Abstract
We report the structures of three intermediates in the O2 activation and insertion reactions of an extradiol ring-cleaving dioxygenase. A crystal of Fe2+-containing homoprotocatechuate 2,3-dioxygenase was soaked in the slow substrate 4-nitrocatechol in a low O2 atmosphere. The X-ray crystal structure shows that three different intermediates reside in different subunits of a single homotetrameric enzyme molecule. One of these is the key substrate-alkylperoxo-Fe2+ intermediate, which has been predicted but not structurally characterized in an oxygenase. The intermediates define the major chemical steps of the dioxygenase mechanism and point to a general mechanistic strategy for the diverse 2-His-1-Carboxylate enzyme family.
Aerobic life is possible because O2 must be activated before it will react rapidly with most biological molecules, thereby preventing indiscriminate oxidation. Oxygenase enzymes have evolved numerous chemical strategies to selectively effect this activation so that the inherent oxidizing power of O2 can be used productively (1, 2). For example, in the non-heme Fe2+ containing extradiol dioxygenase enzymes, which catalyze the pivotal ring-opening step in the aerobic, bacterial degradation pathways for many natural and manmade aromatic compounds (3), substrate binding facilitates oxygen binding and activation. The substrates for these enzymes are analogs of catechol (1,2-dihydroxybenzene), and they bind directly to the iron as a chelate via their adjacent hydroxyl functions (4, 5). The substrate-binding reaction causes a change in the ligation state and electronic properties of the iron so that O2 can bind in an adjacent iron coordination site (4, 6, 7). O2 activation and ring opening then occur without introduction of external reducing equivalents.
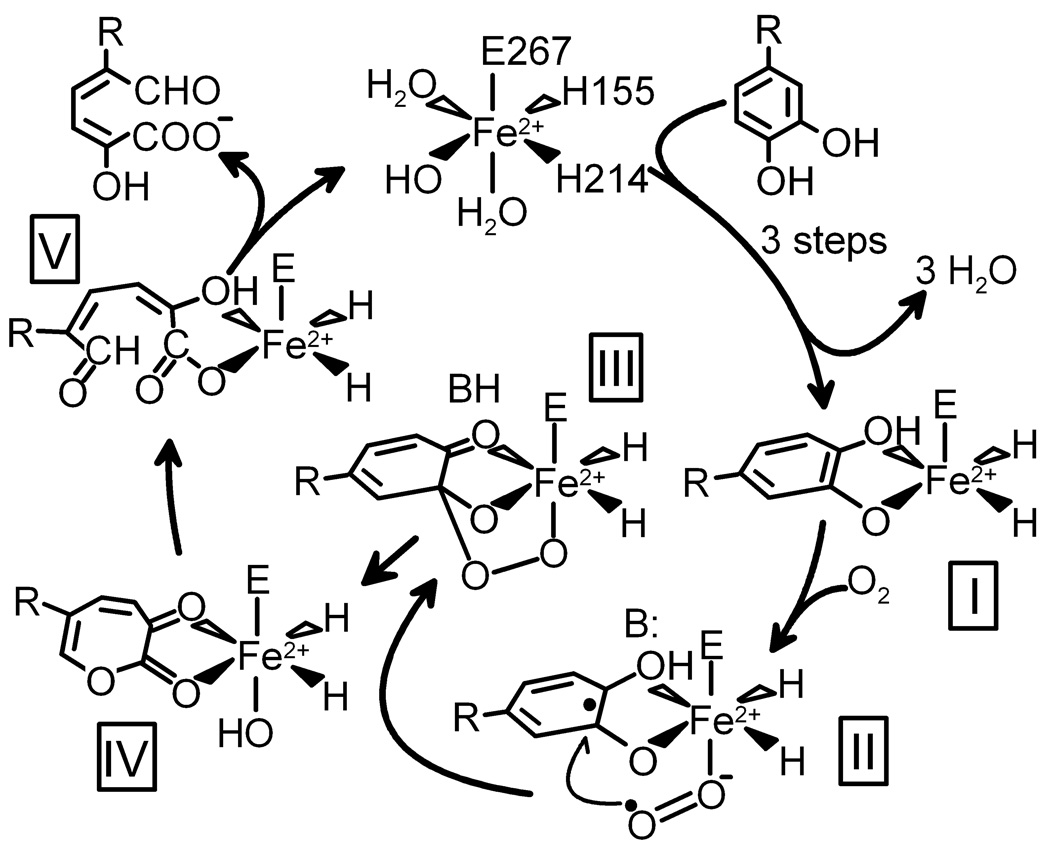
Based on spectroscopic studies of extradiol dioxygenases, we have proposed that the simultaneous binding of substrate and O2 to the iron allows transfer of electron density such that the substrate gains cation radical character, while the O2 becomes a nascent superoxide (Fig. 1, II) (4, 8). Recombination of the incipient radicals would yield an alkylperoxo intermediate (III). A concerted Criegee rearrangement of this intermediate would result in O-O bond cleavage and insertion of one oxygen atom into the aromatic ring to form a lactone intermediate (IV) which would proceed on to the ring-open product (V).
