Abstract
The use of large-scale electrophysiology to obtain high spatiotemporal resolution brain recordings (>100 channels) capable of probing the range of neural activity from local field potential oscillations to single neuron action potentials presents new challenges for data acquisition, storage, and analysis. Our group is currently performing continuous, long-term electrophysiological recordings in human subjects undergoing evaluation for epilepsy surgery using hybrid intracranial electrodes composed of up to 320 micro- and clinical macroelectrode arrays. DC-capable amplifiers, sampling at 32 kHz per channel with 18-bits of A/D resolution are capable of resolving extracellular voltages spanning single neuron action potentials, high frequency oscillations, and high amplitude ultraslow activity, but this approach generates 3 terabytes of data per day (at 4 bytes per sample) using current data formats. Data compression can provide several practical benefits, but only if data can be compressed and appended to files in real-time in a format that allows random access to data segments of varying size. Here we describe a state-of-the-art, scalable, electrophysiology platform designed for acquisition, compression, encryption, and storage of large-scale data. Data are stored in a file format that incorporates lossless data compression using range encoded differences, a 32-bit cyclically redundant checksum to ensure data integrity, and 128-bit encryption for protection of patient information.
Keywords: Quantitative analysis, EEG analysis, Data compression, Range encoding, Data encryption, Cyclic redundancy codes, Multiscale Electrophysiology Format
Introduction
Large-scale electrophysiology recordings are recognized as a powerful tool for systems neurobiology and investigation of normal and pathological brain function {Buzsaki 2004}. Continuous, high, spatial and temporal resolution intracranial electroencephalography (iEEG) and single neuron recordings from humans are being used to investigate cognitive function, e.g. {Kraskov 2007} {Gelbard-Sagiv et al., 2008}. There is accumulating evidence that the bandwidth used for clinical iEEG is inadequate, and that high frequency oscillations can localize epileptogenic brain {Bragin et al., 2002, Gardner et. al., 2007, Worrell 2004, Urrestarazu 2007, Worrell 2008}. These opportunities have not been fully exploited, however, due to limitations in recording and storage technologies that have required scientists and clinicians to limit data acquisition directly by reducing the duration, number of channels, sampling rate or resolution of recordings in order to generate manageable amounts of data. For example, acquisition of “single unit” data (i.e., the extracellular action potentials from individual neurons) routinely requires users to set a fixed voltage threshold prior to the start of recording. When the electrode voltage exceeds this threshold, a limited-duration window of samples surrounding this event is stored and all other samples are discarded. Clearly, post-hoc analysis is then limited to the acquired waveforms, no further data windows can be extracted, and no relationship to other EEG features (e.g. phase, energy) can be generated. A preferred solution would be to record all samples in a compressed file format and then threshold the data offline, allowing the user to optimize detections with regard to the amount of data to analyze. The technology for acquisition of wide bandwidth electrophysiology (high channel count, high input impedance, DC-capable amplifiers, per-channel sampling rate of 32 kHz, 18-bit signal digitization) from high-density hybrid electrode arrays now makes it possible to record the full, physiological range of brain activity, from single neuron action potentials to high amplitude ultraslow field potential oscillations. However, the massive amounts of data produced by these recordings (i.e., “Big Data”) present unique, “biocuration,” or data sharing and interpretation challenges for institutional and laboratory information technology infrastructure {Howe et al., 2008}.
At our institution, large-scale recordings from patients undergoing evaluation for epilepsy surgery are obtained using hybrid electrodes containing microwires and clinical macroelectrodes {Van Gompel 2008a, Van Gompel 2008b, Worrell 2008} (Figure 1). This approach requires the infrastructure to transfer, store and manage up to 40 megabytes per second, 140 gigabytes of data per hour, or 3.3 terabytes per day (at 4 bytes / sample). Using current electrophysiology data storage methods, a typical patient recording (7 days) would require 23 terabytes of disk space for storage. Furthermore, conventional EEG data file formats typically bundle all the recorded channels into a single large file, making data analysis, storage, and transfer all the more unwieldy. Here we describe our approach to acquisition, compression, storage and management of data obtained from large scale electrophysiological studies required for investigation of systems neurobiology of brain function and disease.
Figure 1.
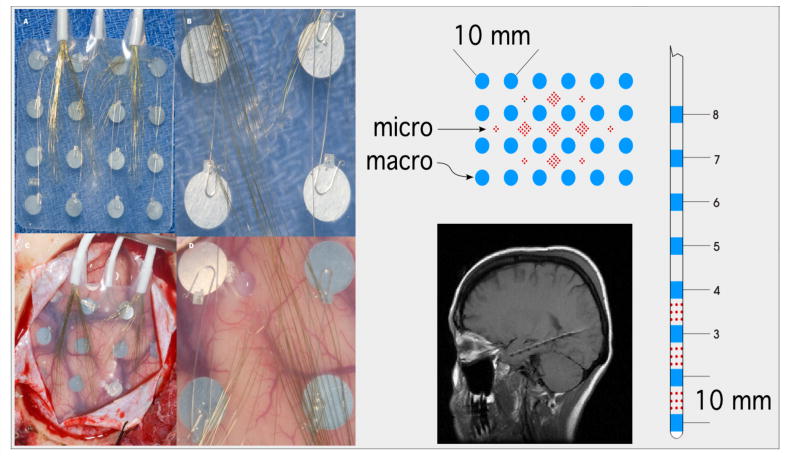
Left) Photographic montage of hybrid subdural grid containi 16 clinical macroelectrodes (4 mm) and 112 microelectrodes. Right) Schematic of hybrid subdural grid and depth electrodes. MRI of hippocampal hybrid depth implant (below).
