Abstract
Intermediates in the reaction cycle of an oxygenase are usually very informative of the chemical mechanism of O2 activation and insertion. However, detection of these intermediates is often complicated by their short lifetime and the regulatory mechanism of the enzyme designed to ensure specificity. Here, the methods used to detect the intermediates in an extradiol dioxygenase, a Rieske cis-dihydrodiol dioxygenase, and soluble methane monooxygenase are discussed. The methods include the use of alternative, chromophoric substrates, mutagenesis of active site catalytic residues, forced changes in substrate binding order, control of reaction rates using regulatory proteins, and initialization of catalysis in crystallo.
Introduction
Oxygen is activated by enzymes in a tightly controlled manner to ensure that the subsequent reactions occur with high specificity. Loss of this control often leads to devastating results for the enzyme and the organism in which it is found. One consequence of this control is that intermediates in the oxygen activation reaction are often short lived, thereby lessening the chance that the overall metabolic reaction will deviate from its prescribed course. This presents a problem for the chemist interested in biological oxygen activation strategies in that the most useful intermediates in understanding this chemistry are those most difficult to detect, trap, and study. The challenge considered in this account is to develop strategies for finding the fleeting species that nature goes to the greatest lengths to hide from view. Here, the strategies for detection of the reactive oxygen species of three non-heme iron-containing families of enzymes are described.
Extradiol Aromatic Ring-Cleaving Dioxygenases
Overview of the Structure and Mechanism
Complex aromatic compounds are converted in bacteria to simple single or double ring compounds, generally containing ortho or para oriented hydroxyl functions.1 The rings of these compounds are opened by dioxygenases, most of which contain mononuclear Fe(II) or Fe(III) in the active site.2 Those that contain Fe(II) cleave catecholic substrates adjacent to the vicinal hydroxyl groups by incorporating both atoms from O2 to yield muconic semialdehyde derivatives. The X-ray crystal structures of these extradiol dioxygenases show that the iron is bound by 2 His and 1 Glu or Asp side chains occupying one face of the iron coordination.3,4 The opposing face is occupied by up to three solvent molecules. This so called “2-His-1-carboxylate facial triad” (2H–D/E) binding motif was first recognized in the extradiol dioxygenase family, but it is now known to be utilized by many types of enzymes that bind Fe(II) and activate O2.5 Although the reactions catalyzed by the 2H–D/E family are very diverse, the oxygen activation process begins in a similar manner in most cases, and it can be appreciated by studying the extradiol dioxygenase mechanism.
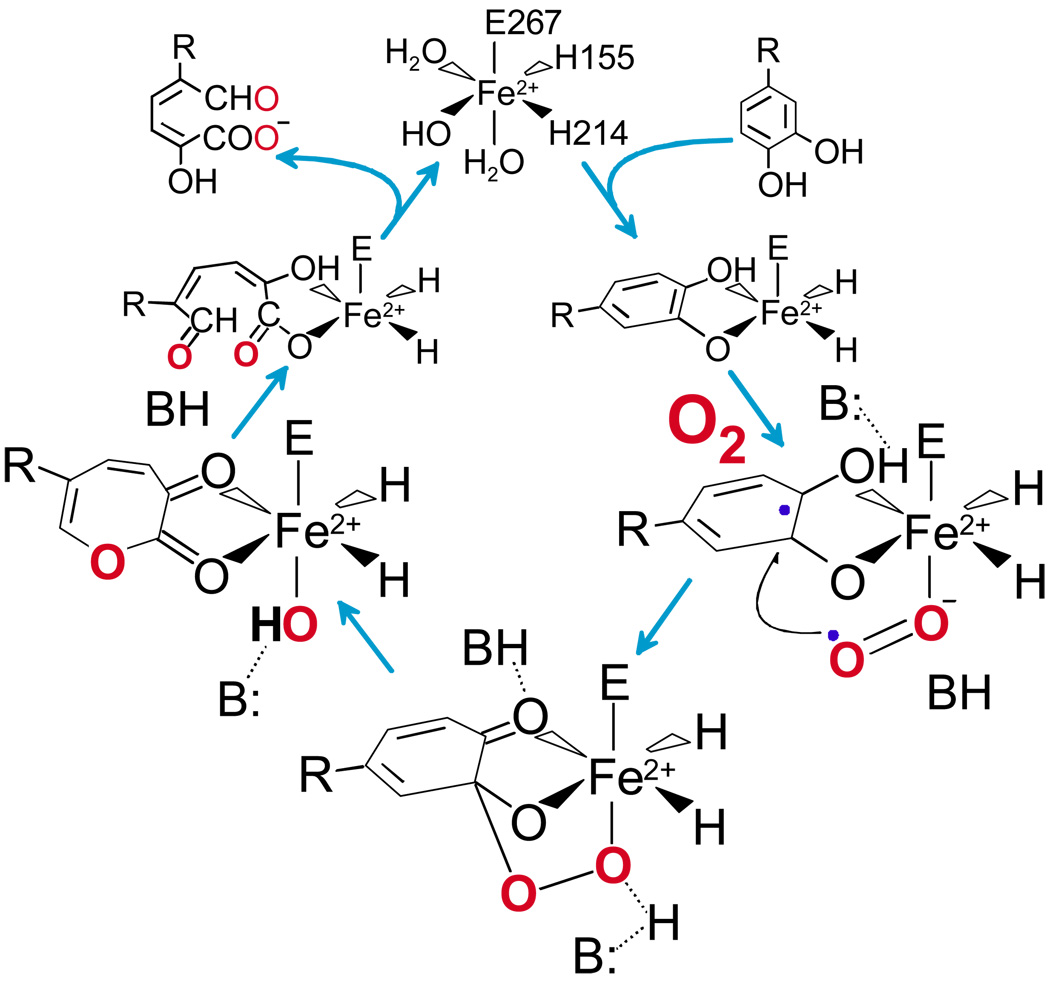
Numerous spectroscopic and X-ray crystallographic studies have shown that catecholic substrates bind directly to the active site Fe(II) as an asymmetric chelate in which only one of the two OH functions is deprotonated.2,4,6–9 This directly displaces two solvents from the “catalytic” face of the facial triad. The third solvent bond to the iron (if present) is weakened or broken by the donation of charge from the anionic catecholic ligand. In our proposed mechanism 6,7 (Figure 1), the binding of the aromatic substrate and O2 to the Fe(II) in adjacent sites is important for two reasons First, it serves to juxtapose the substrates properly so that the oxygen is positioned adjacent to the substrate bond that will be cleaved. Second, the iron provides a conduit for a shift in electron density from the aromatic substrate to the bound oxygen. This simultaneously activates both substrates by giving them radical character so that the subsequent attack of oxygen on the aromatic substrate is spin allowed. The resulting Fe-alkylperoxo intermediate might undergo ring cleavage by a variety of mechanisms, but our initial proposal was for a Criegee rearrangement.6 In this process, the O-O bond of O2 breaks simultaneously with the insertion of one O atom into the ring to yield a lactone. This is subsequently hydrolyzed to yield the bound, ring-open product by the second O atom from the original O2. The rearrangement would be facilitated by active site catalysts that push electron density into the ring or draw density out of the proximal oxygen of O2.
FIGURE 1.
Proposed extradiol dioxygenase mechanism. R = -CH2COOH (HPCA) or –NO2 (4NC) for the studies described here. Single hydroxyl deprotonation during binding results in a bound monoanion in the case of HPCA (shown) and a bound dianion for 4NC.
Kinetic Approaches to the Identification of Intermediates
The mechanistic proposal for the extradiol dioxygenase family predicts the existence of many intermediates, but their discovery was slowed by the lack of an optical spectrum from Fe(II) in the 2H–D/E motif. One way to address this problem was to shift the focus from the iron to the substrate so that intermediates could be detected by changes in a substrate chromophore. The natural substrates for these enzymes are colorless in the visible, but the alternative substrate 4-nitrocatechol (4NC) has a chromophore that shifts dramatically depending upon its state of ionization, its environment, and whether the ring is intact.