Fig. 1.
Proposed reaction mechanism for extradiol ring-cleaving dioxygenases. R = −CH2COOH or −NO2. 4NC binds as the dianion.
The X-ray crystal structures of several extradiol dioxygenases have shown that, as predicted, the substrate does chelate the iron and this creates a vacant site in the iron coordination (5, 9, 10). The structures have also allowed the application of computationally-based theoretical techniques, leading to several new proposals for the mechanism of the O-O bond cleavage and ring opening step(s) (11, 12). However, all of these mechanisms posit the initial formation of an alkylperoxo intermediate, a species that has never been structurally characterized. Here, we have trapped this lynchpin species in a single crystal and structurally characterized it. Remarkably, the O2 adduct precursor and the product complex successor to the alkylperoxo intermediate are found to be simultaneously present in different subunits of the enzyme in the same asymmetric unit, allowing these species to be structurally characterized as well.
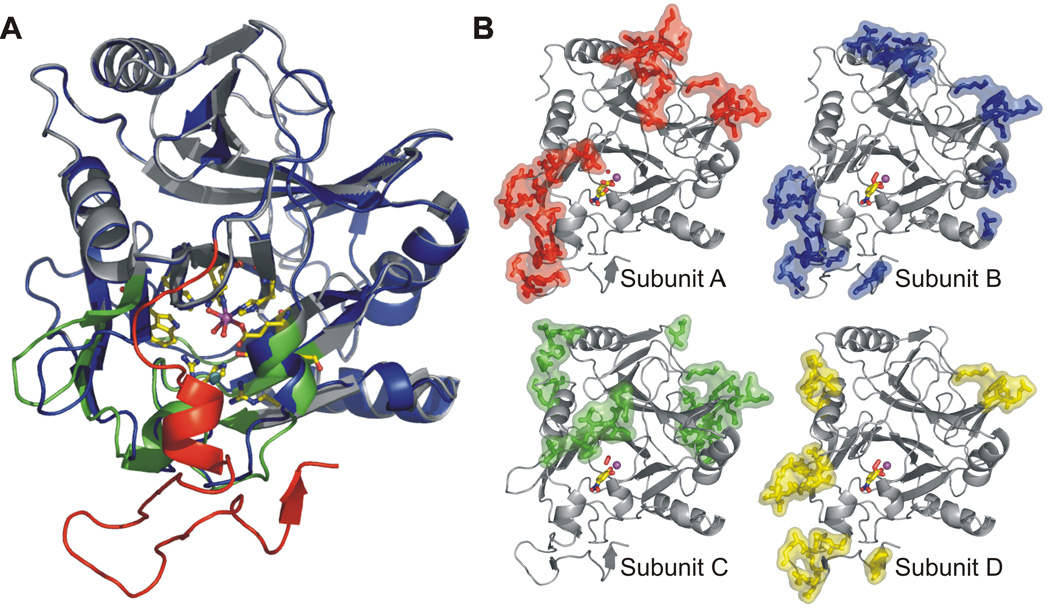
Homoprotocatechuate 2,3-dioxygenase (2,3-HPCD) from Brevibacterium fuscum catalyzes the proximal extradiol ring cleavage of homoprotocatechuate (3,4-dihydroxyphenylacetate, HPCA) to form ring-opened 5-carboxymethyl-2-hydroxymuconic semialdehyde (13). The original structural study of 2,3-HPCD and its complex with HPCA employed a C-terminal deletion variant (14) to facilitate crystallization (10). The full-length enzyme was successfully crystallized only once, but the resulting low resolution structure revealed that the C-terminal forms a “lid” over the substrate-binding pocket (10). In the current study, conditions have been defined to allow reproducible crystallization of the full-length protein in a new space group (P21212) (15). The structures of the new form (1.9 Å resolution) show numerous new intermolecular contacts (Fig. 2) that appear to influence the rates of the catalytic steps so that intermediates are stabilized, as described below.
