SUMMARY
We report here homologous recombination (HR)-mediated gene targeting of two different genes in human iPS and ES cells. HR-mediated correction of a chromosomally-integrated mutant GFP reporter gene reaches efficiencies of 0.14%-0.24% in both cell types transfected by donor DNA with plasmids expressing zinc-finger nucleases (ZFNs). Engineered ZFNs that induce a sequence-specific double-strand-break in the GFP gene enhanced HR-mediated correction by >1400-fold without detectable alterations in stem cell karyotypes or pluripotency. Efficient HR-mediated insertional mutagenesis was also achieved at the endogenous PIG-A locus, with a >200-fold enhancement by ZFNs targeted to that gene. Clonal PIG-A null human ES and iPS cells with normal karyotypes were readily obtained. The phenotypic and biological defects were rescued by PIG-A transgene expression. Our study provides the first demonstration of HR-mediated gene targeting in human iPS cells, and the power of ZFNs for inducing specific genetic modifications in human iPS as well as ES cells.
Keywords: human embryonic stem (ES) cell, induced pluripotent stem (iPS) cell, gene targeting, homologous recombination (HR), zinc finger nucleases (ZFNs)
INTRODUCTION
Human embryonic stem (ES) cells derived from blastocyst-stage embryos can self-renew indefinitely in culture while retaining their pluripotency (Reubinoff et al., 2000; Thomson et al., 1998). Recently, human somatic cells have been successfully reprogrammed into induced pluripotent stem (iPS) cells that exhibit unique characteristics similar to human ES cells (Lowry et al., 2008; Mali et al., 2008; Park et al., 2008; Takahashi et al., 2007; Yu et al., 2007). Patient-specific iPS cells hold enormous promise for personalized cell replacement therapy as well as for research of various human diseases. Full utilization of these human pluripotent stem cells (PSCs) will require the development of efficient methods for performing gene targeting, a sequence-specific and permanent genome modification that exploits the cell’s ability to perform homologous recombination (HR). Gene targeting by HR has played a critical role in genetic studies of various systems including the generation of knockout/knock-in transgenic mouse models using mouse ES cells. The efficiency of HR-mediated gene targeting in human ES cells, however, remains low even 10 years after its first report (Thomson et al., 1998), with only few successful studies published to date using the method that is routinely performed in mouse ES cells (Costa et al., 2007; Davis et al., 2008; Di Domenico et al., 2008; Irion et al., 2007; Ruby and Zheng, 2009; Urbach et al., 2004; Zwaka and Thomson, 2003). Using standard plasmid-based systems, the current HR rate is <10−6 in karyotypically normal human ES cells and other non-transformed mammalian cells. Moreover, to date there has been no report on successful gene targeting in human iPS cells. One reason that gene targeting in human PSCs is more difficult is because they grow poorly as single cells (a practice required for selection of rare targeted clones), compared with mouse ES cells. Thus, strategies that increase the efficiency of gene targeting in human PSCs would improve both the therapeutic and experimental potential of these cells.
Zinc finger nucleases (ZFNs) are engineered sequence-specific nucleases consisting of a customized array of zinc-fingers engineered to bind to a specific DNA sequence and a non-specific DNA endonuclease domain (Durai et al., 2005; Porteus and Carroll, 2005). Each zinc-finger domain recognizes 3–4 bp of DNA and a three-finger ZFN recognizes a 9–10 bp DNA sequence. When two ZFNs bind their cognate target sequence in the proper orientation, the nuclease domain is able to dimerize and create a sequence-specific double-stranded break (DSB). The ZFN-induced DSB can then be repaired with high efficiency by either HR or error-prone non-homologous end-joining (NHEJ) independent of a DNA template for repair. Therefore ZFNs induce a site-specific insertion or deletion at the site of the break after NHEJ, or a defined genetic modification near the site of the DSB by HR with an exogenous donor DNA fragment. ZFNs have been used to make site-specific genomic modifications with high efficiencies in a variety of cell lines and several small organisms (Carroll, 2008). A recent study showed that ZFNs can enhance gene targeting in human ES cells using integrase-defective lentiviral vectors (IDLVs) to deliver ZFN expression cassettes and a HR donor DNA template (Lombardo et al., 2007). Limitations of this study, such as lack of data by Southern blotting for confirming the HR event and excluding random integration of the donor DNA, have been noted by the authors of a recent review (Giudice and Trouson, 2008). Another limitation of this work is that the two human ES cell lines used are prone to karyotypic abnormalities after prolonged culture (Cowan et al., 2004) . Such abnormalities likely contribute to their favorable growth characteristics in culture, and can affect gene-targeting efficiencies. Whether ZFNs might induce chromosomal alternations and changes in growth parameters of targeted human ES cells has not been reported. Although the use of IDLVs reduces the rate of vector integration substantially, it does not eliminate such integration events (Mali et al., 2008; Nightingale et al., 2006).
In this study, we used a virus-free system to perform ZFN-enhanced gene targeting at the endogenous PIG-A gene and at a defective, chromosomally integrated enhanced green fluorescent protein (EGFP) reporter gene in both human ES and human iPS cells. The PIG-A gene is required for the retention of dozens of glycosyl-phosphatidyl-inositol anchored proteins (GPI-APs) on the cell surface and is mutated in hematopoietic stem cells from patients with the blood disorder paroxysmal nocturnal hemoglobinuria (PNH). The defective, chromosomally-integrated EGFP reporter gene we used in our studies requires HR to reconstitute a full-length gene and thereby restore fluorescence. We demonstrate that the transient expression of sequence-specific ZFNs significantly enhanced HR (2400-fold increase) in human ES cells and we were able to readily obtain PIG-A null human ES cells by both HR and mutagenic NHEJ. Importantly, we also show that these ZFNs enhance gene targeting without detrimental effects on either cell karyotypes or pluripotency. Moreover, we also provide the first demonstration that ZFNs can enhance gene targeting in two human iPS cell lines by successfully performing targeted HR events at both the PIG-A locus and a chromosomally-integrated EGFP reporter gene. The present study describes and validates publicly available “open-source” reagents and protocols that will enable researchers to use ZFNs to efficiently create or correct specific mutation at their genes of interest in either human iPS and ES cells.
RESULTS
Using an EGFP gene reporter system to optimize gene targeting in human ES cells
To enhance the efficiency of gene targeting in human ES cells by non-viral vectors, we developed better methods for plasmid delivery and for selection of rare transgenic human ES clones. First, we established an immortalized feeder cell line (W3R) that expresses Wnt3a to promote human ES cell growth and co-expresses 3 drug-resistance genes (Cai et al., 2007). The ability of W3R feeder cells that support human ES cells and are resistant to neomycin, hygromycin B and puromycin allowed us to efficiently select rare clones of transfected human ES cells expressing one of 3 drug-resistant genes: NeoR, HygroR or PuroR. Second, we optimized methods for delivering plasmid DNA into human ES cells using the Amaxa’s improved electroporation method called Nucleofection (Cai et al., 2007). Using this strategy, human ES cells can be transfected with >50% efficiency and stable ES cells can be generated at a rate of 10−5 (Cai et al., 2007; Hohenstein et al., 2008). In this study, we used these optimized methods for plasmid delivery and selection to further improve gene targeting in human ES cells.
To assess the efficiency of HR-mediated gene targeting in human ES cells, we established a mutated GFP gene-based reporter system, similar to one previously described and used in somatic human cells (Porteus, 2006; Porteus and Baltimore, 2003). In this improved version, the EGFP gene was used to achieve a brighter GFP signal as compared to the GFPmut1 gene (Yang et al., 1996) used in the previous studies. A 35-bp DNA fragment containing a stop codon was inserted into the EGFP sequence, 12-bp downstream of a site for which we had previously generated ZFN pairs (Pruett-Miller et al., 2008). This mutated EGFP* cassette was inserted into a lentiviral vector we designated EGIP* (Figure 1A). This EGIP* vector enables the creation of a chromosomally-integrated EGFP* target sequence by lentiviral transduction in a variety of cell types including human ES cells. We also constructed a donor template plasmid called tGFP which contains a truncated EGFP DNA. HR between the transfected tGFP donor and the integrated EGFP* target results in the reconstitution of a full-length EGFP gene without the insertion, and its gene expression restores GFP fluorescence. In 293T cells that can be transfected very efficiently by either Lipofection or Nucleofection, GFP+ correction rate was low (∼7 per 106 cells) when cells were transfected by tGFP alone (Figure S1A). Co-transfection of the same reporter 293T cells with tGFP and two plasmids expressing ZFNs targeting a site in the EGFP gene (Pruett-Miller et al., 2008) led to an increase in the level of GFP+ cells to ∼3% (Figure S1B–C): a >4000-fold enhancement (Figure S1A). The percentage of GFP+ population remained stable over 2 weeks after transfection, indicating minimal cytotoxicity of ZFNs used in these experiments (Figure S1D).