At the core of our approach is a scalable (up to 1024 channels) acquisition system, large-scale storage area network (SAN) database, and a novel electrophysiology file format, called MEF (Multiscale Electrophysiology Format) (Figure 2). MEF achieves significant data size reduction when compared to existing formats (e.g. Neuralynx DMA format, EDF+ {Kemp 1992, Kemp 2003}, Extensible Biosignal Format (EBS) {Hellmann 1992}) using state-of-the-art lossless data compression {Bodden 2002, Martin 1979} and is designed for efficient data transfer, storage and analysis. In addition, MEF satisfies the Health Insurance Portability and Accountability Act (HIPAA) requiring any patient protected health information transmitted over a public network to be encrypted with a minimum 112-bit symmetric encryption [Federal Register 2003]. Sharing electrophysiology data for research purposes requires encryption or elimination of patient information to maintain compliance with HIPAA regulations. Encryption of patient identifying information within the file with an appropriate algorithm represents an elegant solution for maintaining patient confidentiality, while obviating the need for specialized, secure transfer protocols, and reducing the potential to lose relevant information or cause record keeping errors in research data. The existence of data warehouses and the capability to easily and reliably share massive data volumes among researchers has had an enormous impact in genomics and imaging {Howe 2008}, and we anticipate that in the near future advances in human and animal systems neurobiology will be accelerated by the creation of large scale human and animal electrophysiology databases {Lynch 2008}.
Figure 2.
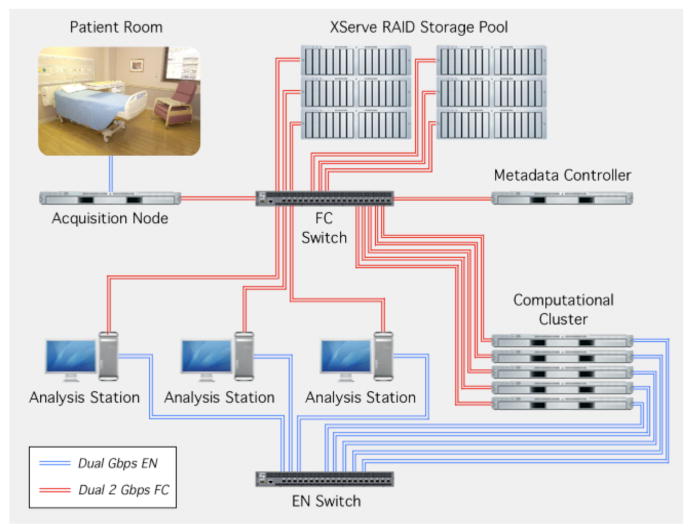
Large scale human electrophysiology acquisition system streams data from the patient's room to the acquisition node via a dedicated dual-Gigabit Ethernet. Data are stored on a 70 terabyte storage pool. Data are accessed via a fiber channel Service Area Network. Large-scale analysis is performed on a dedicated computational cluster.
Methods
Protocol for Large Scale Electrophysiology
The data reported here are from a Mayo Clinic IRB approved investigations of wide-bandwidth electrophysiology recorded from hybrid electrodes in patients undergoing evaluation for epilepsy surgery. The need for intracranial EEG monitoring is a clinical decision made by a multi-disciplinary epilepsy surgery conference with members from neurosurgery, neurology, neuroradiology, and neuropsychology. The location, number and type of intracranial electrodes to be implanted (depth, subdural grid, and strip electrodes) is determined by consensus at the clinical conference. The research protocol involves replacing standard clinical electrodes with custom hybrid electrodes. The only difference between the hybrid and clinical electrodes are the microwire arrays (Figure 1) {Van Gompel 2008a, Van Gompel 2008b, Worrell 2008}. The hybrid depth and subdural electrodes contain standard clinical macroelectrodes and additional microwire arrays (40 um Platinum/Iridium wires spaced 0.5 – 1 mm), and are manufactured by Adtech Medical Instrument Corporation, Racine, WI and PMT Chanhassen, MN US under a 510K.
Platform for collecting and warehousing large-scale human electrophysiology
The capability for collecting, warehousing, and mining wide-bandwidth electrophysiology over multiple spatial scales was originally developed to probe the fine structure of human epileptic brain (Fig 2). A scalable (32 to 320 channels) acquisition platform capable of continuous long-term recording was developed in collaboration with Neuralynx Inc. (http://www.Neuralynx.com). The Digital-Lynx system is unique in that it uses an individual, high resolution, 24 bit A/D converter per channel to directly digitize the electrode signal using a single, DC-coupled, low noise differential amplifier and anti-aliasing filter (low pass 9Khz). All channels are simultaneously sampled at 32 kHz with a DC to 8 kHz signal bandwidth. This high resolution design provides a dynamic input range of ±132 mV with 1 μV resolution (18th bit). All sampled data is packetized and transferred to a PC over a fiber optic data link at 600 Mbits/sec.
The Hybrid Array 40 μm Microwires exhibit a characteristic high impedance (200Kohms to 500Kohms) and the high frequency, weak multi-unit signals (<100 μv) will be degraded by noise and attenuation if not buffered/amplified in close proximity to the brain. An active 32 channel buffered electrode interface was developed for DC-stable microwire/clinical electrode recording and incorporates electrode impedance measurement and patient safety circuitry in a compact package which can be placed on the headwrap. This interface allows individual references for each group of 8 microwires and has proven advantageous for multiple-single unit classification.
The fiber optic connection transfers data to the Neuralynx Cheetah software system. This software package allows for data management, disk file recording of the continuous high resolution sampled data and on-line analysis, processing and display of single unit and scrolling EEG waveforms. The data are then archived to a 70 TB storage area network (SAN) library using a custom file format (MEF) created for efficient data transfer, compression, annotation, archival and retrieval. A SAN is a scalable data storage solution that divides data across multiple hard disk drives to increase data reliability and access speed, and presents the data in such a way that the multiple storage devices appear as a single locally attached volume to the client operating systems. Clinical data are acquired in parallel with an EMU-128 XLTek system (XLTek Inc.). All clinical decisions are based on the clinical XLTek recordings.