Fig. 2.
A) Overlay of monomeric subunit structures in the presence and absence of C-terminus. Full-length enzyme (PDB code 2IG9) is shown in blue, and the C-terminal residues 323 – 362 are shown in red. C-terminus truncated enzyme (PDB code 1F1X, (10)) is shown in gray except for residues in the substrate-binding cleft that exhibit slight structural alterations in the absence of C-terminus, which are shown in green. Active site residues are shown as sticks and mononuclear Fe center, solvent and Cl− are shown as purple, red and cyan spheres, respectively. B) Crystal packing (intermolecular) contacts for subunits of the full-length homotetrameric enzyme 2,3-HPCD in P21212 space group (PDB code 2IGA). Residues involved in contacts (<5Å) with symmetry-related molecules are shown as colored sticks and surfaces.
Transient kinetic studies of 2,3-HPCD in solution have provided inroads into the detection of discrete intermediates in the catalytic cycle (16, 17). The use of an alternative substrate, 4-nitrocatechol (4NC), slows the oxygen activation and insertion steps of the cycle (Fig. 1, I–V) and provides a chromophore that is sensitive to the hydroxyl substituent ionization state, protein environment, and ring cleavage. This approach allowed 6 reaction cycle steps to be detected prior to rate limiting product release. Unfortunately, despite the decreased rates, the anticipated 4NC-Fe2+-O2 and alkylperoxo intermediates were not sufficiently long-lived to directly observe. It has been reported that steps in the reaction cycle of an enzyme in a crystal often occur much more slowly than in solution (18). Accordingly, we show here that when the turnover of 4NC is initiated in a crystal of 2,3-HPCD, the key oxygen activation and reaction intermediates are stabilized (Fig. 3A – C).
Fig. 3.
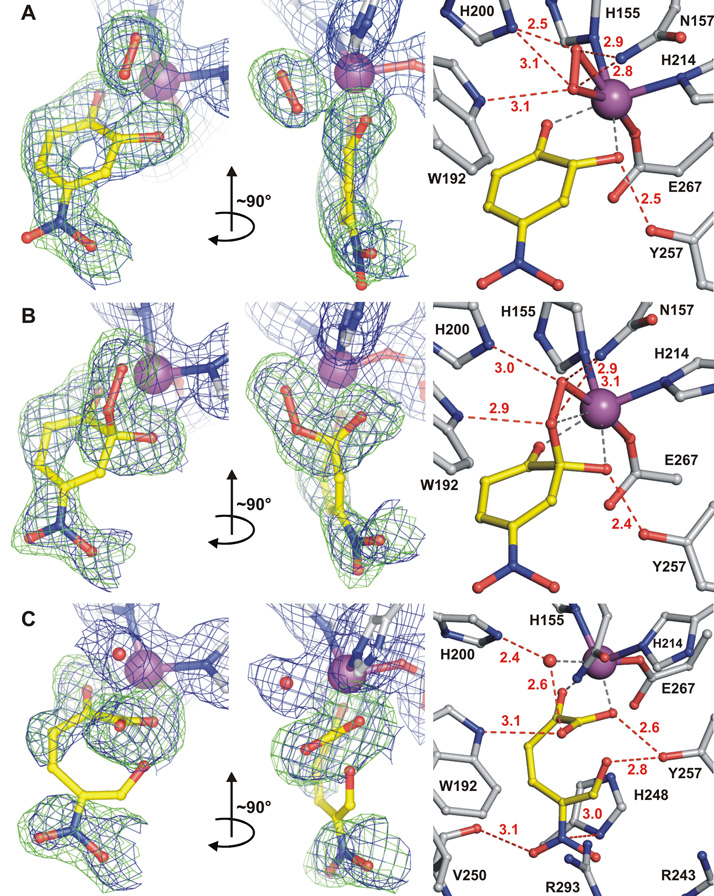
Structures of reactive species trapped in the active sites of 2,3-HPCD during in crystallo reaction with 4NC and O2 (PDB code 2IGA). A) Ternary complex (subunit C) of enzyme with 4NC semiquinone and side-on bound dioxygen species. B) Alkylperoxo intermediate complex (subunit D, similar structure observed in subunit B (not shown)). C) Complex with open-ring product (subunit A). The blue 2Fobs–Fcalc maps are contoured at 1.0 σ (A, B) and 0.75 σ (C). The green Fobs–Fcalc ligand-omit maps were computed