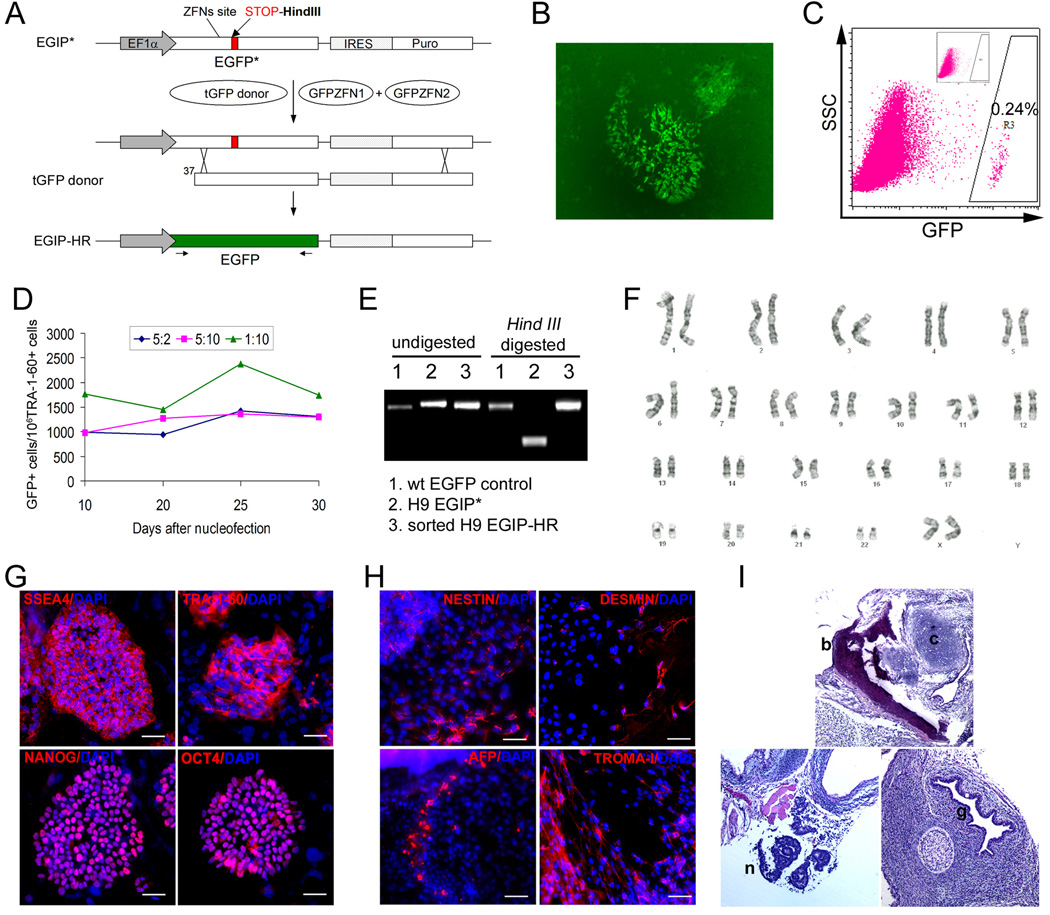
Figure 1. Correction of a mutant EGFP reporter gene by ZFN-mediated gene targeting.
(A) Scheme of the EGFP gene correction strategy. An enhanced GFP gene was mutated (EGFP*) by the insertion of a 35-bp fragment containing a translational stop codon and a Hind III site, positioned 12-bp downstream of the GFP ZFN target site. The defective EGFP(*) transgene was delivered and integrated into human cells by a lentiviral vector called EGIP*. For EGFP gene correction, a repair donor (tGFP) containing 5’-truncated EGFP coding sequence was co-transfected with two plasmids expressing a pair of GFP-targeting ZFNs (GFPZFN1 & GFPZFN2). If HR-mediated repair occurs, expression of the wild-type EGFP gene will be restored. Arrows below the corrected EGFP gene represent primers used to detect the restored full-length EGFP gene.
(B) Fluorescence microscopy of EGFP gene correction with ZFNs in H9-EGIP* human ES cells at day 7 post Nucleofection.
(C) FACS analysis showing that the efficiency of ZFN-mediated EGFP gene correction in H9-EGIP* is as high as 0.24% (gated on TRA-1-60+ human ES cells). Inset shows that no GFP+ cells are detected in 106 cells collected, in the absence of GFP-specific ZFNs.
(D) Numbers of corrected GFP+ cells (per million of TRA-1-60 positive human ES cells) were monitored by FACS over 30 days. Three different ratios of Donor:ZFNs were used at 5:2, 5:10, and 1:10. A ratio (w/w) of 1:10 is equivalent to a molar ratio of 1 donor DNA for 6.1 molecules of each ZFN.
(E) PCR amplification of wild-type EGFP gene using primers indicated in (A). H9-EGIP* template contains a Hind III site therefore PCR product can be digested by Hind III (sample #2). After ZFNs-mediated HR, GFP+ cells were sorted and showed restoration of the wild-type EGFP gene without the Hind III site (samples #1 and #3).
(F) H9 EGIP cells after ZFN-mediated HR show a normal karyotype (46,XY).
(G) H9 EGIP cells after ZFN-mediated HR maintain expression of pluripotency markers such as SSEA4, TRA-1-60, NANOG and OCT4 after expansion by long-term culture. Scale bar = 50 µm.
(H) Immuno-staining of embryoid body (EB) formed by H9 EGIP cells after ZFN-mediated HR. Day 14 EB shows ectoderm (NESTIN), mesoderm (DESMIN), endoderm (AFP) and trophectoderm (TROMA-I) markers. Scale bar = 50 µm.
(I) Teratoma formation of H9 EGIP cells after ZFN-mediated HR. H&E staining indicates in vivo differentiation of ectodermal (neuroepithelial, n), mesodermal (bone, b; cartilage, c) and endodermal (glandular epithelial, g) structures.
We next used the EGFP* HR system in human ES cells after stable integration of the EGIP* reporter vector. Using conditions we had previously established for Nucleofection of human ES cells (Cai et al., 2007), we were able to achieve a transient transfection efficiency of 50–70% for multiple human ES lines each bearing a stably integrated EGIP* reporter vector (data not shown). Following transfection of the tGFP donor DNA (Figure 1A), we measured EGFP gene correction in human ES cells (harboring the EGFP* reporter) with or without co-transfection of the GFP-ZFN expression plasmids. We observed that GFP+ cells at a frequency up to 0.24% with ZFNs compared to <10−6 without ZFNs, a >2400-fold increase in the efficiency of HR in human ES cells (Figure 1B–C). The level of GFP+ cells among undifferentiated (TRA-1–60+) human ES cell population remained stable for at least one month following transfection of ZFN expression plasmids (Figure 1D). Correction of the EGFP* to the wild-type EGFP gene sequence was confirmed by both restriction digestion mapping and direct DNA sequencing (Figure 1E, Figure S2).
Corrected GFP+ human ES cells that had been obtained by ZFN-mediated HR retained normal karyotype, morphology and expression of pluripotency markers even after long-term culture (>50 passages) (Figure 1F–G). Upon induction, they can also differentiate into 3 embryonic germ layers and trophoblast lineages via embryoid body (EB) formation in vitro (Figure 1H). When injected into immuno-deficient mice, the corrected GFP+ cells formed teratomas containing cells representing all three embryonic germ layers (Figure 1I). These data indicate that our gene targeting approach using transiently-expressed ZFNs had no detectable adverse effects on the genome or pluripotency of human ES cells.
Engineering and validation of ZFNs targeted to the endogenous PIG-A gene in human ES cells
Because ZFNs can be engineered to recognize specific target DNA sequences, ZFN-enhanced gene targeting can potentially be used at any endogenous gene of interest. We therefore next wished to test whether ZFNs could enhance gene targeting of an actual endogenous human gene and focused our efforts on PIG-A, a gene we had previously been unable to alter genetically using conventional gene targeting methods. To create PIG-A-specific ZFNs, we used the Oligomerized Pool ENgineering (OPEN) platform recently developed (Maeder et al., 2008), and engineered four ZFN pairs for a site within the PIG-A coding sequence. We chose to target such a sequence because we reasoned that mutation at that site would generate an inactive PIG-A protein.
We validated the four OPEN ZFN pairs targeting the PIG-A gene using a modified version of the EGFP* gene targeting system described above, which contained the PIG-A ZFN target site inserted into the EGFP* gene, an approach similar to what has been used in previous studies (Porteus, 2006; Porteus and Baltimore, 2003; Urnov et al., 2005). We found that all four PIG-A ZFN pairs stimulated gene targeting by greater than 200-fold in 293T cells with the L1/R2 pair showing the highest efficiency (Figure S3). We next tested these validated PIG-A ZFN pairs in human ES cells by determining their ability to induce mutations in the endogenous PIG-A gene by mutagenic NHEJ repair of the ZFN-induced DSB. We used male (XY) human ES cells such as H1 so that only one PIG-A gene (on the X-chromosome) would need to be mutated to generate a phenotypic effect. Null mutants of PIG-A result in a loss of GPI-APs and in turn the resistance of targeted cells to aerolysin-mediated cell killing (Chen et al., 2008). It is important to note that PIG-A and GPI-APs have been shown to be dispensable for the growth of undifferentiated human ES cell in culture (Chen et al., 2008). We utilized the aerolysin selection method to enumerate rare PIG-A null mutations and compared the activities of four PIG-A ZFN pairs in human H1-derived GGFP ES cells. The best pair of ZFNs (L1/R2) identified in the 293T EGFP* reporter assay also generated the greatest number of PIG-A mutant clones, creating 28 aerolysin-resistant colonies per 6×106 cells selected (Figure 2A).