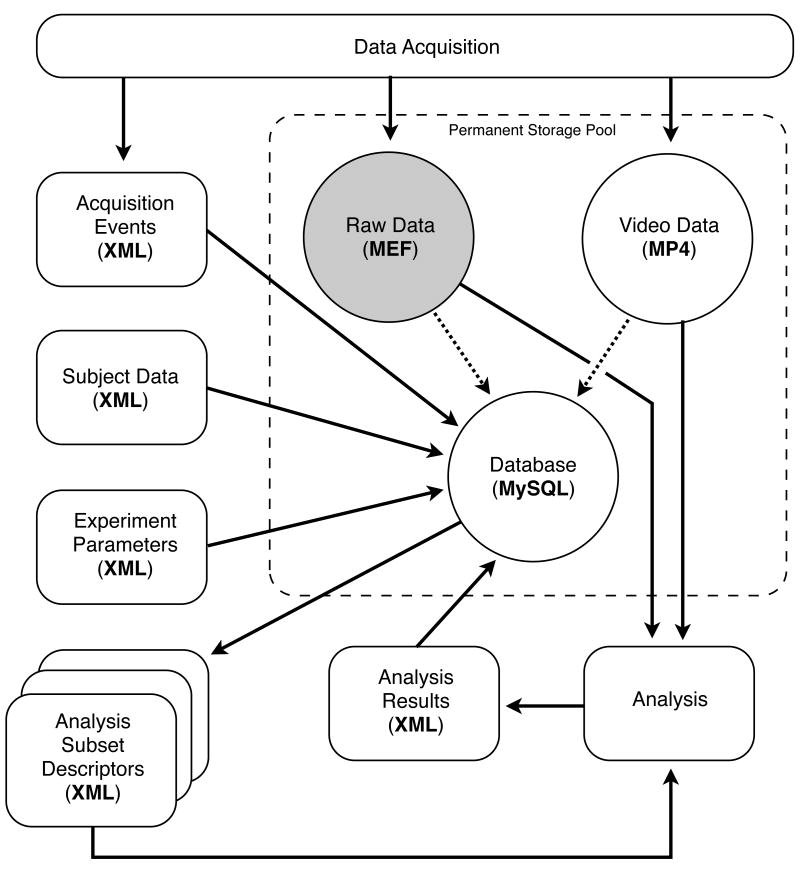
Online event notations and information for all recorded channels are stored in a single separate Extensible Markup Language (XML) event file (Figure 3). The event file and all the associated channel files contain identical 8-byte unique ID (UID) numbers in their unencrypted header blocks in order to validate the association of event and channel files. Event files contain such things as video synchronization records, data annotations, seizure onsets, behavioral state, unit firing, etc. The event file structure consists of variable length XML records, allowing creation of custom event types without disabling existing software that may be unaware of the new event type. This format can also be used to store annotation information related to automated event detections in the file, such as the time of interictal spikes or single-neuron action potentials. We view the XML file as a transient communication medium created for import into a general purpose database (e.g., MySQL), which is better suited to the task of integrating large scale data of various types and providing flexible retrieval options.
Figure 3.
Data flow schematic. Data acquisition creates files stored in a range of data-type-specifc formats. Storing continuously sampled data normally constitutes the largest component of the dataset, which allows data compression to reduce overall storage requirements significantly. Permanent storage of events and metadata in a relational database provides a flexible and reliable storage mechanism that allows subsequent integration of analysis information
Multiscale Electrophysiology Format (MEF)
The Multiscale Electrophysiology Format (MEF) consists of three main parts: 1) a fixed-length 1024-byte header, containing patient information and technical information about the recording, 2) a data section, consisting of a series of encoded data blocks, and 3) a time index section, consisting of three 8-byte element blocks holding block start time, file offset, and sample index values to facilitate rapid random access to the data (Table 1). Each file's header begins with an unencrypted block of data containing the non-private technical information necessary to read and begin decryption (if needed) of the file's header. This data block includes the file's byte order, the file type and version, the length of the header, the encryption algorithm used, and boolean values denoting whether subject and session encryption are used. The next sections of the header employ a dual-tiered encryption scheme, with both sections being encrypted independently. In particular, a “subject” section contains all the subject-identifying data, while a subsequent “session” section contains information regarding data acquisition, such as filter settings and sampling frequency. The session encryption can optionally be applied to the leading coefficients of the statistical model in the data block headers, making the data impossible to decompress without the encryption key. The subject section also contains the session password so that if the subject password is provided, all header information is accessible. If only the “session” password is provided, the subject data remains inaccessible, but the technical details of the recording necessary for data analysis can be decrypted. Subject and session encryption use 128-bit AES encryption [NIST 2001] with passwords chosen by the file's creator. Encryption is not required, and either subject or session encryption, or both, may be omitted if desired.
Table 1.
The MEF file consists of a file header, composed of one unencrypted and two optionally encrypted sections, a data region, composed of sequential data blocks containing block header and compressed data regions, and a block index section, which gives the file offset to each compressed block in the data region.
| File Region | Section | Offset | Length | Contents |
|---|---|---|---|---|
| Header | No Encryption | 0 | 176 | Institution name, encryption algorithm and usage, file version, header length, byte order |
| Subject Encryption | 176 | 160 | Subject first, middle, and last names, and ID number | |
| Session Encryption | 352 | 452 | Number of samples, channel name, recording times, filter settings, maximum block size and length, offset to and number of index entries, max and min recorded values. | |
| Data | Block Header | 1024 | 277 | Compressed block length, difference data length, number of samples in block, block start time, discontinuity flag, block statistics. |
| Compressed Block | Variable | Variable | Encoded Data | |
| Block Indices | Variable | Variable (24- bits per block) | Block start time, file offset to each compressed block, index of first sample in each block. |
The data section of the file (Table 1) consists of recorded samples stored in compressed blocks, the length of which can be specified by the file's creator. Lossless data compression is accomplished via the range encoded differences (RED) algorithm {Bodden 2002, Martin 1979}. Range encoding is a type of integer arithmetic encoding that uses byte-wise scaling to improve encode and decode speeds. RED compression encodes data in two stages: first, differences between sequential samples in the data block are computed; second, the frequency of difference values are computed. The range and frequencies of values in the statistical model are then used to encode values within the block. Differencing time series data efficiently reduces its variance, a property that range encoding benefits significantly from, i.e. as the inherent variance in a signal decreases its compression ratio increases. A 32-bit cyclically redundant checksum (CRC) value {Peterson 1961, Koopman 2002} is calculated from each compressed block and stored as the first entry in the block's header, providing the ability to detect data corruption arising from network transmission errors or disk errors during long-term storage. The block-wise compression scheme used has the advantage that each compressed data block is independent of other blocks in the file. In the event that a particular data block is corrupted due to a disk or network transmission error, the affected block can be removed with no effect on the remaining data. By comparison, a single corrupt value in a difference-encoded file propagates the error to all remaining data in the file. Discontinuities in the recording are indicated by a flag in each block's header, and maximum and minimum recorded values in the block are also stored in the block header to facilitate processing and display. The compressed data blocks are stored with 8-byte alignment to enable direct access to header variables, and to facilitate file recovery if damage to the file results in alignment loss. While data corruption is a low-probability event, the extreme size of these recordings and the fact that we access them repeatedly for different analyses increase the chance of any particular file becoming corrupted. In addition, the size of these files makes it impractical to keep multiple backup copies, making the ability to detect, isolate, and repair data errors all the more important.