by removing ligands from the model and are contoured at +4 σ (A), 3.75 σ (B) and 3 σ (C). Atom color code: gray, carbon (enzyme residues); yellow, carbon (ligands); blue, nitrogen; red, oxygen; purple, iron. Red dashed lines show hydrogen-bonds (Å). Gray dashed lines indicate bonds or potential bonds to iron; distances are given in the text or in Table S3. Additional views of the complexes are given in Fig. S1.
In subunit C (Fig. 3A), 4NC chelates the iron (O24NC−Fe = 2.2 Å, O14NC−Fe = 2.2 Å) (19) in sites opposite Fe2+ ligands His155 and His214, respectively, and a diatomic molecule (see Fig. S4), assigned as oxygen (20), occupies the adjacent site opposite ligand Glu267. The symmetrical, elliptical shape of the electron density of the diatomic molecule and the long distance between C24NC and O2O2 (2.4 Å) suggests that a bond between the 4NC and the bound oxygen has not yet formed. Two aspects of this structure are particularly noteworthy. First, the putative O2 ligand is bound side-on (O1O2−Fe = 2.5 Å and O2O2−Fe = 2.4 Å) rather than the more commonly encountered end-on. Second, the plane of the 4NC ring is puckered rather than planar, which causes the C24NC hydroxyl substituent to move far out of plane, thereby strengthening the hydrogen bond with Tyr257. No deviation from a planar aromatic ring is observed for the anaerobic complex of 2,3-HPCD with substrate (10). The deviation at C24NC from sp2 toward sp3 hybridization is consistent with electron transfer from 4NC to O2 via the iron to give the ring cation-radical character and the O2 superoxide character (see Fig. 1, II). Consequently, the superoxo state is assumed and an O-O distance of 1.34Å was used during refinement (21).
The structures of subunits B and D (Fig. 3B) show that a bond has formed between C24NC and O2O2 (1.4 Å) to generate the alkylperoxo intermediate (see Fig. S5). The ring of 4NC becomes more extensively puckered and C24NC assumes a full sp3 hybridization while both O24NC and O14NC remain coordinated to the iron (2.3 and 2.2 Å, respectively). During refinement of the alkylperoxo ligand, C24NC was restrained to tetrahedral geometry (109.5°) and the O-O bond distance refined to 1.5 Å (22). The bond between O1O2 and the iron is retained (2.1 Å), but O2O2 moves out of bonding distance (2.8 Å) in position to be transferred to the 4NC.