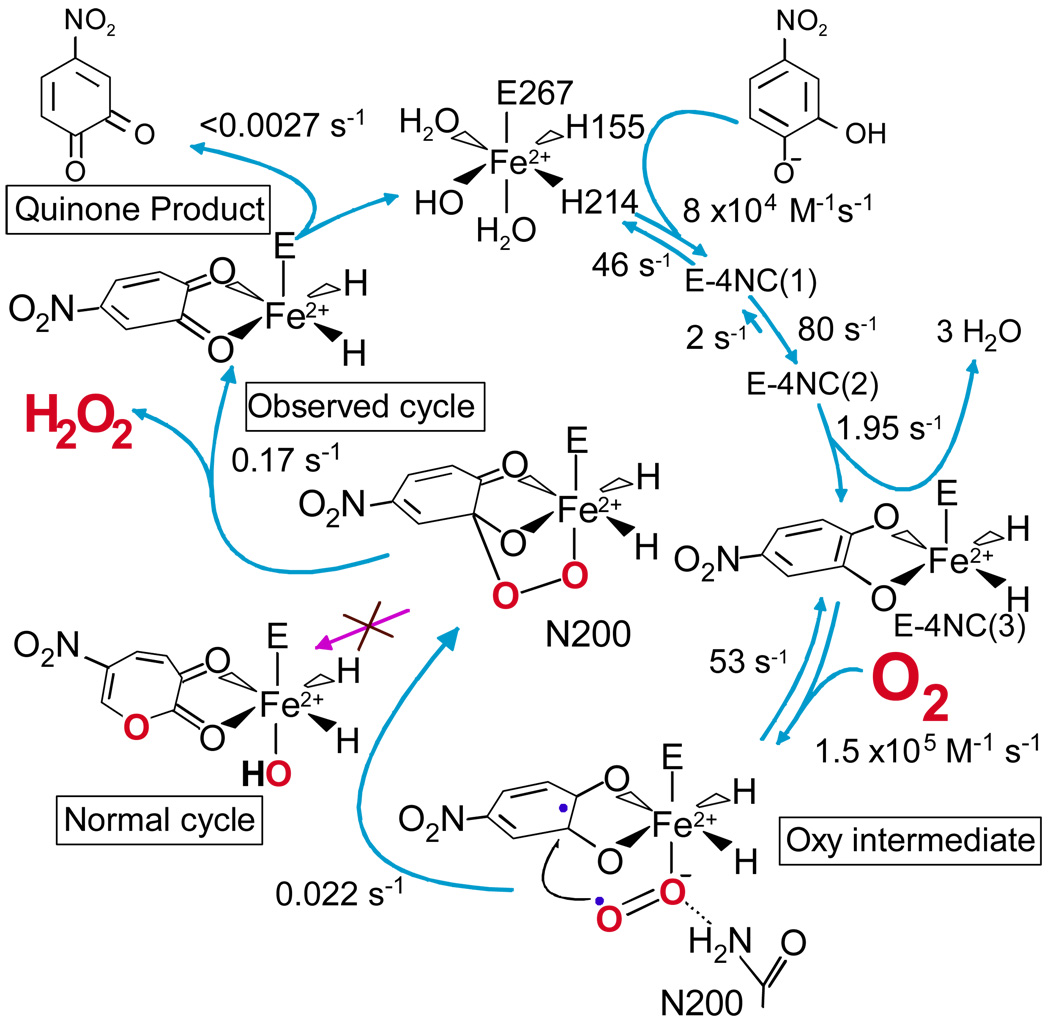
In our search for intermediates, we have used the homotetrameric enzyme homoprotocatechuate 2,3-dioxygenase (HPCD), which cleaves 3,4-dihydroxyphenylacetate (HPCA) between carbons 2 and 3. 4NC is cleaved in the same position at 4% of the HPCA rate. Upon rapid anaerobic mixing of 4NC with HPCD, three kinetically resolvable substrate binding steps were observed as shown in Figure 2.10 Nominally, these are: (i) rapid binding of 4NC monoanion (solution state at pH 7) in the active site, resulting in small spectral intensity changes, (ii) binding 4NC to the Fe(II) and simultaneous deprotonation to cause a substantial spectral shift, and (iii) a further structural reorganization to yield a species termed E-4NC3. The second half of the reaction cycle was investigated by rapidly mixing pre-formed E-4NC3 with O2. Two reaction steps could be directly observed: (i) an intermediate with the ring intact was first converted to the ring open product, and (ii) the product was then released in the overall rate-limiting step. These steps correlate with the final two steps of the postulated cycle shown in Figure 1.
FIGURE 2.
Intermediates discovered in the reaction cycle of the H200N mutant of HPCD when using 4NC as the substrate (rate constants at 4 °C, pH 7.5). The substrate binding steps occur at similar rates for the wild type enzyme. The H200N reaction diverges from the normal cycle after the alkylperoxo intermediate to yield a quinone rather than a ring open product. When using HPCA, the normal ring-open product is obtained. (See references 10 and11 for experimental conditions)
Although the observation of two new intermediates prior to product release was very exciting, the fact that the rate constants for these steps were not O2 concentration dependent suggested that the true O2 binding and activation steps occurred too rapidly to be observed. This was addressed by mutating active site residues with potential to be catalysts for these steps.11 In particular, mutation of His200 (Figure 4 below shows the position of His200) to residues that cannot catalyze acid-base reactions proved to dramatically slow the rates of all the reactions after 4NC is bound. The mutant H200N permitted the observation of two additional steps in the second half of the reaction cycle (Figure 2), both of which exhibited O2 concentration dependence. The rate constant of the faster of these steps had a linear O2 dependence, showing that it is the true O2 association step. The following step was very slow, suggesting that it might be possible to detect the initial oxy-complex of the reaction cycle.
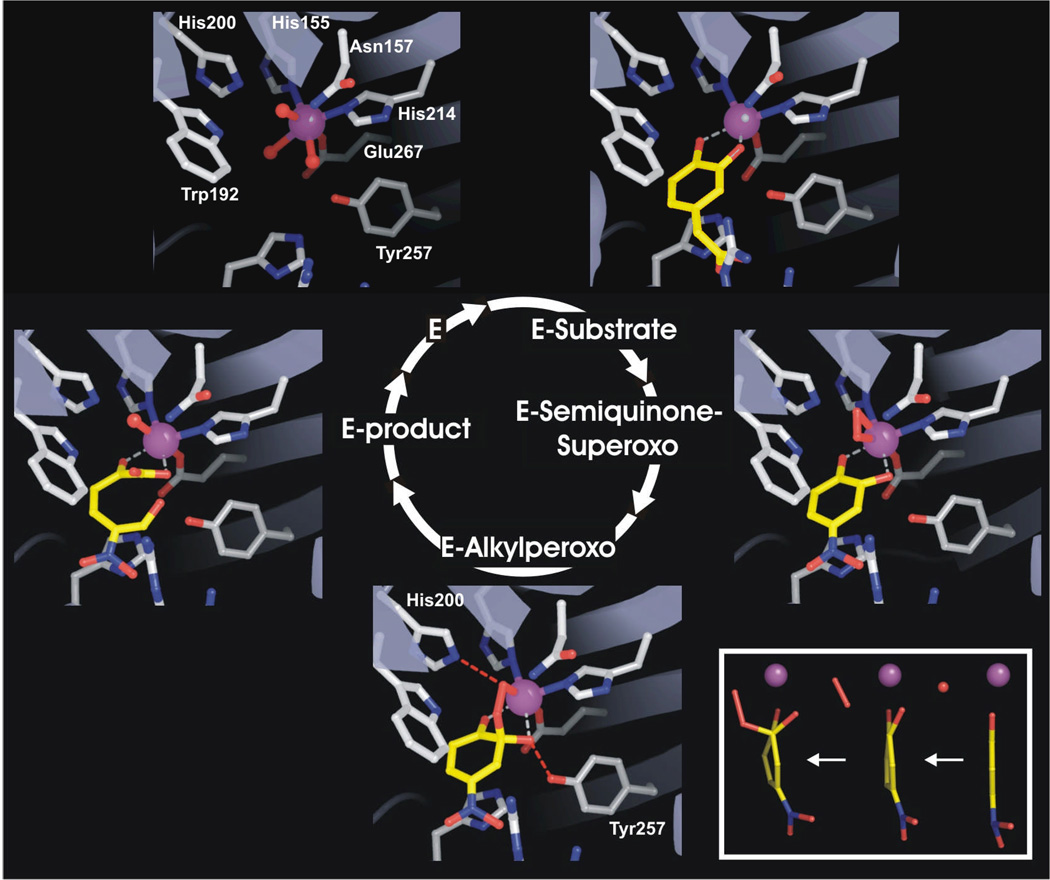
FIGURE 4.
X-ray crystal structure of the intermediates of the HPCD reaction cycle. PDB files: E (1F1X), ES (1Q0C), other intermediates (2IGA). Inset: The aromatic ring of 4NC becomes progressively less planar as the E-semiquinone-superoxo and the alkylperoxo intermediates form.14 The rightmost structure is the oxidized H200E mutant complex (unpublished data).
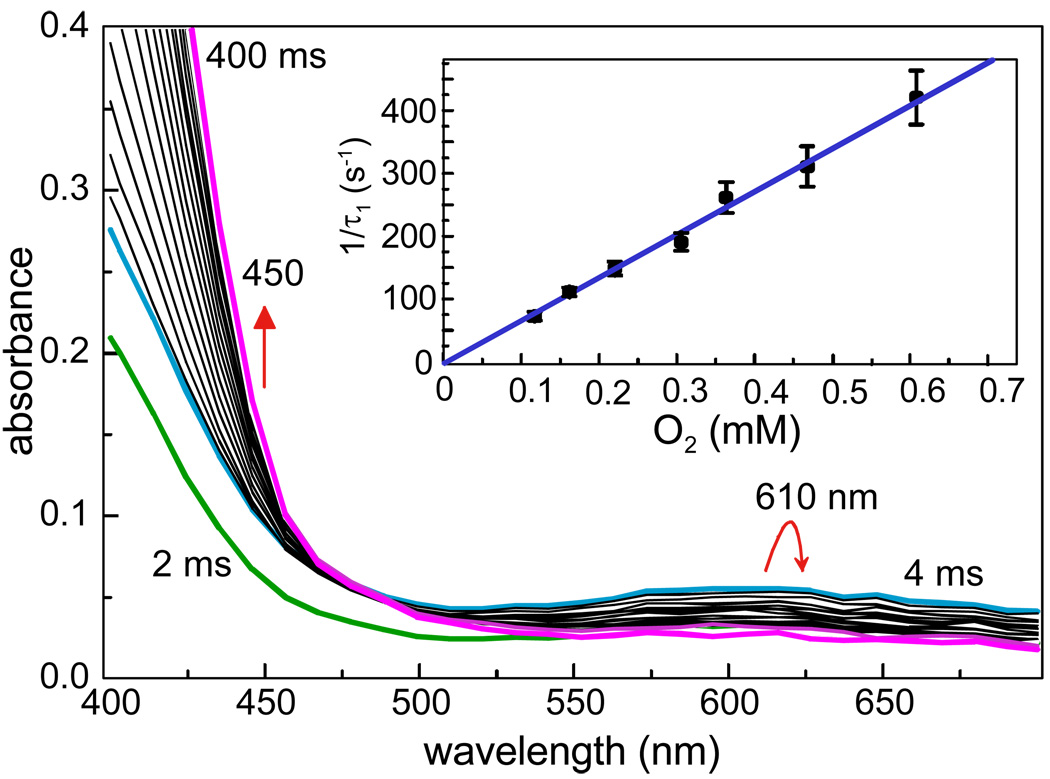
Model compounds for Fe-superoxo and –peroxo species typically exhibit weak optical spectra due to oxygen-to-iron charge transfer.12 In order to search for such a spectroscopic signature, the experiments with the H200N mutant were repeated using the colorless HPCA instead of 4NC. Although faster than the reactions using 4NC, the O2 binding reactions using HPCA could still be observed as shown in Figure 3, now manifested as the rapid formation of a new intermediate absorbing at 610 nm.11 Thus, the first oxy-intermediate of the extradiol dioxygenase family was discovered by the combined use of a slow chromophoric substrate to map the kinetic behavior of the cycle, and then a site directed mutant to stabilize the target species.