Following the compressed data blocks is a series of 8-byte integer triplets encoding the clock time (in microseconds) of the start of each compressed data block, the file offset to the start of each block, and index number of the first sample in each block. These values allow data blocks within the file to be accessed directly based either on a desired time index or recording sample number. Time stamps are stored in Microsecond Coordinated Universal Time (uUTC), which is a variation of standard Unix or Posix UTC time defined by the number of microseconds since midnight January 1, 1970, GMT (also known as “the epoch”). Microseconds are used to provide sufficient temporal resolution for EEG recordings without requiring the use of floating point data types, which are inherently limited in their precision and can cause errors from truncation of the least significant bits.
Results
To date large-scale electrophysiology recordings were obtained from a series of 20 patients using subdural and depth hybrid electrodes. Initial results from patients with hybrid depth electrodes have been previously published {Worrell 2008}. Studies are underway with the the patients implanted with hybrid subdural grid electrodes and will be reported separately. In Figure 4 a representative recording from hybrid depth electrodes implanted into the mesial temporal lobe (amygdala hippocampus) is shown across a wide range of time scales: 10-hours, 10 minutes, and 10 seconds. The single channel of data recorded from a microelectrode demonstrates the long time scale variability seen over the course of hours (Fig 4A), an electrographic seizure discharge (Fig 4B), and extracellular single unit activity (Fig 4C & D). The recordings are notable for the fact that they span neural activity from single neuronal units (10-6 volts) to extracellular fields of almost a 100 millivolts. Microelectrode data were bandpass filtered between 600-6,000 Hz, and action potentials were detected using standard extracellular recording criteria for anatomical location, stability of waveform shape, firing rate (< 2Hz) and multi-modal inter-spike interval distribution. {Bower 2008, Harris 2000}
Figure 4.
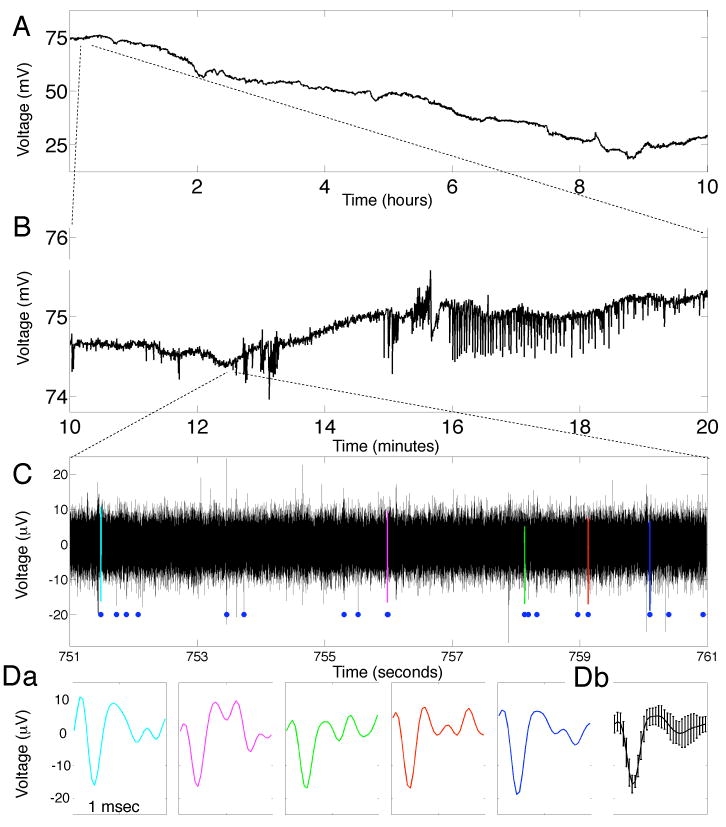
Long-duration, high-frequency, DC-coupled EEG recordings capture all physiologically relevant time scales. A. 10 hours of continuous data from a macroelectrode show a clear DC drift. B. 10 minute, expanded view from A shows a spontaneous seizure approximately 16 min into the recording session. C. 10 second expanded view from B from a microelectrode (bandpass filtered, 600-6,000 Hz) shows action potentials from single neurons. Blue dots show 18 action potentials associated with a single neuron. Da. Expanded view of color-coded action potentials from C showing the similarity of the recorded waveforms. Db. Mean and standard deviation of the 18 action potentials identified in C. Note the dynamic range in both voltage (mV to μV) and time (hours to msec).