The structure in subunit A (Fig. 3C) shows that the aromatic ring of 4NC has been broken between the C24NC and C34NC carbons to form the ring-open product (see Fig 1, V). The product remains bound to the iron via the carboxylate derived from the C24NC carbon and the hydroxyl from the former C14NC carbon. The newly formed carboxylate rotates to the optimal bonding orientation, thereby providing direct evidence for ring fissure. The open-ring structure allows rotation of the bonds that are not fixed by the iron ligand structure. As a result, the electron density for two of the former ring carbons is not observed (see Fig. S2) (23). The aldehyde and nitro substituents of the product interact strongly with the normal anion-binding pocket created by His248, Arg243, and Arg293 allowing electron density to be observed. However, the data do not allow the relative orientations of the aldehyde and nitro groups to be determined with certainty (Fig. S3). The orientation shown in Fig. 3C causes the fewest bad contacts. Solvent occupies the iron ligand position formerly occupied by oxygen, as it does in the structure of the resting enzyme after release of the product.
While the simultaneous presence of three different reaction cycle intermediates in a single enzyme with four independent active sites is not surprising, it was not anticipated that they could be independently observed in an X-ray crystal structure. If they occurred randomly in all of the active sites present in the crystal, only the (uninterpretable) average of all of the structures would be observed. This implies that, in the crystal, the reaction proceeds rapidly only as far as a specific intermediate in the active site located in a given region of the asymmetric unit. Similar observations have been made in a few other enzyme crystals (24). It is likely that the different reactivities of the 2,3-HPCD subunits can be attributed to crystal packing forces which cause minor structural differences (Fig. 2B). This is especially true at the interface between symmetry-related elements involving the residues surrounding the C-terminal lid that covers the active site cavity and alters the interaction of active site residues with the substrate (Fig. 2A) (for further information see Table S2).
The presence of product bound in the active site of one subunit shows that the enzyme in the crystal form used is active (see supporting online material). This suggests that the superoxo and alkylperoxo intermediates observed in the same crystal are relevant to the catalytic process. As such, they offer direct evidence for some of the most important aspects of the mechanistic hypothesis proposed previously (4, 8, 17). Specifically they show that: i) the