FIGURE 3.
Oxy-intermediate formation by the H200N mutant of HPCD using HPCA as the substrate. Inset: O2 concentration dependence of the intermediate formation.11
In Crystallo Approach
A quite different approach to the discovery of intermediates was suggested by the observation that enzymatic reactions often progress more slowly in a crystal.13 A crystal of HPCD was soaked in 4NC and exposed to a very low concentration of O2 prior to flash freezing.14 Remarkably, as illustrated in Figure 4, the 1.9 Å crystal structure showed that three of the four subunits contained different intermediates of the catalytic cycle, allowing them to be structurally characterized.
In the first subunit, the 4NC was chelated to the iron as expected, but a new elliptically shaped electron density was observed in the small molecule binding site of the iron. A molecule of O2 fit the density well, whereas solvent(s) present at full or partial occupancy did not. However, the best evidence that the new ligand is O2, stemmed from examination of the structure of the bound 4NC. This aromatic molecule is expected to be planar, but the structure in the enzyme active site was distinctly buckled with the C2-hydroxyl group moving out of plane (Figure 4, inset). A likely explanation for this is that the 4NC gives up an electron to become a semiquinone. The only diatomic molecule present that can bind to Fe(II) and accept the electron from 4NC is O2. The bond length of each oxygen-Fe(II) bond was the same (∼2.4 Å), showing that the oxygen binds in an unusual side-on configuration. The Fe-O bond lengths were longer than observed for the side-on bound Fe(III)-(hydro)peroxo -complex of naphthalene 1,2-dioxygenase (NDO) (1.99 and 1.74 Å)15 (discussed below), consistent with the formation of an Fe(II)-superoxo species in HPCD.
The structure in the second subunit exhibited continuous electron density from the iron, through the oxygen to the 4NC carbon 2. The oxygen had shifted toward the 4NC so that only the proximal oxygen remained bound to the iron while the distal oxygen assumed the proper orientation for a bond to a tetrahedral carbon. These structural features are consistent with those expected for the alkylperoxo intermediate of the reaction cycle.
The third unique subunit contained the ring-cleaved product complex. Strong electron density near the iron showed the formation of the diagnostic carboxylate function of the product that rotates out of the former plane of the ring to optimize the bond with the iron. In contrast to the iron-bound portions of the product, little electron density was observed for some of the distal atoms. This was expected because of the large increase in flexibility gained after the ring opens.
The observation of different structures in nominally identical subunits apparently derives from both the great decrease in rate caused by reaction in a crystal and crystal packing forces that cause the subunits to take on slightly different structures. These differences are very subtle, suggesting that the enzyme can exert exquisite control over the key oxygen binding, activation, and insertion steps of the catalytic cycle.
Implications for Oxygen Activation
The kinetic and in crystallo studies show that oxygen activation occurs in a step-wise fashion. The ability to bind O2 follows from substrate-initiated solvent release and the transfer of an electron from the substrate to oxygen. The structural studies show that the attacking species is likely to be this Fe(II)-superoxo moiety, yielding an alkylperoxo intermediate. Thus, the attack occurs before the O-O bond is broken. The chemical nature of the O-O bond cleavage and insertion reaction(s) that follows the observed intermediates remains unknown, although the important participation of the perfectly aligned His200 as an active site acid and the inhibitory effects of the electron withdrawing nitro-substituent of 4NC are consistent with the Criegee rearrangement chemistry originally proposed.
Rieske cis-Dihydrodiol Forming Dioxygenases
Overview of the Structure and Reaction
Like the extradiol dioxygenases, the Rieske cis-dihydrodiol forming dioxygenases utilize aromatic compounds as substrates and incorporate both atoms of oxygen from O2.16 However, the reaction differs in that no ring cleavage occurs and the reaction requires two electrons supplied by an NAD(P)H-coupled reductase. There are several subclasses of Rieske dioxygenases differing in the number of electron transfer components and the subunit structure of the oxygenase component. However, all have an oxygenase component that has a Rieske Fe2S2 cluster and a 2H–D/E type mononuclear iron center in the α-subunit. At least three α-subunits compose the complete oxygenase component, and they are spatially arranged to juxtapose the Rieske cluster of one subunit with the mononuclear iron site of the next subunit.17 It is believed these pairs of metal centers form the functional active site. X-ray crystallographic studies show that the substrate binds near the mononuclear iron center, and thus, this is the likely site of oxygen activation and insertion.18
Kinetic Approaches to the Identification of Intermediates
Although the overall stoichiometry of the reaction is two electrons per cis-dihydrodiol formed, there has been debate over the source of these electrons during catalysis and the timing of their delivery. Under one scenario, the Rieske cluster and the mononuclear iron together donate two electrons to O2 bound to the latter. Then, either the resulting Fe(III)-(hydro)peroxo species or the equivalent Fe(V)-oxo-hydroxo species formed by O-O bond cleavage attacks the aromatic substrate.19 A second possibility is that the reductase directly or indirectly donates another electron before the attack on the substrate to yield either an Fe(II)-(hydro)peroxo or an Fe(IV)-oxo-hydroxo species.20
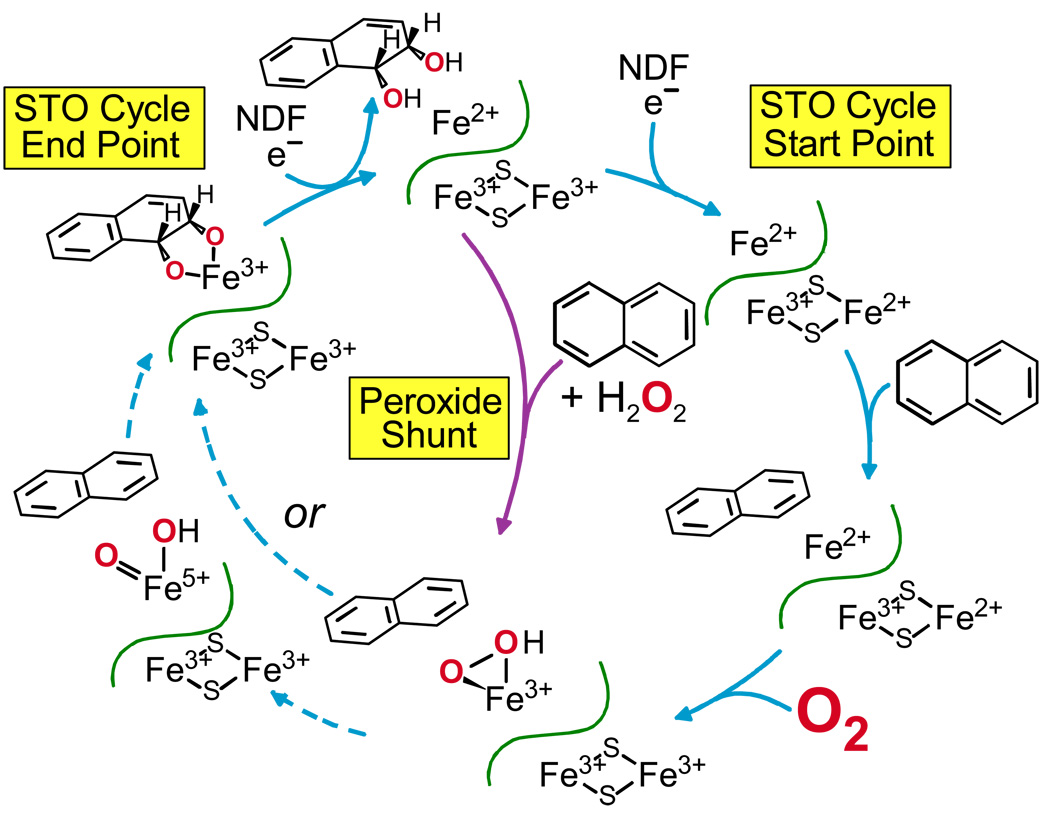
One way to evaluate the timing of electron donation was to carry out a single turnover reaction (Figure 5).19 Using NDO, it was found that fully reduced oxygenase component alone could produce the cis-dihydrodiol product in stoichiometric yield. At the end of the reaction, each of the metal centers was oxidized by one electron, thus it appears that two rather than three electrons are required to activate oxygen. It was found that the enzyme had to be denatured to release the product, fostering the proposal that reduction of the mononuclear iron is required for rapid product release. Thus, the role of the “third electron” is to reduce the mononuclear iron at the end of the reaction to release product and establish the starting state for the next cycle.
FIGURE 5. Single turnover (STO) and peroxide shunt reaction cycles of NDO.19,26.