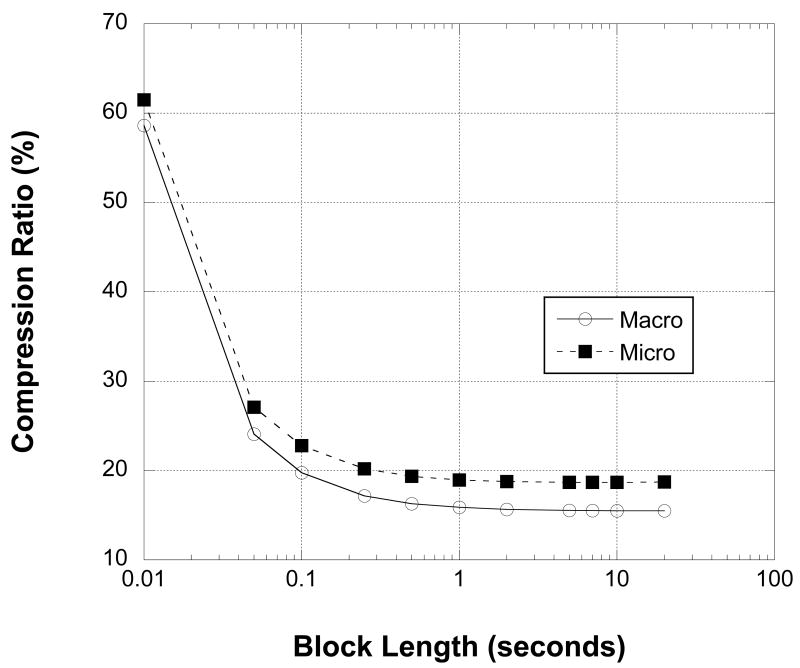
To demonstrate the benefit of the MEF format, a series of randomly-selected 32 kHz macro and microelectrode iEEG channel recordings were compressed using RED compression with varying block lengths. For our tests we defined the theoretical compression ratio as the ratio of the compressed file size (including the header and index block) to the 18-bits of information in each recorded sample in each file. This would be equivalent to comparing our compressed files to an uncompressed file with 18-bits per sample (i.e. 9 bytes for every 4 samples) stored on disk with no sample delimiters and no header. For all channels, the data is compressed to less than 30% of its theoretical size, even with blocks as small as 50 msec, or 1627 samples (Figure 5). Data compression improves markedly as block sizes increase to 1.0 second (32556 samples, in our data), with more modest improvement achieved at larger block sizes.
Figure 5.
Theoretical compression ratios for macro- and microwire 32 kHz channel recordings based on 18 bits of information per sample are plotted against the log of the compressed block length in seconds. Compression ratios tend to improve with longer block lengths and increasing number of samples per block. However, gains beyond 1 sec (32556 samples) are modest and may be outweighed by the advantage of greater direct access to individual time points with smaller blocks.
We also compared the data compression achieved with the MEF file format to real-world recorded data files in widely-used formats. A 32 kHz, 395.8 second iEEG recording with 40 channels in Neuralynx DMA format was converted to MEF format with a 1.0 second block interval, resulting in a net compression ratio of 3.44%, defined as the compressed file size divided by the input file size. For the XLTek file format 32 kHz data was not available, so a 500 Hz data file was used. This file contained 76 recording channels and spanned 65267.5 seconds. Conversion into MEF format with a 10.0 second block interval resulted in a 40.53% net compression ratio. Conversion of 28327.6 sec of data stored in EDF to MEF resulted in a net compression ratio of 8.56% (Table 2). It should be noted that because of the formats' limitations, the XLTek and EDF data files contained only 16 bit sample resolution.
Table 2.
Conversion of other file formats into MEF results in significant file size reduction. XLTek data employs a form of difference encoding, so results are less impressive than for DMA and EDF, which employ no compression.
| Format | Size | Recording Length (sec) | Number of Channels | MEF size | Compression Ratio |
|---|---|---|---|---|---|
| DMA | 7.03 Gb | 395.8 | 40 | 247.6 Mb | 3.44% |
| XLTek* | 4.71 Gb | 65267.5 | 76 | 1.91 Gb | 40.53% |
| EDF* | 1.72 Gb | 28327.6 | 1 | 150.7 Mb | 8.56% |
Our XLTek data was sampled at 500 Hz and our XLTek and EDF data had a 16-bit dynamic range
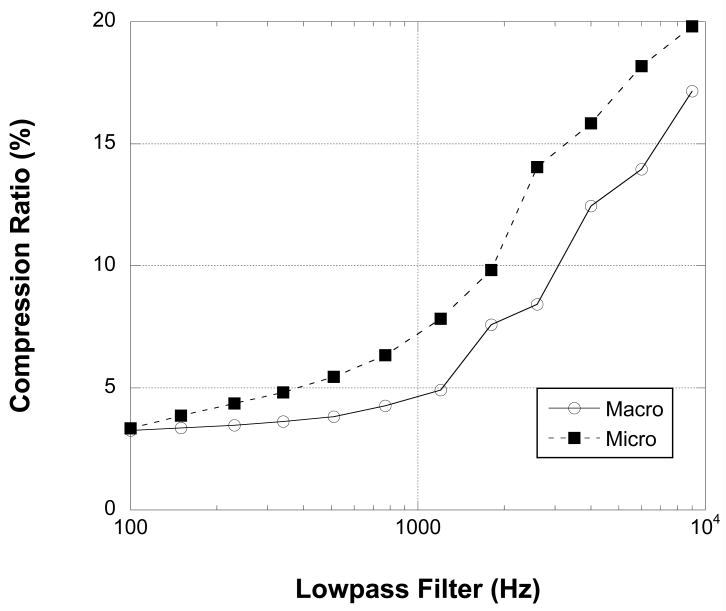
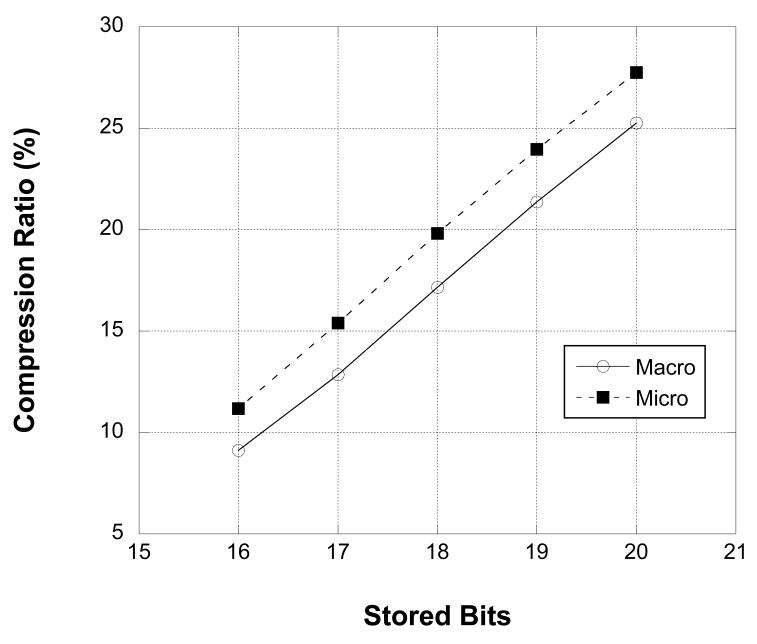
The ability of the RED compression algorithm to adapt to the information content of the recorded signal was tested by low-pass filtering data from a microwire and a clinical macrowire channel with varying cutoff frequencies between 100 and 9000 Hz, maintaining sampling frequency. Figure 6 shows that both files compress to less than 20% of their theoretical size (18 bit sequential samples) with minimal low-pass filtering (9000 Hz) and approach 3% compression at the most aggressive filter levels (100 Hz). Figure 7 shows similarly improved performance by the compression algorithm for the same recorded data as the stored per-sample bit rate is decreased from 20 to 16 bits. Theoretical compression ratios were calculated based on each file's particular bit rate.