aromatic substrate and oxygen are bound to the iron simultaneously in adjacent sites, ii) electron transfer occurs between these two substrates, giving rise to the puckered substrate ring, iii) the attack on oxidized substrate occurs prior to O-O bond cleavage, apparently by the Fe2+-superoxo species, iv) the next cycle intermediate is not a dioxetane as proposed in some early studies (25), and v) the alkylperoxo intermediate that does form is located between the iron and the OH-bearing, C2 carbon of the aromatic substrate.
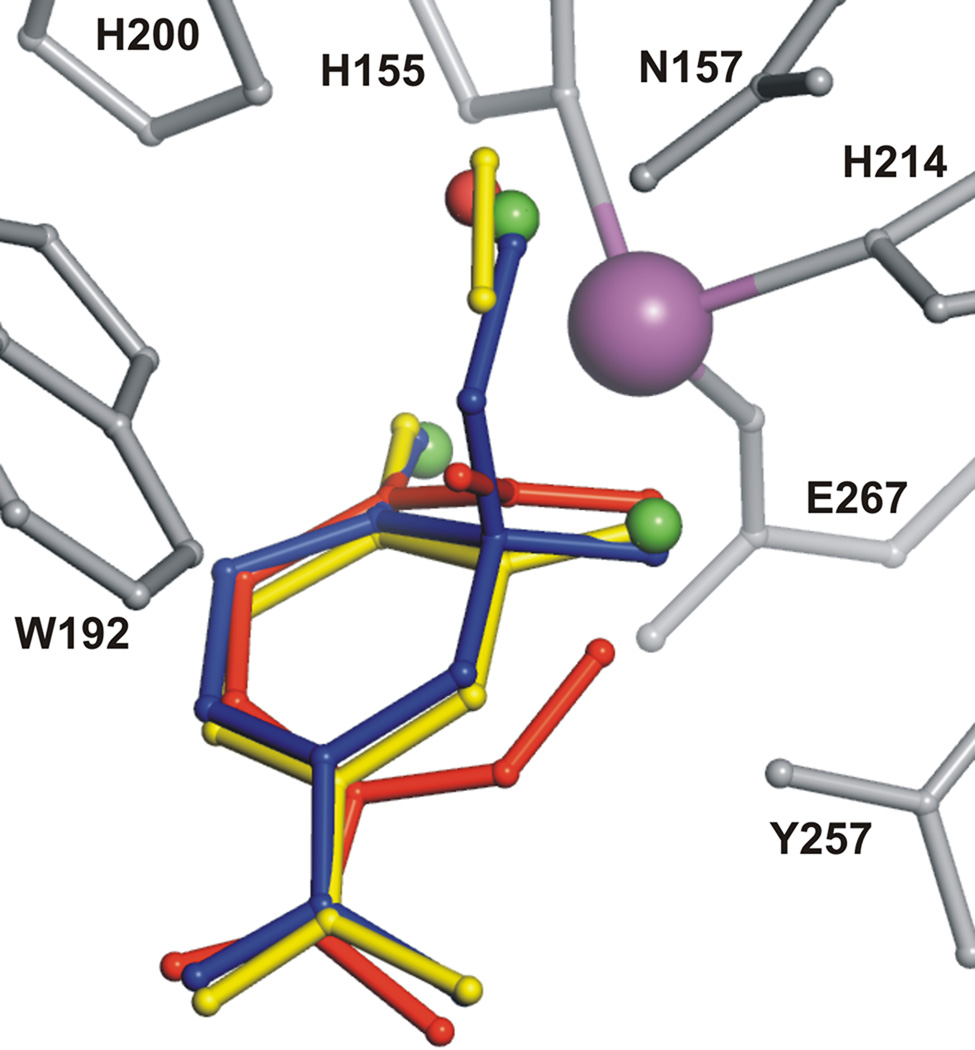
The observation of a side-on rather than an end-on oxy complex was not anticipated because we have shown in the past that the O2 surrogate, NO, which can only form an end-on complex with iron, binds very strongly to the enzyme-substrate complex (6). Thus, there is sufficient room near the diatomic molecule binding site for an end-on O2 complex. Such a complex could lead to, or evolve from, the side-on complex observed. Hydrogen bonding interactions between the bound O2 and the second sphere residues probably play important roles in stabilizing the unusual side-on binding orientation. In particular, His200 and Asn157 interact with dioxygen-derived atoms in both superoxo and alkylperoxo complexes (Fig 3A, B). The side-on binding orientation positions the oxygen-adduct perfectly to react with the substrate at the correct ring carbon (Fig. 4).
Fig. 4.
Structure superposition of Fe-bound ligands observed at different steps of catalytic reaction: solvent in resting state (green), substrate and dioxygen (yellow, Fig. 3A), alkylperoxo intermediate (blue, Fig. 3B), ring-open product and solvent (red, Fig. 3C). Active site residues and Fe are shown in gray and purple, respectively.
It should be noted that the only other structurally characterized O2-adduct among the mononuclear non-heme iron family also exhibits side-on binding (26). This species forms in the Rieske-type cis-diol forming naphthalene 1,2-dioxygenase (NDO) which differs in many structural and mechanistic respects from the ring-cleaving dioxygenase studied here (27, 28). As for 2,3-HPCD, the side-on O2 binding in NDO may be enforced by steric interactions, in that case by close binding of the substrate and an Asn residue. In NDO, two electrons (one from the Fe2+ and the other from a Rieske FeS cluster) are transferred to the O2 in forming the side-on complex, resulting in a peroxo-Fe3+ species. The NDO complex O-O bond (1.47 Å) is as long or longer than that assumed here for the oxygen adduct, and it exhibits shorter bonds to Fe (1.99 and 1.74 Å), consistent with a greater degree of electron transfer to the oxygen and the consequent higher oxidation state of the iron.
