has a financial relationship with a commercial entity that has an interest in the subject of this manuscript.
References
- 1.Thompson WW, Shay DK, Weintraub E, Brammer L, Bridges CB, Cox NJ, Fukuda K. Influenza-associated hospitalizations in the United States. JAMA 2004;292:1333–1340. [DOI] [PubMed] [Google Scholar]
- 2.Thompson WW, Shay DK, Weintraub E, Brammer L, Cox N, Anderson LJ, Fukuda K. Mortality associated with influenza and respiratory syncytial virus in the United States. JAMA 2003;289:179–186. [DOI] [PubMed] [Google Scholar]
- 3.Molinari NA, Ortega-Sanchez IR, Messonnier ML, Thompson WW, Wortley PM, Weintraub E, Bridges CB. The annual impact of seasonal influenza in the US: measuring disease burden and costs. Vaccine 2007;25:5086–5096. [DOI] [PubMed] [Google Scholar]
- 4.Meltzer MI, Cox NJ, Fukuda K. The economic impact of pandemic influenza in the United States: priorities for intervention. Emerg Infect Dis 1999;5:659–671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Meltzer MI, Bridges CB. Economic analysis of influenza vaccination and treatment. Ann Intern Med 2003;138:608. (author reply 608–609). [DOI] [PubMed] [Google Scholar]
- 6.Phipps LP, Essen SC, Brown IH. Genetic subtyping of influenza a viruses using RT-PCR with a single set of primers based on conserved sequences within the HA2 coding region. J Virol Methods 2004;122:119–122. [DOI] [PubMed] [Google Scholar]
- 7.USDA. The Asian bird flu. Available from: http://www.Ars.Usda.Gov/research/docs.Htm?Docid=11425.2006. [accessed 1 May 2009].
- 8.Reuters. Egypt starts pig slaughter, some farmers resist. Available from: http://www.reuters.com/article/GCA-SwineFlu/idUSTRE53T4BA20090430.2009. [accessed 30 April 2009].
- 9.US Dept of HHS. An early detection system for highly pathogenic H5N1 avian influenza in wild migratory birds. Available from: http://www.Pandemicflu.Gov/issues/screening.html.2006. [accessed 1 May 2009].
- 10.US Dept of HHS. National Vaccine Program Office–ongoing influenza defense tactics. Available from: http://www.hhs.gov/nvpo/pandemics/flu4.htm. [accessed 1 May 2009].
- 11.Centers for Disease Control. State-specific influenza vaccination coverage among adults aged >18 years - United States, 200–04 and 2005–06 influenza seasons. MMWR Morb Mortal Wkly Rep 2007;56:953–959. [PubMed] [Google Scholar]
- 12.Nayak DP, Hui EK, Barman S. Assembly and budding of influenza virus. Virus Res 2004;106:147–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Noah DL, Krug RM. Influenza virus virulence and its molecular determinants. Adv Virus Res 2005;65:121–145. [DOI] [PubMed] [Google Scholar]
- 14.Bruns K, Studtrucker N, Sharma A, Fossen T, Mitzner D, Eissmann A, Tessmer U, Roder R, Henklein P, Wray V, et al. Structural characterization and oligomerization of PB1-f2, a pro-apoptotic influenza a virus protein. J Biol Chem 2006;282:353–363. [DOI] [PubMed] [Google Scholar]
- 15.Lee MT, Klumpp K, Digard P, Tiley L. Activation of influenza virus RNA polymerase by the 5′ and 3′ terminal duplex of genomic RNA. Nucleic Acids Res 2003;31:1624–1632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vahlenkamp TW, Harder TC. Influenza virus infections in mammals. Berl Munch Tierarztl Wochenschr 2006;119:123–131. [PubMed] [Google Scholar]
- 17.Olsen B, Munster VJ, Wallensten A, Waldenstrom J, Osterhaus AD, Fouchier RA. Global patterns of influenza a virus in wild birds. Science 2006;312:384–388. [DOI] [PubMed] [Google Scholar]
- 18.Centers for Disease Control. High levels of adamantane resistance among influenza A (H3N2) viruses and interim guidelines for use of antiviral agents - United States, 2005–06 influenza season. MMWR Morb Mortal Wkly Rep 2006;55:44–46. [PubMed] [Google Scholar]
- 19.Centers for Disease Control. CDC issues interim recommendations for the use of influenza antiviral medications in the setting of oseltamivir resistance among circulating influenza A (H1N1) viruses, 2008–09 influenza season. Available from: http://www2a.cdc.gov/han/archivesys/viewMsgV.asp?AlertNum=00279.2009. [accessed 30 April 2009].
- 20.Gaydos JC, Top FH Jr, Hodder RA, Russell PK. Swine influenza A outbreak, Fort Dix, New Jersey, 1976. Emerg Infect Dis 2006;12:23–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Centers for Disease Control. Influenza activity–United States and worldwide, May 18-September 19, 2008. MMWR Morb Mortal Wkly Rep 2008;57;1046–1049. [PubMed] [Google Scholar]
- 22.Lowen AC, Mubareka S, Steel J, Palese P. Influenza virus transmission is dependent on relative humidity and temperature. PLoS Pathog 2007;3:1470–1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Richard SA, Sugaya N, Simonsen L, Miller MA, Viboud C. A comparative study of the 1918–1920 influenza pandemic in Japan, USA and UK: mortality impact and implications for pandemic planning. Epidemiol Infect 2009;Feb 12:1–11 [Epub ahead of print]. [DOI] [PMC free article] [PubMed]
- 24.Hampson AW, Mackenzie JS. The influenza viruses. Med J Aust 2006;185:S39–S43. [DOI] [PubMed] [Google Scholar]
- 25.Stevens J, Blixt O, Tumpey TM, Taubenberger JK, Paulson JC, Wilson IA. Structure and receptor specificity of the hemagglutinin from an H5N1 influenza virus. Science 2006;312:404–410. [DOI] [PubMed] [Google Scholar]
- 26.Kiselar JG, Downard KM. Antigenic surveillance of the influenza virus by mass spectrometry. Biochemistry 1999;38:14185–14191. [DOI] [PubMed] [Google Scholar]
- 27.Noah DL, While EL, Noah JW. Development of high-throughput screening assays for influenza. In: Torrence PF, editor. Combating the threat of pandemic influenza: drug discovery approaches. Hoboken, NJ: John Wiley & Sons, Inc.; 2007. pp. 38–72.
- 28.Centers for Disease Control and Prevention. Pneumonia and influenza (P&I) mortality surveillance. Available from: http://www.cdc.gov/flu/weekly/ [accessed 30 April 2009].