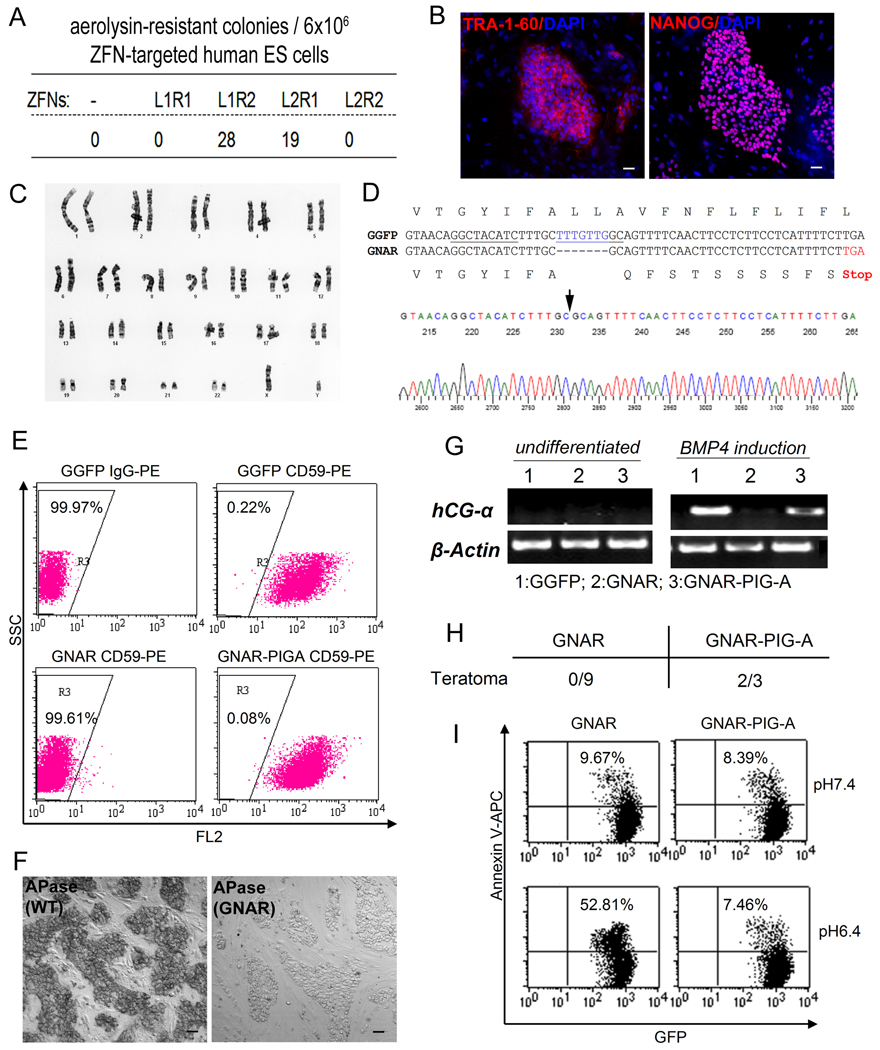
Figure 2. Generation of PIG-A null mutants by ZFN-mediated NHEJ in H1 human ES cells.
(A) Four combinations of ZFN pairs targeting the PIG-A gene were tested for their abilities to induce mutagenic NHEJ in H1 (XY) human ES cells that constitutively express GFP (GGFP). GGFP human ES cells were first transfected by Nucleofection with one of the four ZFN combinations (L1R1, L1R2, L2R1, L2R2) and expanded. 17 days after nucleofection, expanded cells were treated with 0.5 nM aerolysin and allowed to form colonies for 7 days (Chen et al., 2008). The Number of surviving colonies following aerolysin selection was shown.
(B). One of the aerolysin-resistant colonies derived following expression of the L1 and R2 ZFN pairs was isolated and termed as GNAR. After expansion, GNAR cells display unique human ES cell morphology and markers such as TRA-1-60 and NANOG.
(C) GNAR cells exhibit a normal karyotype (46,XY).
(D) DNA sequence in PIG-A exon 6 in GNAR cells, compared with that from parental GGFP (H1) human ES cells. A 7-bp deletion (blue) in the coding sequence was found, resulting in frame shift and a premature translational stop codon (red). The PIG-A ZFN target sites are underlined. A sequencing chromatogram of GNAR cells is shown below the sequence alignment, with an arrow indicating the deletion site.
(E) FACS analysis showing that GNAR cells have lost the expression of GPI-APs such as CD59. This defect can be rescued by introduction of exogenous PIG-A cDNA (GNAR-PIGA). Dot plot was gated on GFP+ human ES cells.
(F) GNAR cells show loss of GPI-APs such as alkaline phosphatase (APase) on the cell surface. Scale bar = 50 µm.
(G) BMP4-induced trophoblast lineage differentiation defect in GNAR cells indicated by loss of hCG-α expression. BMP4 treatment was carried out as described previously (Chen et al., 2008).
(H) Rate of teratoma formation of GNAR ES cells with or without PIG-A transgene expression. Zero out of 9 injections showed teratoma formation monitored up to 4 months. These defects can be largely rescued by exogenous PIG-A cDNA expression.
(I) Differential sensitivities of GNAR and GNAR-PIG-A human ES cells. GNAR cells lack complement inhibitory proteins such as CD59 and CD55, while PIG-A cDNA expression restored these GPI-APs on the cell-surface. Under lower pH (pH=6.4) that activates complement-mediated cell lysis, GNAR showed an significantly increased cell death monitored by Annexin V staining, compared with the GNAR-PIG-A cells or ES cells under normal pH (7.4, upper rows).
One aerolysin-resistant human ES cell clone (called “GNAR”) was chosen for further characterization. GNAR grew similarly to the parental cells from which it was derived, retained a normal karyotype, and displayed morphology and markers unique to human ES cells (Figure 2B–C). DNA sequencing of the targeted region revealed deletion of 7-bp overlapping with the ZFN target sites, resulting in a frame shift mutation and a premature stop codon (Figure 2D). The PIG-A mutation in GNAR cells abolished the cell-surface expression of GPI-APs such as CD59 (Figure 2E) and alkaline phosphatase (APase, Figure 2F) as expected. The loss of GPI-APs in GNAR cells could be rescued by introduction of a PIG-A transgene, confirming that the defect in these cells is solely due to the lack of PIG-A expression (Figure 2E). Similarly, the loss of biological functions by GNAR human ES cells can be restored by the PIG-A transgene expression in 3 different functional assays in vitro and in vivo (Figure 2G–I).
Efficient ZFN-mediated gene targeting of the endogenous PIG-A gene in human ES cells
Using the validated PIG-A ZFN pair L1/R2, we sought to develop a more efficient, precise and generalizable method for gene targeting of the PIG-A gene without the need to use aerolysin selection. Compared with NHEJ, HR is a more precise method to mutate as well as correct a gene in a predetermined fashion. Thus, to inactivate the PIG-A gene by HR-mediated mutagenic insertion, we constructed a donor vector (called “PHD”) in which a PGK-HygroR expression cassette was placed between two PIG-A homology arms (Figure 3A). Successful HR-mediated targeting will result in the insertion of the PGK-HygroR cassette into the coding region in exon 6 of the PIG-A gene, thereby inactivating the PIG-A gene (Figure 3A). By contrast, random insertion of PHD into transcriptionally active regions of the genome will also result in the formation of HygroR ES clones but will not disrupt the PIG-A gene. Thus, random insertions, unlike targeted insertions, will result in the continued cell surface expression of GPI-APs.
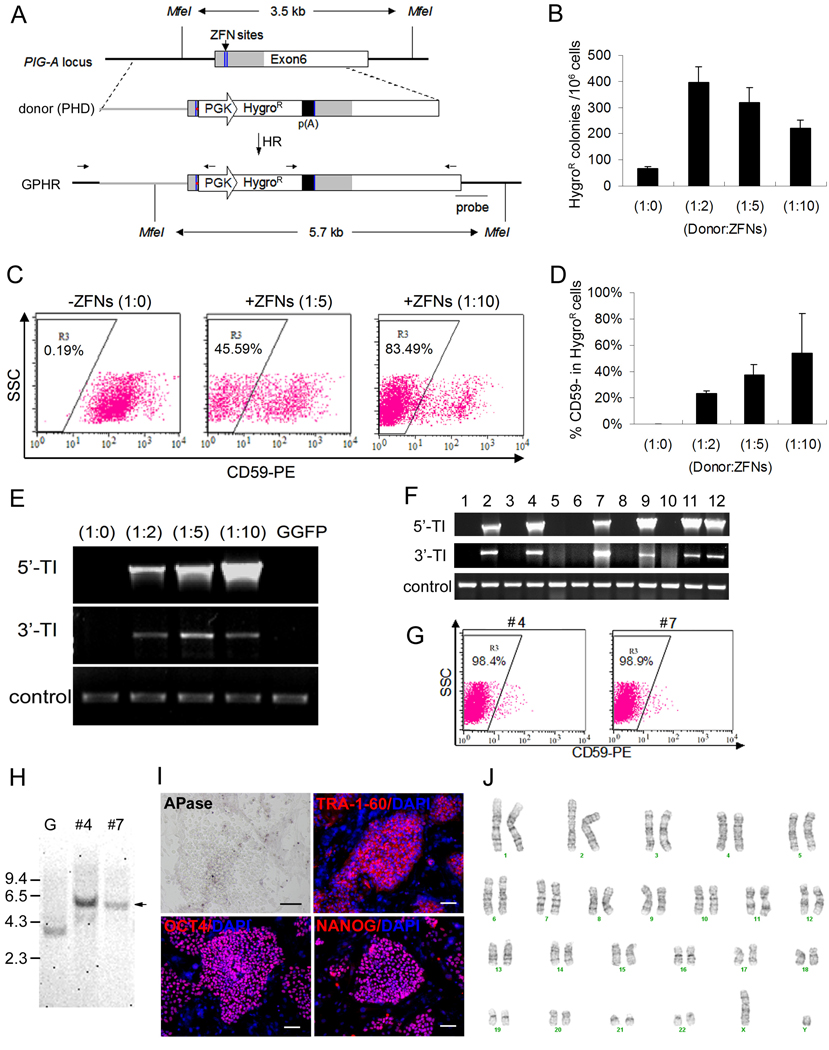
Figure 3. Knockout of the endogenous PIG-A gene in human ES cells by ZFN-mediated homologous recombination (HR).