There are several other indications that the reaction is tightly regulated by conformational changes that respond to substrate or product binding and the oxidation state of the metal centers. For example, when the Rieske cluster of NDO is oxidized, NO binds to the reduced mononuclear iron site to give a characteristic S = 3/2 EPR spectrum.19 In contrast, the equivalent O2 complex apparently does not form since the Mössbauer spectra of aerobic Rieske dioxygenase samples show no evidence for oxidation or an oxygen complex.21 Reduction of the Rieske cluster causes a different result in that neither NO nor O2 can bind to the mononuclear iron.19 Addition of substrate allows both small molecules to bind, and in the case of O2, product formation. These results suggested that the redox state of the Rieske cluster caused a cross-subunit boundary effect that alters the access of small molecules to the mononuclear iron. The presence of substrate appears to make the binding site available. Three different types of experiments suggest how this might occur. MCD studies of the mononuclear iron site in phthalate dioxygenase showed that the binding of substrate near the iron caused the coordination number to change from 6 to 5, presumably opening a site for small molecules.22,23 We showed using pulsed ENDOR techniques, that the distance between the bound substrate and the iron changes when the oxidation state of the Rieske cluster is changed.24 Finally, the recent crystal structure of 2-oxoquinoline 8-monooxygenase showed that when the oxidation state of the Rieske cluster changes, a space for O2 or NO is opened near the mononuclear iron.25
The role of substrate in the O2 activation process was examined using benzoate 1,2 dioxygenase (BZDO). The effect of substrate on the oxidation rate of the Rieske cluster was observed after the fully reduced BZDO oxygenase was rapidly mixed with O2.21 The rate of return of the chromophore of the oxidized cluster showed the apparent rate of electron transfer to the mononuclear iron site. The electron transfer reaction was very slow in the absence of substrate and increased to a rate significantly faster than the enzyme turnover number when substrate was added. It is likely that the electron transfer process itself over the 12 Å separating the metal centers occurs much faster than the reaction monitored by stopped flow. This suggests that the oxygen binding or activation occurs as part of the electron transfer process and is rate limiting. Intriguingly, it was found that changes in the ring substituents of substrate dramatically changed the rate constant for electron transfer. One way to interpret these results is to postulate that substrate directly participates in the oxygen activation process in some manner.
The observation that the two-electron reduced oxygenase component can carry out a single turnover when exposed to O2, suggests that H2O2 might be able to provide both the electrons and the oxygen in a “peroxide shunt” reaction. As shown in Figure 5, central pathway, this is the case for NDO.26 At the end of the reaction, both metal centers were oxidized and product was tightly bound in the active site, effectively limiting the reaction to one turnover. Use of 18O-labeled peroxide showed that both oxygens in the product derived from the peroxide. NDO as isolated, has an oxidized Rieske cluster, but the mononuclear iron is reduced. Since the mononuclear site was oxidized during turnover, a third electron was consumed, opening the possibility that the peroxide shunt utilizes a new mechanism. However, BZDO can be isolated with the mononuclear site in the Fe(III) state21 and gives similar peroxide shunt results, suggesting that peroxide causes adventitious oxidation of the mononuclear iron of NDO.27
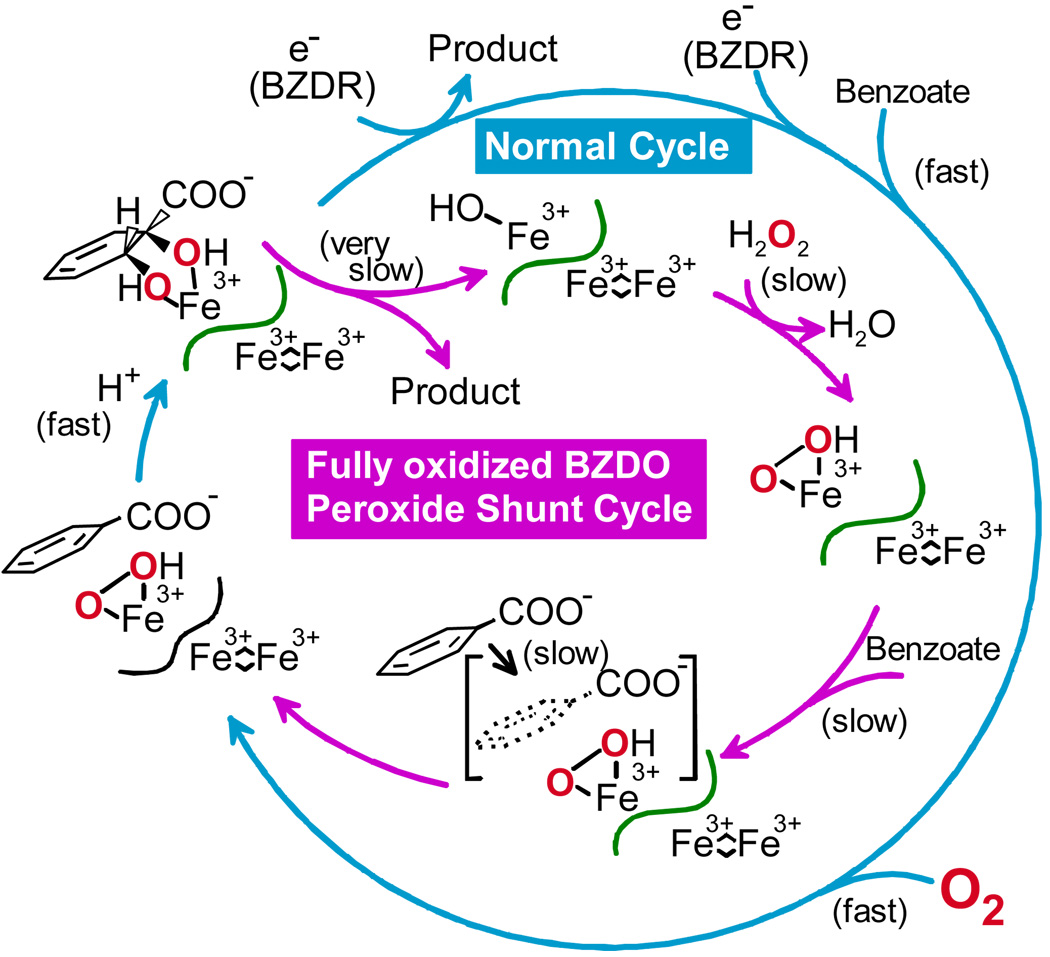
The peroxide shunt reaction of BZDO is similar to that of NDO in yield and specificity, but it is notably slower. Even at high H2O2 concentrations, the formation of product in near stoichiometric yield requires one hour.27 We believe that this is due to the redox coupled regulatory system of the Rieske dioxygenases described above. Substrate used to stabilize BZDO during purification is trapped in the active site due to the Fe(III) oxidation state. Moreover, substrate is in the wrong position in the fully oxidized enzyme to allow small molecule binding. Thus, substrate (or product after a turnover cycle) must slowly dissociate before peroxide can bind to form the activated enzyme (Figure 6).
FIGURE 6.
The peroxide shunt reaction of BZDO is slowed by combination of substrates with incorrect forms of the enzyme.27
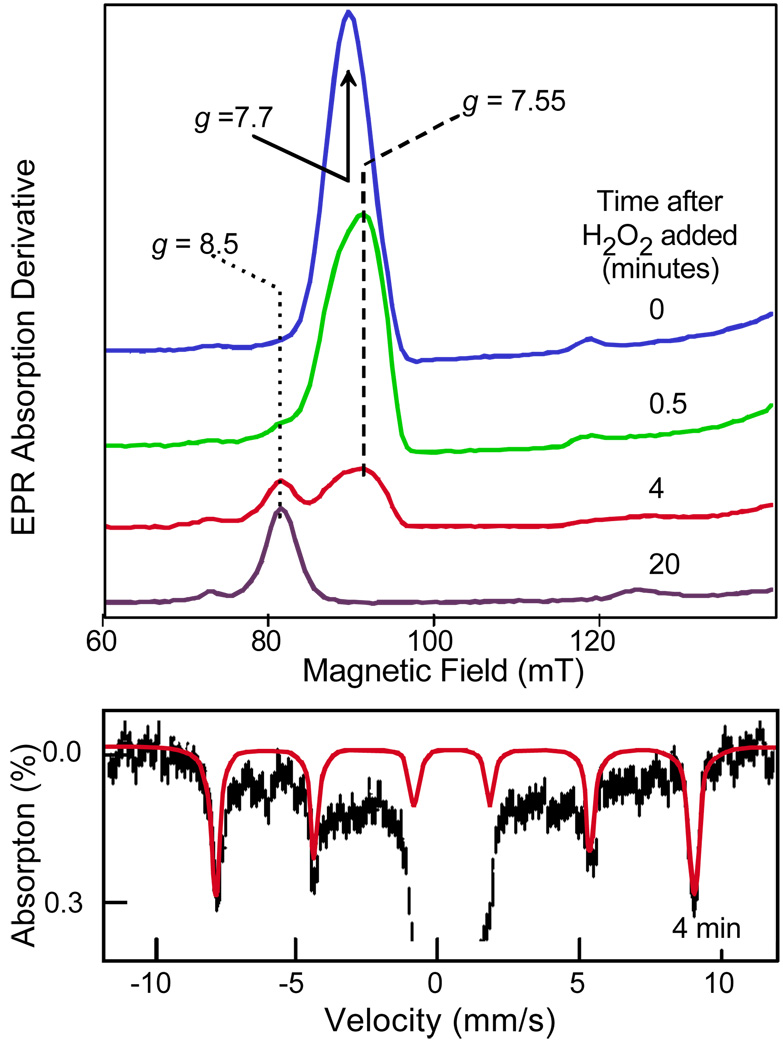
The slow turnover of the BZDO peroxide shunt was exploited to monitor the EPR and Mössbauer spectra of the Fe(III) site during the reaction. As shown in Figure 7, it was found that the intense g = 7.7 resonance from the S = 5/2 Fe(III) site disappeared within two minutes of adding H2O2, but the g = 8.5 signal characteristic of the product complex appeared slowly over the next hour.27 The Mössbauer spectra revealed only Fe(III) throughout the reaction. Further investigation showed that the apparently EPR-silent intermediate had an unusual inverted zero-field splitting causing the ground state to have vanishingly low EPR intensity. Interestingly, EPR and Mössbauer spectra with similar parameters and negative zero-field splitting have been observed for Fe(III)-peroxo model complexes.12,28 Thus, it seems likely that the activated oxygen species of the Rieske dioxygenases is a similar peroxo adduct, although the slightly smaller isomer shift (δ = 0.5 mm/s vs 0.64 mm/s in the models) suggest that it is protonated. It was detected by forcing the formation of the species in a form of the enzyme that does not readily exchange small molecules or substrates in the normal cycle.
