Abstract
Comparative gene mapping of human X-borne genes in marsupials defined an ancient conserved region and a recently added region of the eutherian X, and the separate evolutionary origins of these regions was confirmed by their locations on chicken chromosomes 4p and 1q, respectively. However, two groups of genes, from the pericentric region of the short arm of the human X (at Xp11) and a large group of genes from human Xq28, were thought to be part of a third evolutionary block, being located in a single region in fish, but mapping to chicken chromosomes other than 4p and 1q. We tested this hypothesis by comparative mapping of genes in these regions. Our gene mapping results show that human Xp11 genes are located on the marsupial X chromosome and platypus chromosome 6, indicating that the Xp11 region was part of original therian X chromosome. We investigated the evolutionary origin of genes from human Xp11 and Xq28, finding that chicken paralogs of human Xp11 and Xq28 genes had been misidentified as orthologs, and their true orthologs are represented in the chicken EST database, but not in the current chicken genome assembly. This completely undermines the evidence supporting a separate evolutionary origin for this region of the human X chromosome, and we conclude, instead, that it was part of the ancient autosome, which became the conserved region of the therian X chromosome 166 million years ago.
Sex chromosome pairs are thought to have evolved from ordinary autosomes when one member of the pair acquired a sex determining gene, and progressively degenerated as a result of its genetic isolation (Charlesworth et al. 2005; Graves 2006). Thus, the human X and Y chromosomes differentiated from an ancestral autosome pair after the proto-Y chromosome acquired a testis-determining gene (SRY), around which male-specific genes accumulated. Selection to keep these male-specific genes together progressively restricted recombination with the X chromosome, and the Y degraded to its present small heterochromatic state. In line with this hypothesis, human X-borne genes are found to be autosomal in fish, birds, and even monotreme mammals (Graves 2008).
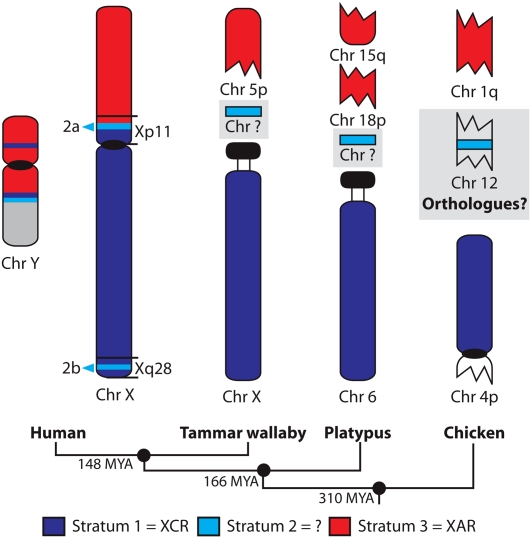
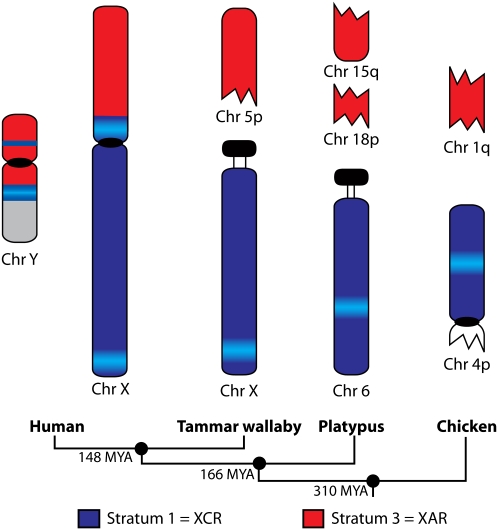
The origin of the mammalian sex chromosomes has been explored by comparing sex chromosomes between distantly related mammal groups. Eutherians (“placental” mammals) diverged from marsupials 148 million years ago (Mya), and these therian mammals diverged from monotreme mammals 166 Mya (Bininda-Emonds et al. 2007). Mapping genes from the human X chromosome in marsupials showed that the entire long arm and pericentric region of the short arm of the human X chromosome is equivalent to the marsupial X chromosome. This therefore constitutes a therian X conserved region (XCR). However, genes located distal to Xp11.23 on the short arm of the human X chromosome mapped to autosomes in marsupials (Graves 1995). This suggested that an autosomal region (X added region [XAR]) was added to the sex chromosomes after the divergence of eutherians from the common ancestor with marsupials 148 Mya and before the eutherian radiation 100 Mya (Fig. 1).