(A) Scheme of PIG-A gene knockout using ZFNs targeting the coding sequence (gray) within exon 6. The DNA sequence of the two half ZFN sites (blue lines) were shown in Figure S10. The HR donor (PHD) vector contains a PGK-HygroR-poly(A) expression cassette flanked by two arms of PIG-A homology sequences (2 kb left + 2kb right). In the PHD vector, a frame shift mutation and stop codon (red dot) upstream of the PGK-HygroR-poly(A) cassette were introduced to replace the spacer sequence between the two ZFN sites. Two sets of PCR primers indicated above the anticipated HR product (GPHR) were used to confirm the junctions of targeted insertion. A DNA probe further downstream of the PIG-A locus and outside the right arm of PHD was used for Southern blot analysis.
(B) After Nucleofection and drug selection, the number of HygroR colonies per million input cells was calculated and compared among different ratios of Donor:ZFNs. The ratio (w/w) of 1:5 is equivalent to 1 donor for 4.2 DNA molecules of each ZFN. Six experiments were performed with the ratio of 1:5 and three experiments for the remaining ratios.
(C) Representative FACS analysis of CD59, a GPI-AP, expressed in the HygroR cell population after gene targeting. Dot plot was gated on GFP+ human ES cells.
(D) Summary of CD59- population in the HygroR cells from two experiments, as analyzed in (C).
(E) PCR analysis of genomic DNA confirms that targeted integration (TI) occurred in HygroR cells only when ZFNs were used and when GPI-AP deficient cells were obtained. TI was detected by primer sets specific for 5’ and 3’ integration junctions as indicated in (A). An arbitrary genomic region (near PIG-A exon 4) was amplified as a positive DNA control.
(F) Twelve GPHR colonies were manually picked after HygroR selection following ZFN-mediated PIG-A knockout in GGFP cells. 6 out of 12 showed targeted integration (TI).
(G) Two of the 12 colonies, #4 and #7, were shown to have nearly complete loss of CD59.
(H) Southern blot analysis of genomic DNA after Mfe I digestion confirmed these two clones (#4 and #7) contain expected targeted insertion at the PIG-A locus. Arrow indicates a longer Mfe I band (5.7-kb, due to the PGK-HygroR insertion) instead of 3.5-kb band in parental GGFP (see Figure 3A). G: GGFP parental human ES cells.
(I) Staining of one of the two clones, #4 (GPHRc4). This clone lacks APase, a GPI-AP, on the cell surface, but maintains undifferentiated human ES cell markers such as TRA-1-60, OCT4 and NANOG. Scale bar = 50 µm. Similar data with clone #7 are shown in Figure S5.
(J) GPHRc4 (clone #4) cells exhibit a normal karyotype (46,XY). Similar data with clone #7 are shown in Figure S5.
We transfected the PHD targeting plasmid into H1-derived GGFP human ES cells in the presence or absence of vectors expressing the L1/R2 ZFN pair to target the endogenous PIG-A locus. Different ratios of donor vs. ZFN expression vector DNA were tested (Figure 3B) and following transfection cells were plated onto HygroR W3R feeder cells and selected for 12 days. ES-like colonies emerged under each transfection condition and the number of colonies was quantified. In the absence of ZFNs, we obtained 67±11 HygroR colonies per 106 input cells at transfection. The formation of HygroR colonies could be due to either random integration or HR. With ZFNs, however, we obtained significantly more HygroR colonies, ranging from 220±54 to 395±105 per 106 input cells (Figure 3B). After quantitation of HygroR ES colonies, all the cells were analyzed for the presence or absence of GPI-APs such as CD59.
When ZFNs were absent, background levels of CD59- cells were detected (0.19%) indicating that most, if not all, HygroR colonies resulted from random insertion of the PHD donor into loci other than PIG-A. In contrast, when ZFNs were present, CD59- cells were present at a high frequency of up to 83% (Figure 3C–D). To confirm the targeted insertion of PHD DNA into the PIG-A locus, we performed PCR analysis using a pair of specific primers that only amplify either the 5’ or 3’ junction of the predicted HR product (Figure 3E). Targeted insertion products were abundantly detected in HygroR cells obtained when ZFNs were used, but not in HygroR cells obtained when ZFNs were absent (Figure 3E).
To create knock-out or knock-in ES cells with a uniform genetic background, it is critical to isolate clonal cell lines following HR to ensure a homogenous population. With the improved HR efficiency we observed with ZFNs, we tested whether we could readily obtain PIG-A knockout clones (called “GPHR”). From 12 HygroR human ES cell colonies, 6 clones (50%) were confirmed to have undergone the predicted HR event at the PIG-A locus as determined by PCR to detect the targeted insertion (Figure 3F) and by FACS to detect the loss of GPI-APs (Figure 3G and Figure S4A). Two out of 6 clones (#4 and #7) showed loss of CD59 in nearly all the cells (Figure 3G), thereby validating their clonality. The clonality and the targeted insertion were further confirmed by Southern blot (Figure 3H). Furthermore, both of these clones showed normal karyotypes, undifferentiated human ES cell morphology, and expression of unique human ES cell markers (Figure 3I–J, Figure S5). As expected, the cell-surface expression of GPI-APs such as APase was absent in these GPHR clones (Figure 3I). The altered phenotypes of these GPHR clones such as GPI-AP deficiency could also be rescued by PIG-A transgene expression (Figure S4B), confirming that the GPI-AP deficiency in GPHR cells was solely due to PIG-A gene knock-out.
Gene targeting in the MP2 human iPS cell line with or without using ZFNs
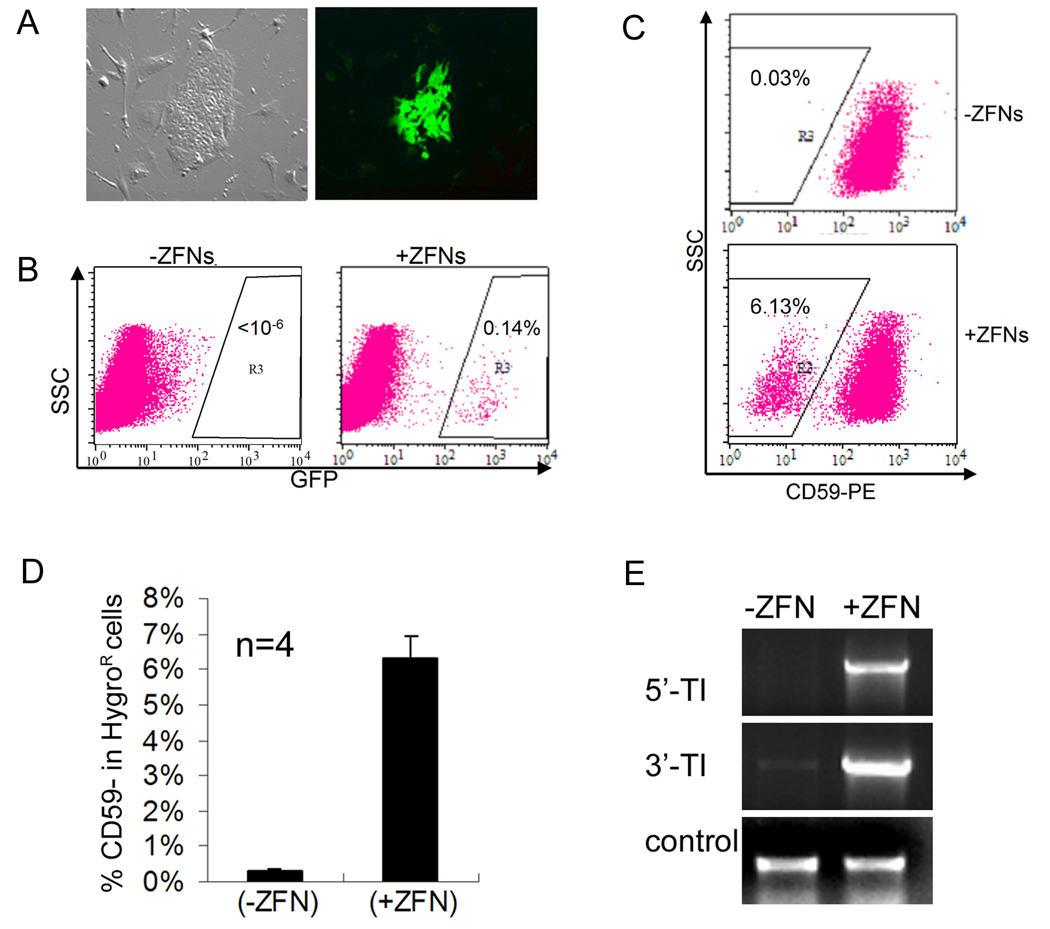
To our knowledge, there has been no published report to date describing gene targeting by any method in human iPS cells. Therefore, we investigated whether HR-mediated gene targeting could be achieved in human iPS cells using the PIG-A and EGFP* systems with or without the aid of ZFNs. The first of the two human iPS cell lines we used is MP2, which has previously been shown to be pluripotent (Mali et al., 2008). Using an optimized Donor:ZFN ratio, we observed GFP+ colonies emerge 5 days following transfection of MP2 iPS cells harboring the mutated EGFP* template (Figure 4A). Gene correction efficiency reached 0.14% when the GFP-specific ZFN expression vectors were used in transfection as compared to <0.0001% when ZFN expression vectors were absent from the transfection (Figure 4B). These data demonstrate that the ZFNs stimulate gene correction by at least 1400-fold in human MP2 iPS cells. Because the mutated EGFP* target is not present in every one of starting MP2 cells (since they are already PuroR and that full selection of the EGIP* integration by puromycin resistance is not feasible), the number is likely an underestimate (Figure 4A–B).