FIGURE 7.
EPR (top) spectra of intermediates in the peroxide shunt of BZDO reacting with benzoate. Mössbauer spectrum and simulation (red) of the nearly EPR silent intermediate after 4 min of reaction. Adapted from reference 27.
In Crystallo Approach
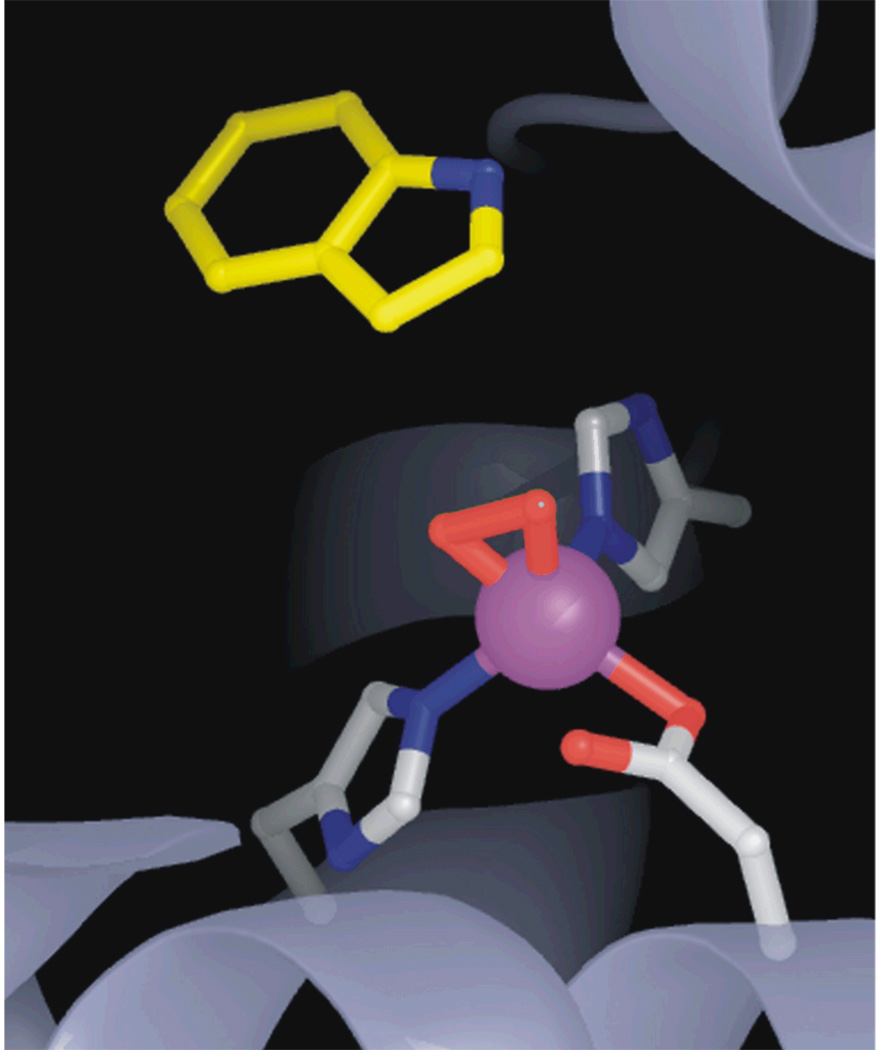
The first X-ray crystal structure of a Rieske dioxygenase was of NDO by Ramaswamy and his collaborators.17 It was subsequently found by this group that incubation of the crystal of fully reduced enzyme with substrate and O2 in a cryosolvent mixture slightly below 0 °C resulted in the formation of an oxy-complex that could be flash frozen and structurally characterized (Figure 8).15 This was the first such complex for the 2H–D/E family, and, like that described above for HPCD, the oxygen bound side-on. Based on the Fe-O bond lengths, the complex is likely to be an Fe(III)-(hydro)peroxo species like that subsequently found in the BZDO peroxide shunt reaction. Thus, the approach of slowing the reaction by conducting it in a crystal allowed a key activated oxygen species to be trapped.
FIGURE 8.
X-ray crystal structure of the oxy-intermediate of NDO (PDB 1O7N)15.
Implications for Oxygen Activation
Although both the extradiol dioxygenases and the Rieske dioxygenases form peroxy intermediates, their mechanisms of oxygen activation are clearly different. The requirement for the input of two external electrons in the Rieske dioxygenase case suggests an activation mechanism more akin to those of cytochrome P450 (P450) or methane monooxygenase (MMO).29 These enzymes activate O2 by forming an Fe(III)-hydroperoxy intermediate that subsequently undergoes heterolytic O-O bond cleavage before reacting with substrates. The kinetic and structural studies of the Rieske dioxygenases show that a similar Fe(III)-(hydro)peroxy species is formed, but it remains unclear whether the O-O bond must break before reaction with aromatic substrates. The Rieske dioxygenases have been shown to carry out many of the reactions of P450 and MMO,30,31 implying O-O bond cleavage before attack. On the other hand, the apparent dependence of the oxygen activation reaction on substrate type described above suggests that a direct attack on substrate by the peroxy adduct as it is formed must also be considered.
Methane Monooxygenase
Overview of the Structure and Mechanism
Methanotrophic bacteria catalyze the oxidation of methane to methanol with the incorporation of one atom from O2. This is the first step in a pathway leading to complete oxidation of CH4 to CO2 that satisfies the carbon and energy needs of the bacterium. Although all methanotrophs can elaborate a membrane bound, copper-containing form of MMO, many type II and X methanotrophs produce exclusively a soluble, iron-containing form (sMMO) in a low copper environment. The sMMO consists of three protein components: a reductase (MMOR), a regulatory protein (MMOB), and an oxygenase (MMOH). MMOH has an (αβγ)2 subunit structure with an bis-μ-hydroxo- dinuclear Fe(III) cluster in the active site in the α-subunit. Kinetic and spectroscopic studies have shown that the oxygen activation and substrate hydroxylation reactions occur at this site.29,32 The O2 is activated through a reductive process that results in O-O bond cleavage and generation of a high-valent iron-oxo reactive species prior to the binding of substrate. Thus, the mechanism of this non-heme enzyme is similar to that of heme-containing P450 in terms of the steps in O2 activation, but differs in the timing of substrate association. This appears to be a manifestation of the overarching need for specificity in methanotrophs.
Kinetic Approaches to the Identification of Intermediates
Early experiments showed that the two-electron reduced form of MMOH in the absence of MMOR and MMOB could turnover once to yield methanol from methane with incorporation of oxygen from O2, directly demonstrating that the diiron cluster is sufficient to activate O2.29,33 The binding of O2 with the reduced diiron cluster of MMOH was found to be accelerated 1000-fold by the addition of MMOB.29 This shifted the rate-limiting step of the single turnover cycle from O2 association to product release, thereby allowing many intermediates in the oxygen activation process to be detected and characterized.