The current studies provide experimental support for many aspects of recent computational investigations of the mechanism, but they also highlight a few inconsistencies. The side-on oxy complex was not predicted in the computational studies (11, 12), which were influenced by the structure of the end-on nitrosyl complex (7). The alkylperoxo intermediate resembles the computed structures in bond lengths and orientations. In particular, it assumes the quasi-axial position relative to the substrate ring suggested by Bugg and Lin to direct extradiol rather than intradiol ring cleavage (29). As is pointed out in computational studies, hydrogen-bonding interactions and proton transfer to O1O2 in the alkylperoxo intermediate are readily supported by the active site structure (Fig. 3A, B) (see supporting online material for a discussion of the particularly strong hydrogen bonds). However, the current structural study shows that the proton donor is likely to be the ideally positioned His200 rather than the suggested water (not present) (12) or Glu267 (located on the other side of the iron) (11). This conclusion is in accord with kinetic studies of 2,3-HPCD His200 mutants that revealed a dramatic decrease in the rate constant for O-O bond cleavage in the absence of an in situ acid catalyst (17).
The extradiol dioxygenase is the archetypal member of the exceptionally broad class of Fe2+ containing enzymes termed the 2-His-1-Carboxylate facial triad family that catalyzes reactions ranging from aromatic biodegradation to the biosynthesis of antibiotics and the building blocks for mammalian connective tissue (9, 30). Many of these enzymes activate O2 during their catalytic cycle, and bind substrates simultaneously to multiple Fe2+ ligand sites as described in the context of extradiol dioxygenases (4). A general mechanistic scenario for this enzyme class emerges from the structural studies presented here in which the dual role of the iron is to properly juxtapose the organic substrate and O2 and to facilitate electron transfer from the former to the latter. After this common initial oxygen activation step, a myriad of reactions can be envisioned.
Supplementary Material
Supporting Online Material
www.sciencemag.org/cgi/content/full/...........
Materials and Methods
Fig. S1–S5
Tables S1–S3
References
References
- 1.Wallar BJ, Lipscomb JD. Chem. Rev. 1996;96:2625–2657. doi: 10.1021/cr9500489. [DOI] [PubMed] [Google Scholar]
- 2.Costas M, Mehn MP, Jensen MP, Que L., Jr. Chem. Rev. 2004;104:939–986. doi: 10.1021/cr020628n. [DOI] [PubMed] [Google Scholar]
- 3.Lipscomb JD, Orville AM. Metal Ions Biol. Syst. 1992;28:243–298. [Google Scholar]
- 4.Arciero DM, Lipscomb JD. J. Biol. Chem. 1986;261:2170–2178. [PubMed] [Google Scholar]
- 5.Senda T, et al. J. Mol. Biol. 1996;255:735–752. doi: 10.1006/jmbi.1996.0060. [DOI] [PubMed] [Google Scholar]
- 6.Arciero DM, Orville AM, Lipscomb JD. J. Biol. Chem. 1985;260:14035–14044. [PubMed] [Google Scholar]
- 7.Sato N, et al. J. Mol. Biol. 2002;321:621–636. doi: 10.1016/s0022-2836(02)00673-3. [DOI] [PubMed] [Google Scholar]
- 8.Shu L, et al. Biochemistry. 1995;34:6649–6659. doi: 10.1021/bi00020a010. [DOI] [PubMed] [Google Scholar]
- 9.Han S, Eltis LD, Timmis KN, Muchmore SW, Bolin JT. Science. 1995;270:976–980. doi: 10.1126/science.270.5238.976. [DOI] [PubMed] [Google Scholar]
- 10.Vetting MW, Wackett LP, Que L, Jr., Lipscomb JD, Ohlendorf DH. J. Bacteriol. 2004;186:1945–1958. doi: 10.1128/JB.186.7.1945-1958.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Deeth RJ, Bugg TDH. J. Biol. Inorg. Chem. 2003;8:409–418. doi: 10.1007/s00775-002-0430-7. [DOI] [PubMed] [Google Scholar]
- 12.Siegbahn PEM, Haeffner F. J. Am. Chem. Soc. 2004;126:8919–8932. doi: 10.1021/ja0493805. [DOI] [PubMed] [Google Scholar]
- 13.Miller MA, Lipscomb JD. J. Biol. Chem. 1996;271:5524–5535. doi: 10.1074/jbc.271.10.5524. [DOI] [PubMed] [Google Scholar]
- 14.Visible residues out of a total of 365: C-terminal-deleted enzyme, 4–322 (PDB 1F1X); original full-length enzyme, 4–359 (PDB 1Q0O); Form reported here, 4–362 (PDB 2IG9, 2IGA).