Figure 4. HR-mediated gene targeting of EGFP and PIG-A genes in female human MP2 iPS cells.
(A) Microscopy of corrected (GFP+) MP2 iPS cells harboring the EGIP* reporter, 5 days after Nucleofection with GFP-specific ZFNs.
(B) FACS analysis of EGFP gene correction in MP2 iPS cells with or without GFP-specific ZFNs. Dot plots were gated on TRA-1-60+ cells.
(C) PIG-A gene targeting in MP2 iPS cells with or without PIG-A specific ZFNs was performed as described in Figure 3A using the 1:5 ratio of Donor:ZFN DNA. After Nucleofection and hygromycin B selection, TRA-1-60+ (human iPS/ES) cells were analyzed for the presence or absence of CD59 (a GPI-AP). Representative FACS dot plots are shown.
(D) Summary of FACS analysis of CD59- MP2 iPS cell populations after PIG-A gene targeting from 4 independent experiments.
(E) PCR confirms the targeted integration into the PIG-A locus in MP2 iPS cells.
Next, we carried out ZFN-mediated gene targeting at the endogenous PIG-A gene in MP2 iPS cells (using the same strategy employed for ES cells, see Figure 3). We anticipated that the frequency with which we would obtain PIG-A deficient MP2 cells would be lower than what we achieved in the human ES cells because MP2 iPS cells are female (XX) in origin and thus contain two copies of the PIG-A gene. It is unclear currently whether the inactivated X-chromosome inherited from the parental IMR90 fibroblasts remains inactivated or is activated in MP2 iPS cells after reprogramming. In the latter case, targeting both alleles of the PIG-A locus is required to generate a null phenotype. Following transfection of MP2 iPS cells with PHD donor DNA with or without PIG-A-specific ZFN expression vectors, we observed HygroR human ES/iPS-like colonies under both conditions. In four independent experiments, we found that on average 6.3±0.7% of HygroR human cells are CD59- when the PIG-A ZFNs were used (Figure 4C–D) compared with background levels (<0.05%) when PIG-A ZFNs were not used. The targeted integration of the PGK-HygroR DNA cassette could also be easily detected in the ZFN-treated cells (Figure 4E).
HR-mediated gene targeting of the endogenous PIG-A gene to generate GPI-AP deficient iPS cells
To better analyze and quantify HR-mediated PIG-A gene targeting, we also performed experiments in a well-characterized male (XY) human iPS cell line (Park et al., 2008). The hFib2-iPS5 cells were derived from dermal fibroblasts of an adult male and shown previously to have a normal karyotype and full pluripotency (Park et al., 2008). Similar to human ES cells in early-passages, the newly-established hFib2-iPS5 cells are delicate to expand and transfect. We optimized conditions for efficient transfection of these cells in the following ways. We initially followed the published method and passaged them as clumps after partial dissociation by collagenase digestion (Park et al., 2007). However, the method yielded poor transfection efficiencies using Nucleofection or other transfection techniques (data not shown). We found that trypsin or accutase treatment generated smaller clumps and single cells that allowed us to achieve higher transient transfection efficiencies (Figure S6), but the survival rate was much lower. We found that treating with a ROCK inhibitor Y27632 before trypsin-mediated cell dissociation significantly improved the colony-forming efficiency of early-passage hFib2-iPS5 (Figure S7), likely due to enhanced survival of single cells/small clumps as found for human ES cells (Watanabe et al., 2007). The addition of Y27632 during and after Nucleofection further helped the attachment/survival of single cells or smaller clumps (<30 cells) in culture after transfection. With these improvements, we transfected hFib2-iPS5 cells with the PHD HR donor with or without the PIG-A ZFN expression vectors as we did for human ES cells. Significantly more HygroR iPS colonies were obtained if Y27632 was present (Figure 5A). When the HygroR iPS cells were analyzed for the loss of GPI-APs such as CD59 (Figure 5B), up to 83% of the HygroR iPS cells were CD59-negative (average 45%±20%, Figure 5C) when ZFNs were used. In contrast, background levels (0.22±0.04%) of CD59-negative cells were found when ZFNs were absent, even though these iPS cells were also HygroR (likely due to the PHD random integration). PCR analysis of harvested HygroR cells confirmed targeted integration of the PHD DNA into the genome of hFib2-iPS5 cells when ZFNs were present (Figure 5D). Without ZFNs, the resulting HygroR iPS cells were neither CD59-negative nor showing the targeted integration (Figure 5D), suggesting that they indeed resulted from random integration of the PHD donor. Therefore, the PIG-A specific ZFNs confer a >200-fold increase in the efficiency of PIG-A gene targeting, equivalent to that observed in human ES cells (Figure 3D).
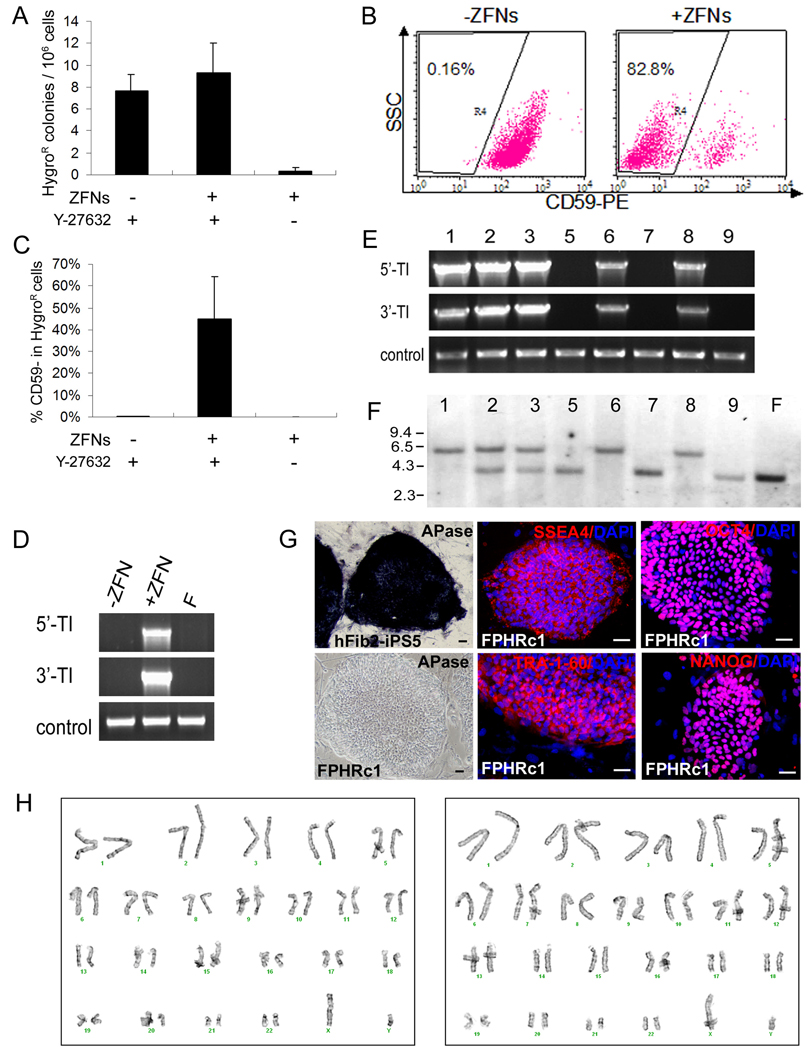
Figure 5. HR-mediated PIG-A gene-targeting in male human iPS cells, generating clonal PIG-A null iPS cells.
(A) PIG-A knockout was performed in hFib2-iPS5 cells derived from adult (male) fibroblasts, using the strategy shown in Figure 3A. A Nucleofection protocol similar to that used for human ES cells was carried out, except ROCK inhibitor (Y27632) was added. Mean and SEM of HygroR colonies per million of input cells from 3 independent experiments are presented.
(B) FACS analysis of PIG-A knockout in hFib2-iPS5 cells with or without ZFNs in the presence of ROCK inhibitor Y27632. TRA-1-60+ (human iPS) cells were gated and analyzed for the presence or absence of CD59. Representative dot plots are shown here.
(C) Comparisons of percentages of CD59-negative cells in HygroR hFib2-iPS5 cells after PIG-A gene targeting using various treatments as described above. N=3
(D) PCR analysis confirms that HygroR hFib2-iPS5 cells contain targeted integration after ZFN-mediated HR treatment. F: parental hFib2-iPS5 cells.
(E) PCR analysis shows targeted integration in five out of eight hFib2-iPS5 colonies we expanded and examined.
(F) Southern blot analysis detects the presence of the targeted (5.7-kb) and the wildtype (3.5-kb) PIG-A alleles (see a diagram in Figure 3A). The expected HR product (the targeted insertion) is found in the same five colonies that are positive by PCR detection (E). Three of them (c1, c6 and c8) appear to be homogeneous PIG-A knockout hFib2-iPS5 clones (FPHR) showing only the targeted allele (5.7-kb). Two (c2 and c3) contains both the targeted and the wildtype alleles. Because the hFib2-iPS5 cells (male, XY) have only one PIG-A allele in the X-chromosome, this is likely due to the fact that colony picked after hygromycin selection is not from a single clone and instead from mixed cells. The remaining 3 HygroR colonies result from off-target insertion showing only the wildtype PIG-A allele as the parental hFib2-iPS5 cells (F). The southern blot data are highly consistent with the pattern of GPI-AP expression in the 8 analyzed colonies (Figure S6B).