Figure 6.
The range-encoded difference algorithm improves its compression ratio as high-frequency information is removed from the recorded data. Compression ratio calculations are based on 18-bits of information in each sample. Reported data represents 2,255,061,204 samples (69,267 seconds) from a macro electrode (white circles) and micro electrode (black diamonds). The relatively low impedance of the macroelectrode compared to the microelectrode yields a lower thermal noise and better overall compression.
Figure 7.
The RED compression algorithm reduces the size of the MEF file as the number of data bits stored is decreased. Percent compression is reported as the ratio of the MEF file size to the theoretical size of the data for each bit rate. Data is reported for 2,255,061,204 samples from a macro electrode and a micro electrode.
The speed of reading and decompressing MEF data was compared to the speed of reading uncompressed raw 32-bit data from disk. Varying lengths of an iEEG data were read from a MEF data file, decompressed, and stored on disk as a binary file of 32-bit integers. Custom software written in C and compiled with the Intel Compiler version 11.0 (Intel Corporation, Santa Clara, CA) was used on an Apple Macintosh computer (Apple Inc., Cupertino, CA) running Mac OS X version 10.5.5 with a 3.2 GHz Intel Xeon 8-Core processor and 32Gb of RAM to read the raw data from disk, and to read the corresponding MEF file from disk and decompress the data into 32-bit integers in memory. The MEF decompression was single-threaded, removing any potential advantage to the machine's multiple processors. As shown in Figure 8, reading plus decompression is faster than reading uncompressed data. Further improvement can be achieved by multithreading the data decompression. (Figure 9, 325,560,000 samples read.) Data block header encryption was not used in these examples, but it typically adds 0.5% to the encoding time.
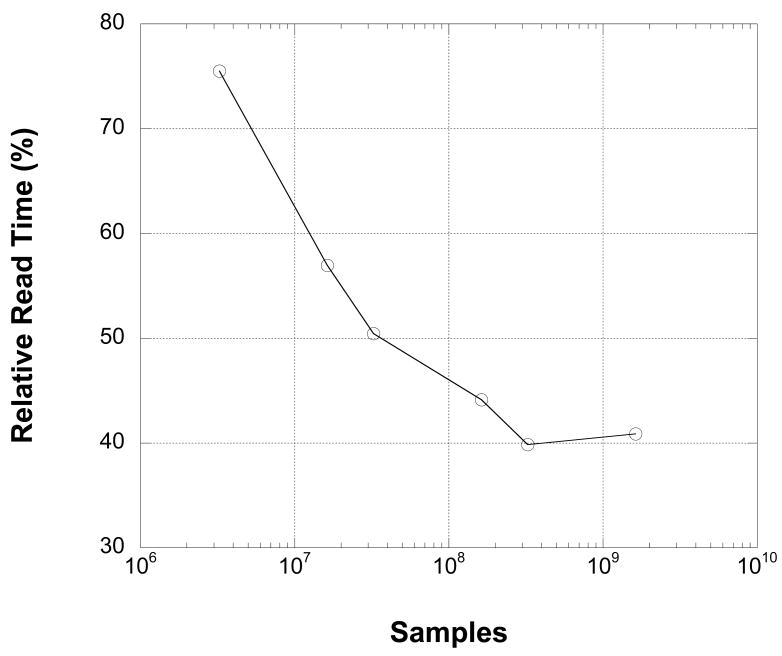
Figure 8.
Reading and decompressing the MEF data from disk is faster than reading raw 32-bit integer data from disk. Data are reported as the percentage of the raw 32-bit integer read time required to read and decode the corresponding MEF file for the given number of samples. Raw and MEF read times were measured using one processor thread on an Apple Macintosh with a 3.2 GHz Intel processor and 32 Gb of RAM.
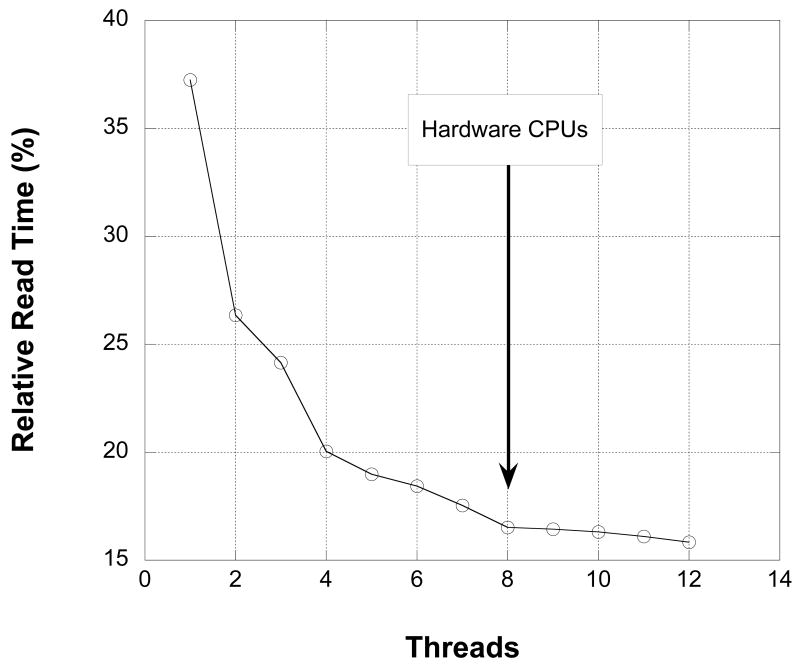
Figure 9.