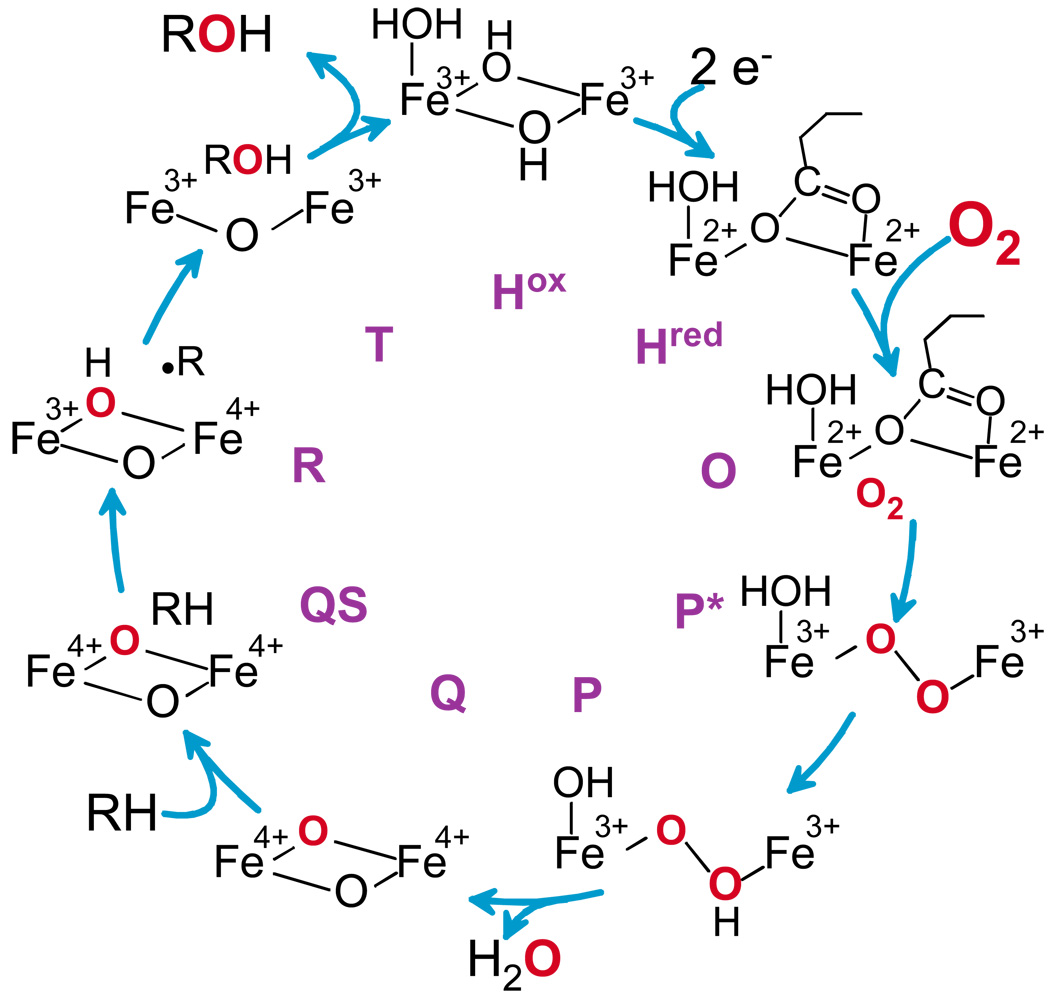
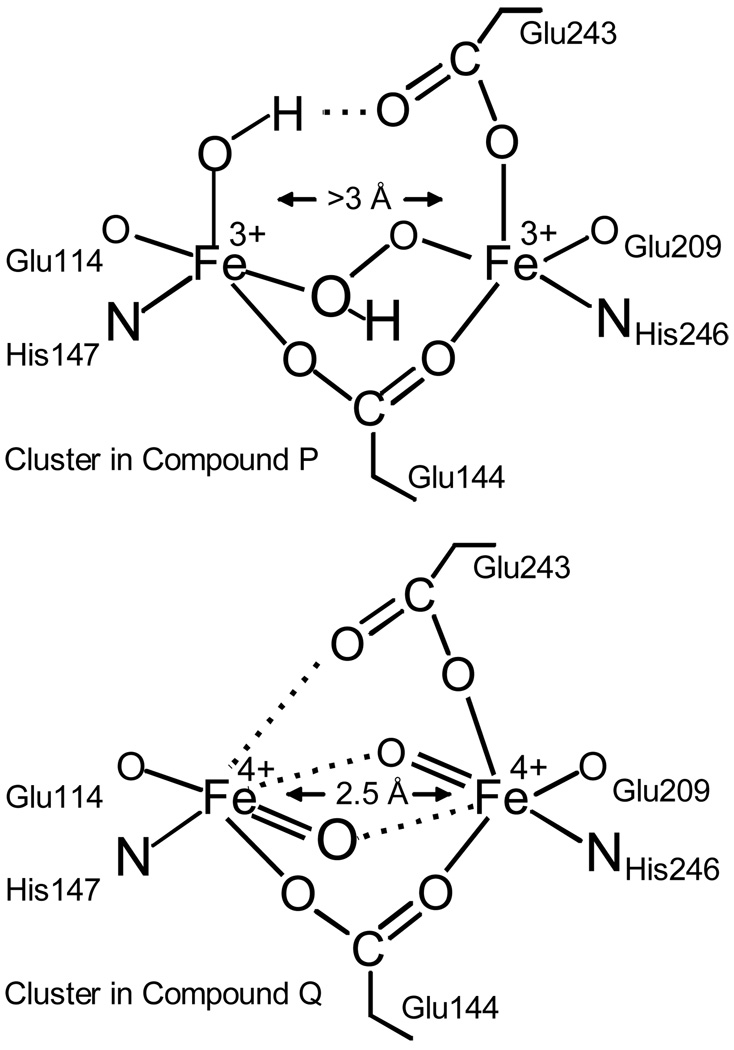
For the sMMO from Methylosinus trichosporium OB3b, analysis of the O2 concentration dependence of the transient kinetic phases detected in the O2 binding process showed that two intermediates must occur as illustrated in Figure 9.29 Initially, O2 binds rapidly and effectively irreversibly to the diferrous enzyme (but not the cluster) to yield an intermediate termed O in which both irons remain Fe(II). Then, the oxygen complex with the diiron cluster forms, resulting in oxidation of the irons to yield an Fe(III)Fe(III)-peroxo (or possibly superoxo) intermediate termed P*. Donation of a proton yields the Fe(III)Fe(III)-hydroperoxo species P.34 Finally, donation of a second proton results in O-O bond cleavage to yield the unique Fe(IV)Fe(IV) bis-μ-oxo species Q.33 Each step in this process occurs more slowly than the last, allowing each intermediate to accumulate. P and Q have been trapped, allowing the oxidation state of their diiron cluster and their overall structure (Figure 10) to be studied using spectroscopic approaches.29,32,35 Each has an optical chromophore that allows their reaction with substrates to be monitored.
FIGURE 9.
Intermediates in the reaction cycle of sMMO.
FIGURE 10.
Proposed structure of sMMO P and Q from spectroscopic studies. Although P is clearly a peroxy adduct, its precise structure has not been definitively established.
Q decay is linearly accelerated by increasing concentrations of nearly all substrates, in particular methane, confirming it as the reactive species.33,36 The ability of Q to directly oxidize substrates was further indicated by two experiments. First, the reaction with nitrobenzene resulted in the formation of nitrophenol in the active site (intermediate T) at the same rate as Q decay.33 Second, a deuterium kinetic isotope effect (KIE) of 50 was observed for Q decay reaction with methane.29
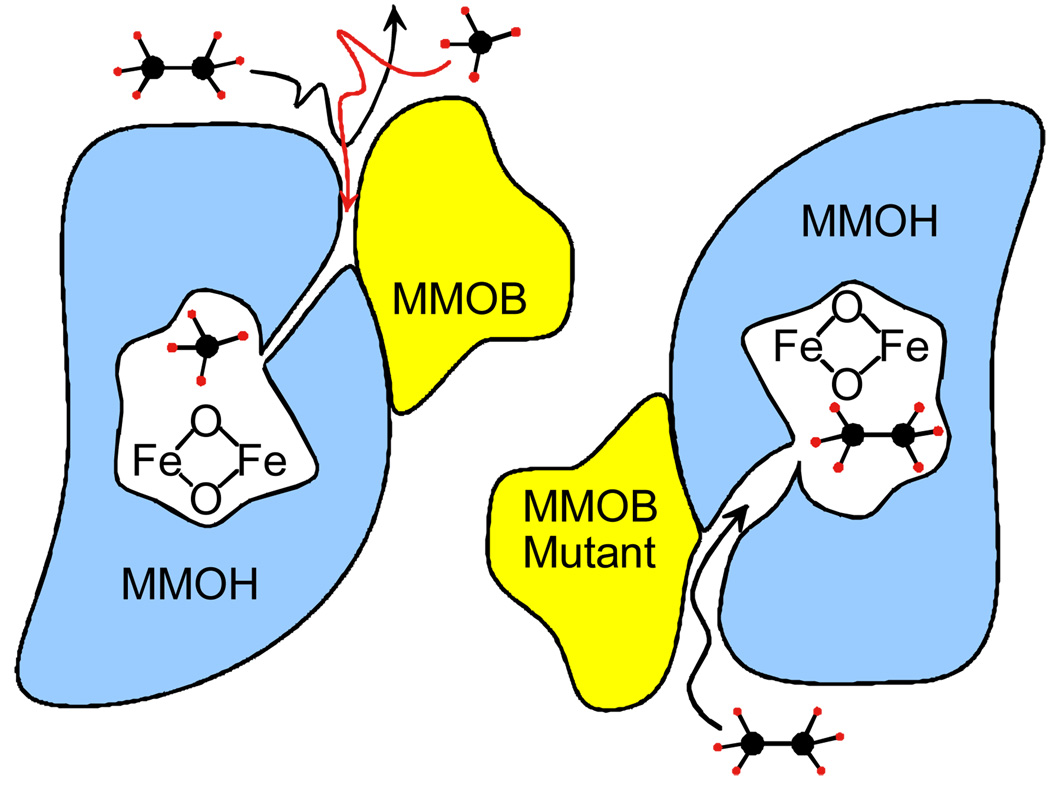
Analysis of the temperature dependence of the Q decay reaction with substrates suggested that it occurs in two steps, nominally, substrate binding (QS) followed by transfer of oxygen to form bound product.36 Significantly, the binding reaction appears to occur slowly in comparison to that typically observed for enzyme reactions. This makes the binding reaction rate limiting for all saturated hydrocarbon substrates except methane (Figure 11, left). Consequently, methane is the only one of these substrates that exhibits a deuterium KIE in the Q decay reaction. As expected, the similarly small and stable fluoromethanes also exhibit a large KIE for this reaction. 37 The binding rate is affected by the formation of a complex between MMOH and MMOB.29,36,38 Mutations in the MMOB surface were found to significantly increase the apparent binding rate of large substrates and the dissociation rate of large products (Figure 11, right).39 Thus, it was possible to unmask the deuterium KIE for the Q reaction with ethane using an MMOB mutant in which four key surface residues were simultaneously changed.40
FIGURE 11.