(G) Staining of the FPHRc1 (PIG-A knockout) iPS clone shows loss of APase (a GPI-AP) on the cell surface, compared to the parental hFib2-iPS5 cells, but maintains the expression of pluripotency markers (TRA-1-60, SSEA-4, OCT4 and NANOG). Scale bar = 50 µm. (H) FPHRc1 (left) and FPHRc6 (right) clones retain a normal karyotype (46,XY).
To molecularly characterize the targeting events, we randomly picked 9 HygroR iPS colonies from treated cells after co-transfection of PHD and ZFN-expression plasmids. We successfully expanded cells from 8 colonies and analyzed them by PCR to detect the targeted insertion (Figure 5E), by Southern blot to confirm HR at the PIG-A locus (Figure 5F), and by FACS analysis to confirm the loss of GPI-APs due to a PIG-A null mutation (Figure S6). Five out of the 8 hFib2-iPS5 PIG-A HR (FPHR) colonies (62.5%) we analyzed had the targeted insertion at the PIG-A locus (Figure 5E–F). Three of them (colonies c1, c6 and c8) contained only the targeted PIG-A allele and homogenously lacked GPI-AP expression (Figure 5F and Figure S6), suggesting a clonal origin for each picked colony. We found that the donor DNA is present only in the targeted locus in these 3 homogenous FPHR clones (Figure S8A). By a more sensitive PCR-mediated method, we did not detect the presence of the ZFN DNA in the genome of the 3 clones (Figure S8B). The homogenous iPS cells such as clone FPHR c1 lost the expression of GPI-APs, but retained undifferentiated iPS/ES cell morphology and the expression of pluripotency markers (Figure 5G). The differentiation potential of FPHRc1 iPS cells was similar to the PIG-A null human ES cells based on the in vitro assays described in Figure 2 (data not shown). Two randomly selected iPS clones (c1 and c6) retained a normal karyotype after HR-mediated gene targeting and expansion (Figure 5H). In summary, successful correction of the mutated EGFP gene and mutation of the endogenous PIG-A gene by HR show that ZFNs can stimulate gene targeting in human iPS cells by at least several hundred fold. This degree of stimulation is sufficient to enable the isolation of clonal populations of iPS cells with the desired genetic modification.
DISCUSSION
The importance of making a specific genetic modification in human PSCs has been widely recognized (Giudice and Trounson, 2008). The creation or correction of a specific mutation by HR-mediated gene targeting ensures permanent genetic alterations, a capability that will be of great utility in stem cell research and for potential applications of human PSCs in therapeutic uses. In 2003 and 2004, two landmark papers demonstrated the feasibility of gene targeting by HR in human ES cells (Urbach et al., 2004; Zwaka and Thomson, 2003). Four more papers (Costa et al., 2007; Davis et al., 2008; Di Domenico et al. 2008; Irion et al., 2007) in the past 2 years confirmed HR-mediated gene targeting in human ES cells using a plasmid-based delivery method. These studies collectively demonstrated that HR is feasible in human ES cells, but is technically challenging because of the low rate (<10−6) using standard targeting strategies. These technical limitations have limited the broad use of targeting in human PSCs and a general approach that increases the efficiency of targeting would increase the range of experimental and potentially therapeutic uses of these cells.
One strategy to increase the rate of gene targeting in human pluripotent stem cells is to use highly specific ZFNs. ZFN-induced DSBs at specific sequences have been used to stimulate targeting efficiencies in a wide range of cell types and organisms (Carroll, 2008; Cathomen and Joung 2008), and were recently shown to stimulate gene targeting in two human ES cell lines (Lombardo et al., 2007). The strength and weakness of this pioneering paper that used ZFNs (efficiently delivered by IDLV vectors) to achieve gene targeting in multiple cell types including human ES cells have been discussed comprehensively (Giudice and Trounson, 2008). Another limitation of this study is that the two human ES cell lines used, HUES1 and HUES3, are prone to have karyotypic abnormalities after prolonged culture (Cowan et al., 2004). Moreover, an analysis of whether ZFNs might induce unwanted chromosomal rearrangements and changes in growth parameters was absent (Lombardo et al., 2007). In the present study, we have demonstrated that: 1) ZFNs can be used to stimulate gene targeting in human PSCs, including iPS cells, without inducing karyotypic changes or changes in their pluripotent potential; 2) defined phenotypic changes can be rationally created by the use of ZFN-mediated gene targeting; 3) clonal cell lines with targeted genetic modifications can be derived and expanded; and 4) non-viral methods of inducing gene targeting using ZFNs can be used in human PSCs. These results are discussed in more detail below.
We used a virus-free DNA delivery system and successfully targeted two different genes in karyotypically normal human ES and iPS cells. Our studies were performed using novel ZFNs targeting a disease-related gene (PIG-A) as well as the previously described ZFNs targeting the EGFP gene (Pruett-Miller et al., 2008). Using optimized ratios of donor DNA and ZFN–expressing plasmid vectors as well as an improved method of DNA delivery, our protocol allowed us to achieve high efficiencies (stimulations of 200–2000 fold over the rate without ZFNs) of HR-mediated gene targeting in either creating a mutation in the endogenous PIG-A gene or correcting a chromosomally-integrated mutant EGFP gene in both human ES and iPS cells. Despite using homology arms (a total of 2 kb) that would be considered too short using standard targeting strategies (Di Domenico et al., 2008; Suzuki et al., 2008), we achieved a gene correction rate of 0.14–0.24% in human ES and iPS cells co-transfected with a pair of GFP-targeted ZFNs (Figure 1 and Figure 4). Similarly, the gene targeting efficiency at the endogenous PIG-A gene was also high when a pair of ZFNs was used together with a donor vector containing short (and thus suboptimal) homology arms (a total of 4 kb). This approach should be applicable to any gene of interest for which a pair of highly active and specific ZFNs is generated, and with other methods for more efficient DNA delivery (Lombardo et al., 2007; Suzuki et al., 2008).
We found that both human ES and iPS cells that had been targeted using ZFNs had no karyotypic abnormalities and did not exhibit any alteration in their pluripotent properties, even after extended expansion to accumulate sufficient cells required for comprehensive cellular and molecular analyses. In addition, phenotypic and biological defects of PIG-A-null human ES cells GNAR (created by ZFN-mediated NHEJ) and GPHR (created by ZFN-mediated HR) could be rescued by expression of a PIG-A transgene. Together, these data suggest that undesired genome alternations by a pair of high-quality and validated ZFNs are minimal. However, we note that acute ZFN-mediated cell toxicity could be observed when ZFNs used at a higher concentration with the Nucleofection (Figure 3B) or the Lipofectamine 2000 transfection methods (data not shown). Long-term and off-target effects of ZFNs remain to be fully characterized. Although major alteration of genome due to off-target DSBs usually results in cell death and elimination of affected cells, minor changes such as small deletions and additions could be carried on in the surviving population. It has been difficult to locate in the whole genome a small alternation induced by off-target DSBs. It is now possible to detect such abnormality by comparing parental and targeted cell lines using advanced technologies such as genome-wide, high-resolution SNP analysis and ideally whole-genome (deep) DNA sequencing. In addition to using ZFNs that possess high-quality zinc finger DNA-binding domains (as was done in this report), two other strategies to reduce off-target effects of ZFNs have been described. The first one uses modifications of the nuclease domain to minimize homo-dimerization of ZFNs which may lead to cleavage at off-target sites (Miller et al., 2007; Pruett-Miller et al., 2008; Szczepek et al., 2007). The second one uses small molecules to attenuate the extent and timing of ZFN expression (Pruett-Miller et al., 2009). It will be interesting to determine the degree to which these strategies minimize the off-target effects and toxicity of ZFNs when they are utilized with ZFNs in human PSCs. The ability to expand a few desirable ES or iPS clones unlimitedly alleviates the need to achieve perfection in every single cell using ZFN-mediated HR. Gene targeting at the level of human iPS and ES cells with unlimited self-renewal capacity also allows for the full characterization of genomic integrity before the use of the selected iPS and ES clones that have undergone gene targeting events. Taken together, the studies reported here demonstrate that high-quality ZFNs made by using the recently described OPEN platform can induce highly efficient gene targeting in human ES and iPS cells with minimal toxicity and karyotypic alterations. The OPEN method was developed by the Zinc Finger Consortium (http://www.zincfingers.org) and accounts for context-dependent DNA-binding activities of zinc fingers. OPEN ZFNs have been previously used to modify three human genes (Maeder et al., 2008), five zebrafish genes (Foley et al., 2009), and plant genes (Maeder et al., 2008; Townsend et al., 2009). The results of this report extend the number of endogenous human genes modified by OPEN ZFNs, and demonstrate that OPEN ZFNs can be used in more than just somatic human cells. Importantly, the reagents and protocols needed to make ZFNs by the OPEN method are publicly available and therefore any interested academic laboratory can use the approach to engineer customized ZFNs for their target gene of interest in a variety of cell types such as human PSCs.