- 15.Materials and methods and structure statistics (Table S1) are available as supporting material on Science online.
- 16.Groce SL, Miller-Rodeberg MA, Lipscomb JD. Biochemistry. 2004;43:15141–15153. doi: 10.1021/bi048690x. [DOI] [PubMed] [Google Scholar]
- 17.Groce SL, Lipscomb JD. Biochemistry. 2005;44:7175–7188. doi: 10.1021/bi050180v. [DOI] [PubMed] [Google Scholar]
- 18.Hajdu J, et al. Nat. Struct. Biol. 2000;7:1006–1012. doi: 10.1038/80911. [DOI] [PubMed] [Google Scholar]
- 19.Nomenclature: The superscript denotes the molecular origin of the atom. 4NC hydroxyl substituents are termed O14NC and O24NC based on the carbon with which they are associated. O2 atoms are termed O1O2 and O2O2 where O1 is that which remains bound to the iron in the alkylperoxo intermediate.
- 20.No diatomic molecule that can bind to Fe2+ other than O2 is present in the crystallization medium. Attempts to fit the electron density without a diatomic molecule, with only a single atom, or with two solvents were unsuccessful (Fig. S4).
- 21.At the current resolution (1.95 Å), diatomic bond lengths between 1.2 and 1.4 Å could be used to fit the electron density. Thus, the state of reduction of the bound O2 could not be determined from the density alone.
- 22.Although no appropriate model complex for an Fe2+ alkylperoxo species has been reported, a Mn(II) chelate complex exhibits an O-O bond length of 1.411 Å, similar to that of free H2O2. Komatsuzaki H, et al. Inorg. Chem. 1998;37:6554–6555. doi: 10.1021/ic980509j.
- 23.The electron density is low for the three carbons closest to the nitro substituent supporting the presence of the more flexible, ring-open product in the active site. Product from 4NC was soaked into a crystal of 2,3-HPCD, and it assumed the same structure (Fig S2).
- 24.Hogg M, Wallace SS, Doublie S. EMBO J. 2004;23:1483–1493. doi: 10.1038/sj.emboj.7600150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hayaishi O, Katagiri H, Rothberg S. J. Am. Chem. Soc. 1955;77:5450–5451. [Google Scholar]
- 26.Karlsson A, et al. Science. 2003;299:1039–1042. doi: 10.1126/science.1078020. [DOI] [PubMed] [Google Scholar]
- 27.Wolfe MD, Parales JV, Gibson DT, Lipscomb JD. J. Biol. Chem. 2001;276:1945–1953. doi: 10.1074/jbc.M007795200. [DOI] [PubMed] [Google Scholar]
- 28.Carredano E, et al. J. Mol. Biol. 2000;296:701–712. doi: 10.1006/jmbi.1999.3462. [DOI] [PubMed] [Google Scholar]
- 29.Bugg TDH, Lin G. Chem. Commun. 2001:941–952. [Google Scholar]
- 30.Hegg EL, Que L., Jr. Eur. J. Biochem. 1997;250:625–629. doi: 10.1111/j.1432-1033.1997.t01-1-00625.x. [DOI] [PubMed] [Google Scholar]
- 31.Acknowledgments. The authors thank A. R. Pearson, B. J. Johnson, C. M. Wilmot, and D. H. Ohlendorf for their guidance and critical discussions during the course of this work, and J. C. Nix for technical assistance in data collection. This work was supported by NIHGMS GM24689. We are grateful for beam time and assistance with X-ray data collection at the Lawrence Berkeley Laboratory Advanced Light Source (ALS), and for facilities and computer support from the Minnesota Supercomputing Institute. Coordinates have been deposited in the Protein Data Bank (PDB) [www.rcsb.org/pdb] as entries 2IG9 (full-length enzyme) and 2IGA (enzyme reacted with 4NC and O2).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Online Material
www.sciencemag.org/cgi/content/full/...........
Materials and Methods
Fig. S1–S5
Tables S1–S3
References