Figure 1.
Previously proposed scheme for the origin of the human X and Y chromosomes. The three evolutionary strata (Kohn et al. 2004) and the recently added (XAR, red) and conserved (XCR, dark blue) regions of the human X and Y chromosomes are shown. The XAR and XCR regions are shown on the chromosomes (Chr) of the tammar wallaby, platypus, and chicken, but the location of stratum 2 (light blue) in the tammar wallaby and platypus is unknown. The existence of homologs of stratum 2 genes on chicken chromosome 12 is evidence for a separate evolutionary origin for this stratum (Kohn et al. 2004). Divergence times given between each of the groups are millions of years ago.
Recently it was discovered that genes from XCR, as well as the XAR, are all autosomal in platypus, a monotreme mammal; XCR genes map to chromosome 6 and XAR genes map to chromosomes 15 and 18 (Veyrunes et al. 2008). Thus, the platypus chromosome 6 is entirely homologous with the marsupial X chromosome, and represents the autosome from which the therian X and Y chromosomes diverged (Fig. 1). This implies that the origin of the human sex chromosomes must be after the therians diverged from a common ancestor with monotremes (166 Mya).
The separate origins of the XCR and XAR regions were also confirmed by mapping chicken orthologs of human XCR genes to the short arm of chicken chromosome 4p (0–20 Mb), and XAR genes to a block on chicken chromosome 1q (104–122 Mb) (Nanda et al. 2000; Graves and Shetty 2001; Ross et al. 2005). However, more recent detailed comparisons of the location of chicken orthologs of human X-borne genes were inconsistent with this simple interpretation. Two groups of genes, one from the pericentric region of the short arm of the human X (at Xp11 between 46.8 and 57.9 Mb) and a group distal on the long arm (at Xq28 between 152.4 and 153.5 Mb), were found to be located on chicken chromosome 12 and a number of microchromosomes (Fig. 1; Kohn et al. 2004). In the zebrafish and the pufferfish, these human Xp11 and Xq28 genes co-localized principally on zebrafish linkage group 8 and pufferfish chromosome 9. These groups of genes also co-localized on at least three scaffolds in the frog (scaffold_154, scaffold_456, and scaffold_507). This suggested that the Xp11 and Xq28 gene groups were a part of a single group of genes with a separate evolutionary origin to the genes located on chicken chromosomes 1q and 4p (Kohn et al. 2004). The hypothesis of this third evolutionary block on the eutherian X is supported by independent evidence from comparisons of divergence between X and Y paralogs (Lahn and Page 1999).
Kohn et al. (2004) designated the three evolutionary regions strata 1, 2, and 3 (Fig. 1). Stratum 1 genes are located on human Xq, the marsupial X chromosome, platypus chromosome 6q, and chicken chromosome 4p. Stratum 2 genes, from within human Xp11 (stratum 2a) and Xq28 (stratum 2b), are located on chicken chromosome 12 and various microchromosomes. Stratum 3 genes, from human Xp11.23-Xpter, map to chicken chromosome 1q, and are autosomal in marsupials and monotremes (Kohn et al. 2004).
Here we map several human stratum 2a genes from Xp11 in two model marsupials, the tammar wallaby (Macropus eugenii) and the Brazilian short-tailed gray opossum (Monodelphis domestica), and in a monotreme mammal, the platypus (Ornithorhynchus anatinus). We also examine the relationship between stratum 2 genes (stratum 2a from Xp11 and stratum 2b from Xq28) and their homologs (both orthologs and paralogs) in the human, rat, marsupial, and chicken genomes, to characterize the origins of these human X chromosome genes.
Results
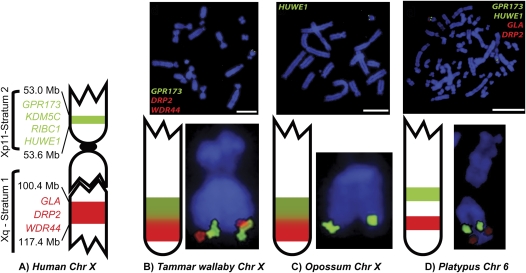
The GPR173, KDM5C, RIBC1, and HUWE1 genes (the GPR173–HUWE1 region) lie within stratum 2a at Xp11 on the human X chromosome (Fig. 2A). We explored the origin of this region by physical mapping of marsupial and monotreme BAC clones containing the GPR173-HUWE1 region to metaphase chromosomes. We also compared the location of Xp11 stratum 2a genes with two other genes (DRP2 and WDR44) from stratum 1 of the human X (Xq22) in marsupials and a monotreme.