Our report provides the first demonstration of human iPS cells modified by HR and demonstrates the feasibility and efficiency of an approach that can be used for creating or correcting specific mutations. Using the endogenous PIG-A gene and a chromosomally integrated GFP gene as targets, we demonstrated that our improved protocol of delivering gene targeting donor and ZFN-expressing plasmids works efficiently for human iPS as well as human ES cells. A major difference we observed is that early-passage iPS cells, especially those passaged by collagenase as cell clumps, survived poorly if they are harvested by trypsin as smaller cell clumps or single cells required by the Nucleofection method. However, treating iPS cells by adding a ROCK inhibitor Y27632 before trypsin-mediated cell dissociation and during the Nucleofection significantly improved the survival of transfected human iPS cells and the formation of HygroR colonies (Figure 5A). The overall HR efficiency measured by the PIG-A gene targeting in the newly established iPS cell lines is still lower than that in the well-adapted human ES lines such as the H1/GGFP line. This is due to at least in part the poor colony-forming efficiency of hFib2-iPS5 cells, which is >8–9 fold lower than that of H1/GGFP human ES cells (Figure S7). Despite of the current limitation in cell culture resulting in a lower number of total HygroR colonies per million of input hFib2-iPS5 cells, we are still able to generate a cell population containing >40% PIG-A deficient cells after drug-selection and to isolate true PIG-A knockout iPS clones from this XY iPS cell line. Together with the data using a different human iPS line (MP2), we showed for the first time that an endogenous gene can be efficiently targeted in human iPS cells, demonstrating the potential of using gene targeting in research and clinical applications of disease-specific human iPS cells. Possible future uses of gene targeting in human iPS cells include: 1) Increasing the efficiency of creating conditional alleles using the Cre-Lox system (Irion et al., 2007); 2) Correcting disease-causing mutations in X-linked recessive or autosomal recessive diseases; 3) Using gene targeting to create better models of human disease in human iPS cells; 4) Using serial gene targeting by HR to create precise bi-allelic modifications in autosomal genes or X-linked genes in female cells; and 5) Using HR to correct disease causing mutations in patient-specific iPS cells and using the genetically corrected cells to cure a disease as recently demonstrated in a mouse model (Hanna et al., 2007).
Previous studies have demonstrated even genes that are not transcriptionally active in undifferentiated human ES cells such as MIXL1, CCR5 and Fezf2 can be also targeted (Davis et al., 2008; Lombardo et al., 2007; Ruby and Zheng, 2009), as has been observed in numerous cases of mouse ES cell studies. Enhancement of ZFNs on gene targeting of CCR5 has been previously demonstrated in the two human ES cell lines used (Lombardo et al., 2007). Thus, with the aid of ZFNs that greatly enhance the HR efficiency in human ES and iPS cells, we are optimistic that we will also be able to achieve gene targeting in iPS cells that are not expressed in the undifferentiated state. The experimental systems we described here, i.e., the validated HR donor constructs and ZFNs for two genes (EGFP and PIG-A) with simply phenotypic readouts, will further help investigators to optimize gene targeting in human iPS and ES cells with or without a pair of ZFNs. Thus, our present study provides a solid foundation for researchers interested in efficient correcting or mutating a specific endogenous gene in both human iPS and ES cells.
EXPERIMENTAL PROCEDURES
Culture of human ES and iPS cells
Human H1 and H9 ES cell lines were obtained from WiCell Research Institute and cultured as previously described (Chen et al., 2008; Yu et al., 2008). GGFP derived from H1 ES cells with stable GFP expression by the lentiviral transduction (Dravid et al., 2005) was used in PIG-A gene targeting. MP2 iPS and hFib2-iPS5 cells were passaged by collagenase and cultured as previously described (Mali et al., 2008; Park et al., 2008).
Engineering of PIG-A specific ZFNs
ZFNs specific for a target site within the PIG-A coding sequence (Figure S10) were engineered using the OPEN method as previously described (Maeder et al., 2008). The top two of 3-zinc-finger arrays for each half-site that exhibited the highest levels of transcriptional activation in the bacterial two-hybrid system were chosen for use as ZFNs. To construct ZFN expression vectors for mammalian cells, XbaI-BamHI fragments harboring the engineered zinc finger arrays were cloned into XbaI and BamHI-digested pST1374 and fused to the endonuclease domain in the vector (Maeder et al., 2008). A pair of ZFNs targeting the GFP gene was previously described (Pruett-Miller et al., 2008). The target site and amino acid sequences of these two GFP-specific ZFNs are also shown in Figure S9. They are expressed from a similar expression vector. Requests for OPEN ZFN reagents and protocols should be directed to JKJ.
EGFP gene correction in 293T cells
First, EGFP was mutated with either a 35-bp insertion (Figure S2) or a 41-bp insertion containing PIG-A ZFN sites (Figure S3) and inserted into a lentiviral vector with IRES-PuroR cassette (Figure 1A). After transduction and puromycin selection, surviving cells containing EGIP* (termed 293T–EGIP* or 293T–EGIP*PIGA) were used for EGFP gene targeting. 2.5×105 293T–EGIP* or 293T–EGIP*PIGA cells were plated in one 12-well coated with 0.1% gelatin one day before lipofection. At the day of transfection, 1 µg of tGFP donor with various amounts of ZFNs expression vectors were transfected using standard Lipofectamine 2000 protocol (Invitrogen). GFP+ cells started to show up at 2.5-day after transfection and were monitored by FACS and microscopy thereafter.
EGFP gene correction in human ES and iPS cells
H9 human ES cells were transduced with EGIP* (35-bp insertion) lentivirus and selected with puromycin (1 µg/ml) and therefore, the starting population harbors EGIP* template. 5×106 H9-EGIP* cells grown on MEF feeder cells were harvested at ∼80% confluence with brief trypsin digestion and pipetting. Then the cells were resuspended in 100µl Amaxa mouse ES nucleofection buffer with various ratios of tGFP donor:ZFNs (e.g., 1 : 10=1 µg : 10 µg) and transfected using the Nucleofector II and preset programs such as A-23 (Amaxa). Cells were replated 1:1 onto MEFs afterwards and GFP+ cells began to emerge 4–5 days later. For MP2 iPS cells that are already puromycin-resistant, no puromyin selection was done and therefore only a portion of the starting population have the EGIP* template. 3×106 mixed MP2-EGIP* cells (without puromycin-mediated transgene selection) were used for EGFP gene correction with same Nucleofection settings as for H9-EGIP* cells.
PIG-A gene targeting in human ES and iPS cells
GGFP human ES cells, MP2 iPS or hFib2-iPS5 cells (3–5×106) were transfected with 4 µg of PIG-A targeting donor (PHD) and various amounts of the L1/R2 ZFN pair (Figure S10). Transfected cells were then replated onto W3R feeders (Cai et al., 2007) and selected with conditioned medium containing hygromycin (10 µg/ml) for 12 days beginning from day 4 post-nucleofection. In hFib2-iPS5 cell gene targeting experiments, 10 µM ROCK inhibitor Y27632 (Watanabe et al., 2007) was added for 1-hour prior to and 24-hours post Nucleofection. Hygromycin-resistant colonies were counted after selection and single colonies were manually picked and expanded.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Sarah Dowey and the Johns Hopkins Hospital Prenatal Cytogenetics Laboratory for professional karyotyping. We also thank 3 anonymous reviewers for suggestions that help us in revision. This work is funded in part by the Stem Cell Research Foundation (S2005-026), Johns Hopkins Institute for Cell Engineering and the NIH grant R01 HL073781 to LC. JZ is supported by a Maryland Stem Cell Research Postdoctoral Fellowship Grant. JKJ’s group is supported by the NIH (R01GM069906, R21RR024189 and R21HL091808), the Cystic Fibrosis Foundation and the MGH Pathology Service. MHP’s lab is supported by the State of Texas, NIH RO1 HL079295 and from a Career Development Award from the Burroughs-Wellcome Fund.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Cai L, Ye Z, Zhou BY, Mali P, Zhou C, Cheng L. Promoting human embryonic stem cell renewal or differentiation by modulating Wnt signal and culture conditions. Cell Res. 2007;17:62–72. doi: 10.1038/sj.cr.7310138. [DOI] [PubMed] [Google Scholar]
- Carroll D. Progress and prospects: Zinc-finger nucleases as gene therapy agents. Gene Ther. 2008;15:1463–1468. doi: 10.1038/gt.2008.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cathomen T, Joung KJ. Zinc-finger nucleases: the next generation emerges. Mol Ther. 2008;16:1200–1207. doi: 10.1038/mt.2008.114. [DOI] [PubMed] [Google Scholar]
- Chen G, Ye Z, Yu X, Zou J, Mali P, Brodsky RA, Cheng L. Trophoblast differentiation defect in human embryonic stem cells lacking PIG-A and GPI-anchored cell-surface proteins. Cell Stem Cell. 2008;2:345–355. doi: 10.1016/j.stem.2008.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa M, Dottori M, Sourris K, Jamshidi P, Hatzistavrou T, Davis R, Azzola L, Jackson S, Lim SM, Pera M, et al. A method for genetic modification of human embryonic stem cells using electroporation. Nat Protoc. 2007;2:792–796. doi: 10.1038/nprot.2007.105. [DOI] [PubMed] [Google Scholar]