Molecular sieve model for the methane selectivity of MMOH. Adapted from reference 42.
Q is one of the most powerful oxidizing species in nature based on its ability to break the C-H bond of methane. Nevertheless, it appears to react rather slowly due to the encapsulating enzyme structure, which severely restricts substrate access to the activated oxygen. We speculate that nature has established this complex system to allow the enzyme to selectively oxidize methane, the only growth substrate for the methanotroph.39,41 The size restriction into the active site gives a kinetic advantage to methane. Moreover, the large KIE found for methane (and not other substrates even when the access restrictions are eased) suggests that quantum tunneling is also enhanced for methane C-H bond cleavage to further increase its kinetic advantage.36,40–42
Implications for Oxygen Activation
The discovery of Q and the ease with which its kinetic behavior can be monitored has allowed the study of the O2 activation process in unprecedented detail. The stepwise nature of the process leading to the breaking of the O-O bond after protonation is very clear from the intermediates observed during the formation of Q. The chemical nature of the subsequent oxygen insertion process is less clear, but the ability to experimentally and computationally approach it has fostered many new ideas and theories. Our original proposal for this reaction followed from that for P450 in which Q would abstract a hydrogen atom from the substrate to yield a substrate radical and an Fe(III)Fe(IV)-OH enzyme intermediate (R).29 Other proposals32 based on computations and the difficulty of observing rearranged products from diagnostic radical clock substrates include: (i) Formation of a methane carbon-iron bond intermediate, (ii) concerted oxygen insertion, and (iii) formation of a bound radical intermediate with a very short lifetime. Although the nature of the reaction remains controversial, one experiment that derives from the need for the enzyme to protect the reactive oxygen of Q gives some additional insight. The MMOB surface mutant T111Y appears to remove most restrictions to entry of methane and ethane into the MMOH active site. Moreover, the large deuterium KIE characteristic of tunneling is lost in the methane reaction. As a result, the reaction of Q with normal and deuterated forms of these substrates was observed without the usual protective effects of the enzyme structure. In this experiment, the resulting Polanyi plot relating bond energy to reaction rate is similar to that expected for reactions known to occur by hydrogen atom abstraction as proposed for P450 and in our original proposal for MMO.41
Conclusions
Oxygen activation by the three non-heme iron containing oxygenases described here proceeds along distinctly different mechanistic paths. Accordingly, quite different strategies were required in each case to seek out and characterize the intermediates carrying the activated form of oxygen. For extradiol dioxygenases, a combination of mutagenesis and a slow substrate were sufficient to stabilize the oxy intermediate. Knowledge of the regulatory mechanism of Rieske dioxygenases was required to define a method to approach the oxy-intermediate in a way that forced the structure of the enzyme itself stabilized the species. Finally, an understanding of nature’s requirement to protect the reactive oxygen species of sMMO and the regulatory protein that controls the process allowed this species to trapped and characterized. In each case, some form of peroxo adduct was an intermediate in the activation process. However, the means by which these species are formed and their fate leads to the diversity of reaction outcomes represented in this set of enzymes. Finally, the studies described here show that use of enzyme crystals to slow and then visualize activated oxygen species is a powerful new approach with potential applications throughout this exciting field.
Acknowledgments
This work was supported by NIH grants GM24689 and GM40466.
Biographies
Elena G. Kovaleva was born in Moscow, Russia and received her B.S. degree from the University of Maryland (1998) and her Ph.D. from the University of Iowa (2004). She is currently a postdoctoral associate at the University of Minnesota interested in kinetic and in crystallo approaches to enzyme mechanism.
Matthew B. Neibergall was born in Council Bluffs, Iowa and received his B.S. degree from Bethel University (1998) and his Ph.D. from the University of Minnesota (2006). He is currently a postdoctoral associate at the University of Minnesota interested in spectroscopic characterization of reactive intermediates and the regulation of catalysis by metalloenzymes.
Sarmistha Chakrabarty was born in Rishra, India and received her B.S. (1997) and M.S. (1999) degrees from Visva Bharati University, Shantiniketan, India. She is currently a graduate student at the University of Minnesota studying the mechanism of Rieske dioxygenases.
John D. Lipscomb was born in Wilmington, Delaware and received his B.A. in chemistry from Amherst College (1969) and M.S. (1971) and Ph.D. (1974) degrees in Biochemistry from the University of Illinois. He completed postdoctoral studies at the Gray Freshwater Biological Institute in 1977 and assumed a position in the Biochemistry Department of the University of Minnesota where he now has the rank of Professor. He studies the mechanisms of oxygen activation by metalloenzymes.
References
- 1.Dagley S. Biochemistry of Aromatic Hydrocarbon Degradation in Pseudomonads. In: Sokatch JR, editor. The Bacteria. Vol. 10. New York, N.Y: Academic Press; 1986. pp. 527–556. [Google Scholar]
- 2.Lipscomb JD, Orville AM. Mechanistic Aspects of Dihydroxybenzoate Dioxygenases. In: Sigel H, Sigel I, editors. Metal Ions in Biological Systems. Vol. 28. New York: Marcel Dekker; 1992. pp. 243–298. [Google Scholar]
- 3.Han S, Eltis LD, Timmis KN, Muchmore SW, Bolin JT. Crystal Structure of the Biphenyl-Cleaving Extradiol Dioxygenase from a PCB-Degrading Pseudomonad. Science. 1995;270:976–980. doi: 10.1126/science.270.5238.976. [DOI] [PubMed] [Google Scholar]
- 4.Senda T, Sugiyama K, Narita H, Yamamoto T, Kimbara K, Fukuda M, Sato M, Yano K, Mitsui Y. Three-Dimensional Structures of Free Form and Two Substrate Complexes of an Extradiol Ring-Cleavage Type Dioxygenase the BphC Enzyme from Pseudomonas sp. Strain KKS102. J. Mol. Biol. 1996;255:735–752. doi: 10.1006/jmbi.1996.0060. [DOI] [PubMed] [Google Scholar]
- 5.Hegg EL, Que L., Jr The 2-His-1-Carboxylate Facial Triad. An Emerging Structural Motif in Mononuclear Non-Heme Iron(II) Eur Enzymes. J. Biochem. 1997;250:625–629. doi: 10.1111/j.1432-1033.1997.t01-1-00625.x. [DOI] [PubMed] [Google Scholar]
- 6.Arciero DM, Lipscomb JD. Binding of 17 O-Labeled Substrate and Inhibitors to Protocatechuate 4,5-Dioxygenase-Nitrosyl Complex. Evidence for Direct Substrate Binding to the Active Site Fe2+ of Extradiol Dioxygenases. J. Biol. Chem. 1986;261:2170–2178. [PubMed] [Google Scholar]
- 7.Shu L, Chiou Y-M, Orville AM, Miller MA, Lipscomb JD, Que L., Jr X-Ray Absorption Spectroscopic Studies of the Fe(II) Active Site of Catechol 2,3-Dioxygenase. Implications for the Extradiol Cleavage Mechanism. Biochemistry. 1995;34:6649–6659. doi: 10.1021/bi00020a010. [DOI] [PubMed] [Google Scholar]
- 8.Vetting MW, Wackett LP, Que L, Jr, Lipscomb JD, Ohlendorf DHCrystallographic, Comparison ofManganese- Iron-Dependent Homoprotocatechuate 2,3-Dioxygenases. J. Bacteriol. 2004;186:1945–1958. doi: 10.1128/JB.186.7.1945-1958.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vaillancourt FH, Barbosa CJ, Spiro TG, Bolin JT, Blades MW, Turner RFB, Eltis LD. Definitive Evidence for Monoanionic Binding of 2,3-Dihydroxybiphenyl to 2,3-Dihydroxybiphenyl 1,2-Dioxygenase from UV Resonance Raman Spectroscopy, UV/Vis Absorption Spectroscopy, and Crystallography. J. Am. Chem. Soc. 2002;124:2485–2496. doi: 10.1021/ja0174682. [DOI] [PubMed] [Google Scholar]
- 10.Groce SL, Miller-Rodeberg MA, Lipscomb JD. Single-Turnover Kinetics of Homoprotocatechuate 2,3-Dioxygenase. Biochemistry. 2004;43:15141–15153. doi: 10.1021/bi048690x. [DOI] [PubMed] [Google Scholar]