Multithreading the RED decompression on a multi-processor computer provides a significant speed increase. 325,560,000 samples were read on an 8-processor system with 32 Gb of RAM. Values are expressed as a percentage of the time required to read an identical number of 32-bit samples from an uncompressed raw data file.
Discussion
Systems neurobiological data acquisition has always forced scientists and clinicians to “trade off” one or more aspects of recording to stay within the capabilities of recording equipment and to produce files containing manageable volume of data. These considerations have limited the utility of such data to the questions that originally motivated the acquisition of the data. Current recording technology coupled with MEF file format uncouples data acquisition from storage and analysis constraints, allowing systems neurobiologists to acquire, store and manipulate all physiologically relevant data. While the MEF format is flexible enough to be used with other block-wise compression algorithms, including lossy algorithms if desired, RED encoding offers significant advantages for lossless compression of time-series data. Principal among these advantages are the algorithm's high lossless compression rate and its computational speed. An additional advantage is the algorithm's ability to adapt to the statistical variation in the raw data, which is particularly useful in non-stationary signals such as EEG (Cranstoun 2002), resulting in improved compression ratios in filtered or slowly varying data without requiring changes to the algorithm. The variable block length further permits the user to balance the overall file compression rate versus quick access to specific time points within the file. We typically store our 32 kHz recordings with a block size of 1.0 seconds (32556 samples), although reasonably good compression should be obtainable above 2000 samples per block at most sampling frequencies.
With the increasing processor speed of modern computers, the limiting factor in the speed of data-intensive procedures increasingly becomes access to the data on the computer's disk drive. While data compression does increase the computational load in accessing EEG recordings, the reduction in the size of the data on disc results in a net speed increase, provided the compression algorithm is not overly computationally intensive. Data compression will become more important as hospitals increasingly use electronic patient records and data networks in routine clinical practice. This problem is more pronounced when transferring files across the internet, for example between institutions, where data transfer rates can be significantly slower. Similarly data compression has become more important in research studies as collaborators share data between labs and institutions. Prior EEG data compression studies suggest a correlation between the complexity of the compression algorithm used and the compression ratio achieved. {Antoniol 1997} However computational speed is required to permit real-time compression during data recording, as well as to facilitate display and processing of previously recorded data.
The MEF file structure has been designed to facilitate data storage, transmission, access and processing despite the large number of electrophysiological samples involved. The block structure of the data makes the file resilient to minor file damage during storage or transmission, as only the damaged block(s) will be lost, while the remaining data blocks are unaffected. The index data portion of the file can be reconstructed from the block data if damaged, or if it is practically difficult to construct the indices during recording. The index data permits rapid random access to individual data blocks during viewing or processing, regardless of the length of the overall file. Sampling frequencies are channel specific, making the MEF format suitable for any time series data, including scalp EEG, polysomnography, electrocardiography, and analytic transforms of recorded data, in addition to intracranial EEG. Other data types are possible as well as long as they can be stored as 24-bit or smaller integer time series. Additional data size reduction can be achieved as well in hybrid array recordings by downsampling the macroelectrode signals. The MEF format is equally applicable to human and animal recordings, and header fields have been designed to accommodate either type of subject. The ability to encrypt patient information is fully compliant with HIPPA standards, and thus facilitates data sharing by removing the burden of data deidentification otherwise required. Large scale data also presents challenges for data analysis. The MEF data format divides channels into separate files composed of independent blocks to facilitate parallel processing. The index data section at the end of each file facilitates rapid random access to any point in the file based on either time (uUTC time), or sample number.
The format specification, C source code, Java classes, and Matlab functions to generate and read MEF files have been made freely available under the GNU open-source software license in the hope that this will facilitate widespread use of this file format (http://mayoresearch.mayo.edu/mayo/research/msel/). In addition, Neuralynx Inc recording equipment will now be capable of saving recordings directly to MEF format. (http://www.neuralynx.com)
Conclusions
Systems electrophysiology can require recording from a large number of electrodes and over a wide dynamic range. In this paper we described a human electrophysiology platform capable of recording from 320 electrodes (scalable to 1024 channels) and with a per channel sampling rate of 32 kHz. The practical challenges of managing the massive data volumes generated with high spatiotemporal electrophysiology are significant, but the data compression, information encryption, 32-bit CRC, and block index structure incorporated in MEF data files are important tools for addressing these challenges. Range encoded difference compression reduced the size of recorded data files to less than 20% of the 18 bits per sample encoded at a one second block size, while increasing the speed at which recorded data can be accessed. 128-bit AES encryption meets the patient information privacy restrictions imposed on clinical data by HIPAA regulations. The 32-bit cyclically redundant checksum detects any data corruption that may occur, and MEF's block-wise approach to compression limits the effects of data errors to the data block in which errors occur. The MEF index table provides ready access to any arbitrary point in the recorded data, specified by either the time of the recorded segment or the sequential index of the recorded samples. Software libraries to read, write, and process MEF data are freely available. The system described here is scalable and can be tailored to the electrophysiological questions of interest, without necessitating a trade off dictated by data volume and management.