- Cowan CA, Klimanskaya I, McMahon J, Atienza J, Witmyer J, Zucker JP, Wang S, Morton CC, McMahon AP, Powers D, et al. Derivation of embryonic stem-cell lines from human blastocysts. N Engl J Med. 2004;350:1353–1356. doi: 10.1056/NEJMsr040330. [DOI] [PubMed] [Google Scholar]
- Davis RP, Ng ES, Costa M, Mossman AK, Sourris K, Elefanty AG, Stanley EG. Targeting a GFP reporter gene to the MIXL1 locus of human embryonic stem cells identifies human primitive streak-like cells and enables isolation of primitive hematopoietic precursors. Blood. 2008;111:1876–1884. doi: 10.1182/blood-2007-06-093609. [DOI] [PubMed] [Google Scholar]
- Di Domenico AI, Christodoulou I, Pells SC, McWhir J, Thomson AJ. Sequential genetic modification of the hprt locus in human ESCs combining gene targeting and recombinase-mediated cassette exchange. Cloning Stem Cells. 2008;10:217–230. doi: 10.1089/clo.2008.0016. [DOI] [PubMed] [Google Scholar]
- Dravid G, Ye Z, Hammond H, Chen G, Pyle A, Donovan P, Yu X, Cheng L. Defining the role of Wnt/beta-catenin signaling in the survival, proliferation, and self-renewal of human embryonic stem cells. Stem Cells. 2005;23:1489–1501. doi: 10.1634/stemcells.2005-0034. [DOI] [PubMed] [Google Scholar]
- Durai S, Mani M, Kandavelou K, Wu J, Porteus MH, Chandrasegaran S. Zinc finger nucleases: custom-designed molecular scissors for genome engineering of plant and mammalian cells. Nucleic Acids Res. 2005;33:5978–5990. doi: 10.1093/nar/gki912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foley JE, Yeh JR, Maeder ML, Reyon D, Sander JD, Peterson RT, Joung JK. Rapid mutation of endogenous zebrafish genes using zinc finger nucleases made by Oligomerized Pool ENgineering (OPEN) PLoS ONE. 2009;4:e4348. doi: 10.1371/journal.pone.0004348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giudice A, Trounson A. Genetic modification of human embryonic stem cells for derivation of target cells. Cell Stem Cell. 2008;2:422–433. doi: 10.1016/j.stem.2008.04.003. [DOI] [PubMed] [Google Scholar]
- Hanna J, Wernig M, Markoulaki S, Sun CW, Meissner A, Cassady JP, Beard C, Brambrink T, Wu LC, Townes TM, et al. Treatment of sickle cell anemia mouse model with iPS cells generated from autologous skin. Science. 2007;318:1920–1923. doi: 10.1126/science.1152092. [DOI] [PubMed] [Google Scholar]
- Hohenstein KA, Pyle AD, Chern JY, Lock LF, Donovan PJ. Nucleofection mediates high-efficiency stable gene knockdown and transgene expression in human embryonic stem cells. Stem Cells. 2008;26:1436–1443. doi: 10.1634/stemcells.2007-0857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irion S, Luche H, Gadue P, Fehling HJ, Kennedy M, Keller G. Identification and targeting of the ROSA26 locus in human embryonic stem cells. Nat Biotechnol. 2007;25:1477–1482. doi: 10.1038/nbt1362. [DOI] [PubMed] [Google Scholar]
- Lombardo A, Genovese P, Beausejour CM, Colleoni S, Lee YL, Kim KA, Ando D, Urnov FD, Galli C, Gregory PD, et al. Gene editing in human stem cells using zinc finger nucleases and integrase-defective lentiviral vector delivery. Nat Biotechnol. 2007;25:1298–1306. doi: 10.1038/nbt1353. [DOI] [PubMed] [Google Scholar]
- Lowry WE, Richter L, Yachechko R, Pyle AD, Tchieu J, Sridharan R, Clark AT, Plath K. Generation of human induced pluripotent stem cells from dermal fibroblasts. Proc Natl Acad Sci U S A. 2008;105:2883–2888. doi: 10.1073/pnas.0711983105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeder ML, Thibodeau-Beganny S, Osiak A, Wright DA, Anthony RM, Eichtinger M, Jiang T, Foley JE, Winfrey RJ, Townsend JA, et al. Rapid “open-source” engineering of customized zinc-finger nucleases for highly efficient gene modification. Mol Cell. 2008;31:294–301. doi: 10.1016/j.molcel.2008.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mali P, Ye Z, Hammond HH, Yu X, Lin J, Chen G, Zou J, Cheng L. Improved efficiency and pace of generating induced pluripotent stem cells from human adult and fetal fibroblasts. Stem Cells. 2008;26:1998–2005. doi: 10.1634/stemcells.2008-0346. [DOI] [PubMed] [Google Scholar]
- Miller JC, Holmes MC, Wang J, Guschin DY, Lee YL, Rupniewski I, Beausejour CM, Waite AJ, Wang NS, Kim KA, et al. An improved zinc-finger nuclease architecture for highly specific genome editing. Nat Biotechnol. 2007;25:778–785. doi: 10.1038/nbt1319. [DOI] [PubMed] [Google Scholar]
- Nightingale SJ, Hollis RP, Pepper KA, Petersen D, Yu XJ, Yang C, Bahner I, Kohn DB. Transient gene expression by nonintegrating lentiviral vectors. Mol Ther. 2006;13:1121–1132. doi: 10.1016/j.ymthe.2006.01.008. [DOI] [PubMed] [Google Scholar]
- Park IH, Zhao R, West JA, Yabuuchi A, Huo H, Ince TA, Lerou PH, Lensch MW, Daley GQ. Reprogramming of human somatic cells to pluripotency with defined factors. Nature. 2008;451:141–146. doi: 10.1038/nature06534. [DOI] [PubMed] [Google Scholar]
- Porteus MH. Mammalian gene targeting with designed zinc finger nucleases. Mol Ther. 2006;13:438–446. doi: 10.1016/j.ymthe.2005.08.003. [DOI] [PubMed] [Google Scholar]
- Porteus MH, Baltimore D. Chimeric nucleases stimulate gene targeting in human cells. Science. 2003;300:763. doi: 10.1126/science.1078395. [DOI] [PubMed] [Google Scholar]
- Porteus MH, Carroll D. Gene targeting using zinc finger nucleases. Nat Biotechnol. 2005;23:967–973. doi: 10.1038/nbt1125. [DOI] [PubMed] [Google Scholar]
- Pruett-Miller SM, Connelly JP, Maeder ML, Joung JK, Porteus MH. Comparison of zinc finger nucleases for use in gene targeting in mammalian cells. Mol Ther. 2008;16:707–717. doi: 10.1038/mt.2008.20. [DOI] [PubMed] [Google Scholar]
- Pruett-Miller SM, Reading DW, Porter SN, Porteus MH. Attenuation of zinc finger nuclease toxicity by small-molecule regulation of protein levels. PLoS Genet. 2009;5:e1000376. doi: 10.1371/journal.pgen.1000376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reubinoff BE, Pera MF, Fong CY, Trounson A, Bongso A. Embryonic stem cell lines from human blastocysts: somatic differentiation in vitro. Nat Biotechnol. 2000;18:399–404. doi: 10.1038/74447. [DOI] [PubMed] [Google Scholar]
- Ruby KM, Zheng B. Gene Targeting in an HUES line of human embryonic stem cells via electroporation. Stem Cells. 2009 doi: 10.1002/stem.73. Published online March 29 2009; DOI: 10.1002/stem.73. [DOI] [PubMed] [Google Scholar]
- Suzuki K, Mitsui K, Aizawa E, Hasegawa K, Kawase E, Yamagishi T, Shimizu Y, Suemori H, Nakatsuji N, Mitani K. Highly efficient transient gene expression and gene targeting in primate embryonic stem cells with helper-dependent adenoviral vectors. Proc Natl Acad Sci U S A. 2008;105:13781–13786. doi: 10.1073/pnas.0806976105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szczepek M, Brondani V, Buchel J, Serrano L, Segal DJ, Cathomen T. Structure-based redesign of the dimerization interface reduces the toxicity of zinc-finger nucleases. Nat Biotechnol. 2007;25:786–793. doi: 10.1038/nbt1317. [DOI] [PubMed] [Google Scholar]
- Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, Yamanaka S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- Thomson JA, Itskovitz-Eldor J, Shapiro SS, Waknitz MA, Swiergiel JJ, Marshall VS, Jones JM. Embryonic stem cell lines derived from human blastocysts. Science. 1998;282:1145–1147. doi: 10.1126/science.282.5391.1145. [DOI] [PubMed] [Google Scholar]
- Townsend JA, Wright DA, Winfrey RJ, Fu F, Maeder ML, Joung JK, Voytas DF. High Frequency Modification of Plant Genes Using Engineered Zinc Finger Nucleases. Nature. 2009 doi: 10.1038/nature07845. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urbach A, Schuldiner M, Benvenisty N. Modeling for Lesch-Nyhan disease by gene targeting in human embryonic stem cells. Stem Cells. 2004;22:635–641. doi: 10.1634/stemcells.22-4-635. [DOI] [PubMed] [Google Scholar]
- Urnov FD, Miller JC, Lee YL, Beausejour CM, Rock JM, Augustus S, Jamieson AC, Porteus MH, Gregory PD, Holmes MC. Highly efficient endogenous human gene correction using designed zinc-finger nucleases. Nature. 2005;435:646–651. doi: 10.1038/nature03556. [DOI] [PubMed] [Google Scholar]
- Watanabe K, Ueno M, Kamiya D, Nishiyama A, Matsumura M, Wataya T, Takahashi JB, Nishikawa S, Muguruma K, Sasai Y. A ROCK inhibitor permits survival of dissociated human embryonic stem cells. Nat Biotechnol. 2007;25:681–686. doi: 10.1038/nbt1310. [DOI] [PubMed] [Google Scholar]
- Yang TT, Cheng L, Kain SR. Optimized codon usage and chromophore mutations provide enhanced sensitivity with the green fluorescent protein. Nucleic Acids Res. 1996;24:4592–4593. doi: 10.1093/nar/24.22.4592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, Vodyanik MA, Smuga-Otto K, Antosiewicz-Bourget J, Frane JL, Tian S, Nie J, Jonsdottir GA, Ruotti V, Stewart R, et al. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318:1917–1920. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- Yu X, Zou J, Ye Z, Hammond H, Chen G, Tokunaga A, Mali P, Li YM, Civin C, Gaiano N, et al. Notch signaling activation in human embryonic stem cells is required for embryonic, but not trophoblastic, lineage commitment. Cell Stem Cell. 2008;2:461–471. doi: 10.1016/j.stem.2008.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwaka TP, Thomson JA. Homologous recombination in human embryonic stem cells. Nat Biotechnol. 2003;21:319–321. doi: 10.1038/nbt788. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.