- 11.Groce SL, Lipscomb JD. Aromatic Ring Cleavage by Homoprotocatechuate 2,3-Dioxygenase: Role of His200 in the Kinetics of Interconversion of Reaction Cycle Intermediates. Biochemistry. 2005;44:7175–7188. doi: 10.1021/bi050180v. [DOI] [PubMed] [Google Scholar]
- 12.Roelfes G, Vrajmasu V, Chen K, Ho RYN, Rohde J-U, Zondervan C, La Crois RM, Schudde EP, Lutz M, Spek AL, Hage R, Feringa BL, Münck E, Que L., Jr End-on and Side-on Peroxo Derivatives of Non-Heme Iron Complexes with Pentadentate Ligands: Models for Putative Intermediates in Biological Iron/Dioxygen Chemistry. Inorg. Chem. 2003;42:2639–2653. doi: 10.1021/ic034065p. [DOI] [PubMed] [Google Scholar]
- 13.Hajdu J, Neutze R, Sjogren T, Edman K, Szoke A, Wilmouth R, Wilmot CM. Analyzing Protein Functions in Four Dimensions. Nature Struct. Biol. 2000;7:1006–1012. doi: 10.1038/80911. [DOI] [PubMed] [Google Scholar]
- 14.Kovaleva EG, Lipscomb JD. Crystal Structures of Fe2+ Dioxygenase Superoxo, Alkylperoxo, and Bound Product Intermediates. Science. 2007;316:453–457. doi: 10.1126/science.1134697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Karlsson A, Parales JV, Parales RE, Gibson DT, Eklund H, Ramaswamy S. Crystal Structure of Naphthalene Dioxygenase: Side-on Binding of Dioxygen to Iron. Science. 2003;299:1039–1042. doi: 10.1126/science.1078020. [DOI] [PubMed] [Google Scholar]
- 16.Gibson DT. Microbial Metabolism of Aromatic Hydrocarbons and the Carbon Cycle. In: Hagedorn SR, Hanson RS, Kunz DA, editors. Microbial Metabolism and the Carbon Cycle. Chur, Switzerland: Harwood Academic Publishers; 1988. pp. 33–58. [Google Scholar]
- 17.Kauppi B, Lee K, Carredano E, Parales RE, Gibson DT, Eklund H, Ramaswamy S. Structure of an Aromatic-Ring-Hydroxylating Dioxygenase-Naphthalene 1,2-Dioxygenase. Structure. 1998;6:571–586. doi: 10.1016/s0969-2126(98)00059-8. [DOI] [PubMed] [Google Scholar]
- 18.Carredano E, Karlsson A, Kauppi B, Choudhury D, Parales RE, Parales JV, Lee K, Gibson DT, Eklund H, Ramaswamy S. Substrate Binding Site of Naphthalene 1,2-Dioxygenase: Functional Implications of Indole Binding. J. Mol. Biol. 2000;296:701–712. doi: 10.1006/jmbi.1999.3462. [DOI] [PubMed] [Google Scholar]
- 19.Wolfe MD, Parales JV, Gibson DT, Lipscomb JD. Single Turnover Chemistry and Regulation of O2 Activation by the Oxygenase Component of Naphthalene 1,2-Dioxygenase. J. Biol. Chem. 2001;276:1945–1953. doi: 10.1074/jbc.M007795200. [DOI] [PubMed] [Google Scholar]
- 20.Tarasev M, Ballou DP. Chemistry of the Catalytic Conversion of Phthalate into Its cis-Dihydrodiol During the Reaction of Oxygen with the Reduced Form of Phthalate Dioxygenase. Biochemistry. 2005;44:6197–6207. doi: 10.1021/bi047724y. [DOI] [PubMed] [Google Scholar]
- 21.Wolfe MD, Altier DJ, Stubna A, Popescu CV, Münck E, Lipscomb JD. Benzoate 1,2-Dioxygenase from Pseudomonas putida: Single Turnover Kinetics and Regulation of a Two-Component Rieske Dioxygenase. Biochemistry. 2002;41:9611–9626. doi: 10.1021/bi025912n. [DOI] [PubMed] [Google Scholar]
- 22.Gassner GT, Ballou DP, Landrum GA, Whittaker JW. Magnetic Circular Dichroism Studies on the Mononuclear Ferrous Active Site of Phthalate Dioxygenase from Pseudomonas cepacia Show a Change of Ligation State on Substrate Binding. Biochemistry. 1993;32:4820–4825. doi: 10.1021/bi00069a017. [DOI] [PubMed] [Google Scholar]
- 23.Pavel EG, Martins LJ, Ellis WR, Jr, Solomon EI. Magnetic Circular Dichroism Studies of Exogenous Ligand and Substrate Binding to the Non-Heme Ferrous Active Site in Phthalate Dioxygenase. Chemistry & Biology. 1994;1:173–183. doi: 10.1016/1074-5521(94)90007-8. [DOI] [PubMed] [Google Scholar]
- 24.Yang T-C, Wolfe MD, Neibergall MB, Mekmouche Y, Lipscomb JD, Hoffman BM. Substrate Binding to NO-Ferro-Naphthalene 1,2-Dioxygenase Studied by High-Resolution Q-Band Pulsed 2H-ENDOR Spectroscopy. J. Am. Chem. Soc. 2003;125:7056–7066. doi: 10.1021/ja0214126. [DOI] [PubMed] [Google Scholar]
- 25.Martins BM, Svetlitchnaia T, Dobbek H. 2-Oxoquinoline 8-Monooxygenase Oxygenase Component: Active Site Modulation by Rieske-[2Fe-2S] Center Oxidation/Reduction. Structure. 2005;13:817–824. doi: 10.1016/j.str.2005.03.008. [DOI] [PubMed] [Google Scholar]
- 26.Wolfe MD, Lipscomb JD. Hydrogen Peroxide-Coupled cis-Diol Formation Catalyzed by Naphthalene 1,2-Dioxygenase. J. Biol. Chem. 2003;278:829–835. doi: 10.1074/jbc.M209604200. [DOI] [PubMed] [Google Scholar]
- 27.Neibergall MB, Stubna A, Mekmouche Y, Münck E, Lipscomb JD. Hydrogen Peroxide Dependent cis-Dihydroxylation of Benzoate by Fully Oxidized Benzoate 1,2-Dioxygenase. Biochemistry. 2007;46 doi: 10.1021/bi700120j. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Neese F, Solomon EI. Detailed Spectroscopic and Theoretical Studies on [Fe(EDTA)(O2)]3−: Electronic Structure of the Side-on Ferric-Peroxide Bond and Its Relevance to Reactivity. J. Am. Chem. Soc. 1998;120:12829–12848. [Google Scholar]
- 29.Wallar BJ, Lipscomb JD. Dioxygen Activation by Enzymes Containing Binuclear Non-Heme Iron Clusters. Chem. Rev. 1996;96:2625–2657. doi: 10.1021/cr9500489. [DOI] [PubMed] [Google Scholar]
- 30.Resnick SM, Lee K, Gibson DT. Diverse Reactions Catalyzed by Naphthalene Dioxygenase from Pseudomonas sp. Strain NCIB 9816. J. Ind. Microbiol. Biotechol. 1996;17:438–457. [Google Scholar]
- 31.Chakrabarty S, Austin RN, Deng D, Groves JT, Lipscomb JD. Radical Intermediates in Monooxygenase Reactions of Rieske Dioxygenases. J. Am. Chem. Soc. 2007;129:3514–3515. doi: 10.1021/ja068188v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Baik M-H, Newcomb M, Friesner RA, Lippard SJ. Mechanistic Studies on the Hydroxylation of Methane by Methane Monooxygenase. Chem. Rev. 2003;103:2385–2419. doi: 10.1021/cr950244f. [DOI] [PubMed] [Google Scholar]
- 33.Lee S-K, Nesheim JC, Lipscomb JD. Transient Intermediates of the Methane Monooxygenase Catalytic Cycle. J. Biol. Chem. 1993;268:21569–21577. [PubMed] [Google Scholar]
- 34.Lee S-K, Lipscomb JD. Oxygen Activation Catalyzed by Methane Monooxygenase Hydroxylase Component: Proton Delivery During the O-O Bond Cleavage Steps. Biochemistry. 1999;38:4423–4432. doi: 10.1021/bi982712w. [DOI] [PubMed] [Google Scholar]
- 35.Shu L, Nesheim JC, Kauffmann K, Münck E, Lipscomb JD, Que L., Jr An Fe(IV)2O2 Diamond Core Structure for the Key Intermediate Q of Methane Monooxygenase. Science. 1997;275:515–518. doi: 10.1126/science.275.5299.515. [DOI] [PubMed] [Google Scholar]
- 36.Brazeau BJ, Lipscomb JD. Kinetics and Activation Thermodynamics of Methane Monooxygenase Compound Q Formation and Reaction with Substrates. Biochemistry. 2000;39:13503–13515. doi: 10.1021/bi001473l. [DOI] [PubMed] [Google Scholar]
- 37.Beauvais LG, Lippard SJ. Reactions of the Diiron(IV) Intermediate Q in Soluble Methane Monooxygenase with Fluoromethanes. Biochem. Biophys. Res. Commun. 2005;338:262–266. doi: 10.1016/j.bbrc.2005.08.220. [DOI] [PubMed] [Google Scholar]
- 38.Zhang J, Wallar BJ, Popescu CV, Renner DB, Thomas DD, Lipscomb JD. Methane Monooxygenase Hydroxylase and B Component Interactions. Biochemistry. 2006;45:2913–2926. doi: 10.1021/bi052256t. [DOI] [PubMed] [Google Scholar]
- 39.Wallar BJ, Lipscomb JD. Methane Monooxygenase Component B Mutants Alter the Kinetics of Steps Throughout the Catalytic Cycle. Biochemistry. 2001;40:2220–2233. doi: 10.1021/bi002298b. [DOI] [PubMed] [Google Scholar]
- 40.Brazeau BJ, Wallar BJ, Lipscomb JD. Unmasking of Deuterium Kinetic Isotope Effects on the Methane Monooxygenase Compound Q Reaction by Site-Directed Mutagenesis of Component B. J. Am. Chem. Soc. 2001;123:10421–10422. doi: 10.1021/ja016632i. [DOI] [PubMed] [Google Scholar]
- 41.Brazeau BJ, Lipscomb JD. Key Amino Acid Residues in the Regulation of Soluble Methane Monooxygenase Catalysis by Component B. Biochemistry. 2003;42:5618–5631. doi: 10.1021/bi027429i. [DOI] [PubMed] [Google Scholar]
- 42.Zheng H, Lipscomb JD. Regulation of Methane Monooxygenase Catalysis Based on Size Exclusion and Quantum Tunneling. Biochemistry. 2006;45:1685–1692. doi: 10.1021/bi051605g. [DOI] [PubMed] [Google Scholar]