Acknowledgments
The authors acknowledge the contributions of Lammert Bies, Arturo Campos, Andrew Gardner, and PK Niyaz. This work was supported by the National Institutes of Health (Grant K23 NS47495) and by an Epilepsy Therapy Development Project grant from the Epilepsy Foundation of America.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Antoniol G, Tonella P. EEG data compression techniques. IEEE Transactions on Biomedical Engineering. 1997 Feb;44(2):105–14. doi: 10.1109/10.552239. [DOI] [PubMed] [Google Scholar]
- Bodden E, Clasen M, Kneis J. Proseminar Datenkompression 2001. University of Technology Aachen; 2002. Arithmetic Coding in a nutshell. [Google Scholar]
- Bower MR, Buckmaster PS. Changes in granule cell firing rates precede locally recorded spontaneous seizures by minutes in an animal model of temporal lobe epilepsy. J Neurophysiol. 2008 May;99(5):2431–42. doi: 10.1152/jn.01369.2007. [DOI] [PubMed] [Google Scholar]
- Bragin A, Mody I, Wilson CL, Engel J., Jr Local generation of fast ripples in epileptic brain. J Neurosci. 2002 Mar 1;22(5):2012–21. doi: 10.1523/JNEUROSCI.22-05-02012.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buzsaki G. Large-scale recording of neural ensembles. Nat Neurosci. 2004 May;7(5):446–51. doi: 10.1038/nn1233. [DOI] [PubMed] [Google Scholar]
- Cranstoun SD, Ombao HC, von Sachs R, Guo W, Litt B. Time-frequency spectral estimation of multichannel EEG using the Auto-SLEX method. IEEE Trans Biomed Eng. 2002 Sep;49(9):988–96. doi: 10.1109/TBME.2002.802015. [DOI] [PubMed] [Google Scholar]
- Gelbard-Sagiv H, Mukamel R, Harel M, Malach R, Fried I. Internally generated reactivation of single neurons in human hippocampus during free recall. Science. 2008 Oct 3;322(5898):96–101. doi: 10.1126/science.1164685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner A, Worrell G, Marsh E, Dlugos DJ, Litt B. Human and automated detection of high-frequency oscillation in clinical intracranial EEG recordings. J Clin Neurophysiol. 2007;118:1134–43. doi: 10.1016/j.clinph.2006.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris KD, Henze DA, Csicsvari J, Hirase H, Buzsáki G. Accuracy of tetrode spike separation as determined by simultaneous intracellular and extracellular measurements. J Neurophysiol. 2000 Jul;84(1):401–14. doi: 10.1152/jn.2000.84.1.401. [DOI] [PubMed] [Google Scholar]
- Health Insurance Reform: Security Standards; Final Rule. Federal Register. 2003 Feb;68(34):8334–81. [PubMed] [Google Scholar]
- Hellmann G, Kuhn M, Prosch M, Spreng M. Extensible biosignal (EBS) file format: simple method for EEG data exchange. Electroencephalogr Clin Neurophysiol. 1996 Nov;99(5):426–31. doi: 10.1016/s0013-4694(96)96502-5. [DOI] [PubMed] [Google Scholar]
- Howe D, Costanzo M, Fey P, Gojobori T, Hannick L, Hide W, Hill DP, Kania R, Schaeffer M, Pierre S, Twigger S, White O, Yon Rhee S. Big data: The future of biocuration. Nature. 2008 Sep 4;455(7209):47–50. doi: 10.1038/455047a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemp B, Olivan J. European data format ‘plus’ (EDF+), an EDF alike standard format for the exchange of physiological data. Clin Neurophysiol. 2003 Sep;114(9):1755–61. doi: 10.1016/s1388-2457(03)00123-8. [DOI] [PubMed] [Google Scholar]
- Kemp B, Värri A, Rosa AC, Nielsen KD, Gade J. A simple format for exchange of digitized polygraphic recordings. Electroencephalogr Clin Neurophysiol. 1992 May;82(5):391–3. doi: 10.1016/0013-4694(92)90009-7. [DOI] [PubMed] [Google Scholar]
- Koopman P. 32-Bit Cyclic Redundancy Codes for Internet Applications. The International Conference on Dependable Systems and Networks. 2002 June;459 [Google Scholar]
- Kraskov A, Quiroga RQ, Reddy L, Fried I, Koch C. Local field potentials and spikes in the human medial temporal lobe are selective to image category. J Cogn Neurosci. 2007 Mar;19(3):479–92. doi: 10.1162/jocn.2007.19.3.479. [DOI] [PubMed] [Google Scholar]
- Lynch C. Big data: How do your data grow? Nature. 2008 September 4;455:28–29. doi: 10.1038/455028a. [DOI] [PubMed] [Google Scholar]
- Martin GNN. Range encoding: an algorithm for removing redundancy from a digitised message. Video & Data Recoding Conference; Southampton. 1979. [Google Scholar]
- NIST. Federal Information Processing Standards Publication 197. Springfield, VA: NTIS; Nov, 2001. Announcing the ADVANCED ENCRYPTION STANDARD (AES) [Google Scholar]
- Peterson WW, Brown DT. Cyclic Codes for Error Detection. Proceedings of the IRE. 1961 January;49:228. [Google Scholar]
- Urrestarazu E, Chander R, Dubeau F, Gotman J. Interictal high-frequency oscillations (100-500 Hz) in the intracerebral EEG of epileptic patients. Brain. 2007 Sep;130(Pt 9):2354–66. doi: 10.1093/brain/awm149. [DOI] [PubMed] [Google Scholar]
- Van Gompel JJ, Worrell GA, Bell ML, Patrick TA, Cascino GD, Raffel C, Marsh WR, Meyer FB. Intracranial electroencephalography with subdural grid electrodes: techniques, complications, and outcomes. Neurosurgery. 2008a Sep;63(3):498–505. doi: 10.1227/01.NEU.0000324996.37228.F8. [DOI] [PubMed] [Google Scholar]
- Van Gompel JJ, Stead SM, Giannini C, Meyer FB, Marsh WR, Fountain T, So E, Cohen-Gadol A, Lee KH, Worrell GA. Phase I trial: safety and feasibility of intracranial electroencephalography using hybrid subdural electrodes containing macro- and microelectrode arrays. Neurosurg Focus. 2008b Sep;25(3):E23. doi: 10.3171/FOC/2008/25/9/E23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worrell GA, Gardner AB, Stead SM, et al. High-frequency oscillations in human temporal lobe: simultaneous microwire and clinical macroelectrode recordings. Brain. 2008 Apr;131(Pt 4):928–37. doi: 10.1093/brain/awn006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worrell GA, Parish L, Cranstoun SD, Jonas R, Baltuch G, Litt B. High-frequency oscillations and seizure generation in neocortical epilepsy. Brain. 2004;127:1496–506. doi: 10.1093/brain/awh149. [DOI] [PubMed] [Google Scholar]