Figure 2.
Localization of BAC clones to male marsupial and female monotreme chromosomes. (A) The human X chromosome with the distance from the terminus of the short arm of the stratum 1 and 2 genes given in megabases (Mb). (B) Localization of GPR173 (green), DRP2 (red), and WDR44 (red) to tammar wallaby Xq. (C) Localization of HUWE1 to opossum Xq. (D) Localization of GPR173 (green), HUWE1 (green), and GLA/DRP2 (red) to platypus chromosome 6q. Scale bar, 10 μm.
Localization of human Xp11 stratum 2a genes in marsupials
Tammar wallaby BAC clones were obtained by screening the AGI tammar wallaby BAC library with either PCR products or overgos as probes for genes of interest (Supplemental Table S1). The identity of each BAC clone was confirmed by sequencing of the PCR product amplified from it (Supplemental Table S1).
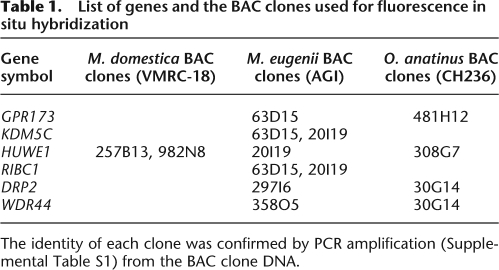
Tammar wallaby BAC clones were identified containing GPR173 and KDM5C (Table 1). The GPR173 gene hybridized only to 63D15, but the KDM5C gene hybridized to both the 63D15 and 20I19 BAC clones, suggesting that the BAC clones overlapped. This was confirmed by PCR amplification; GPR173 sequences were amplified only from 63D15, whereas KDM5C sequences were amplified from both 63D15 and 20I19. The two BAC clones were then tested for the presence of two other genes from the GPR173–HUWE1 region. The RIBC1 PCR product was successfully amplified from both the 63D15 and 20I19 BAC clones, and the HUWE1 PCR product was amplified from only the 20I19 BAC clone. These results suggest that within the two overlapping tammar wallaby BAC clones, each of ∼250 kb, there lies four genes in the same order as found in human: GPR173–KDM5C–RIBC1–HUWE1. The two tammar wallaby BAC clones containing the GPR173–HUWE1 region were then localized in the tammar wallaby by fluorescence in situ hybridization (FISH). Both clones hybridized to the tip of the long arm of the tammar wallaby X chromosome (Fig. 2B).
Table 1.
List of genes and the BAC clones used for fluorescence in situ hybridization
The identity of each clone was confirmed by PCR amplification (Supplemental Table S1) from the BAC clone DNA.
The GPR173–HUWE1 region was also examined in the opossum (Monodelphis domestica) using sequences in the public database. The Ensembl v50 (July 2008) database positioned GPR173 and KDM5C homologs at ∼72.2 Mb, close to the tip of the long arm of the opossum X chromosome (∼79.3 Mb). RIBC1 and HUWE1 were present in an unanchored contig so two opossum BAC clones containing all or part of opossum HUWE1 (Table 1) were localized by FISH to the telomere of the long arm of the opossum X chromosome (Fig. 2C). Thus, these four genes, GPR173, KDM5C, RIBC1, and HUWE1, colocalize to the tip of the X chromosome in these distantly related marsupial species.
Characterization of the distal end of the long arm of the marsupial X chromosome
The location of the GPR173–HUWE1 region close to the end of the X chromosome in two marsupials led us to investigate whether the entire end of the marsupial X was conserved. Examination of the Ensembl v50 (July 2008) opossum assembly showed that the 2-Mb region (71.6–73.6 Mb) with orthology to human Xp11, including the GPR173 and KDM5C genes, lies 5.7 Mb from the telomere of the long arm of the opossum X chromosome. This distal 7.7 Mb region (71.6–79.3 Mb) contains orthologs of both Xp11 stratum 2a genes and Xq22.1–22.2 stratum 1 genes (including GLA, DRP2, and WDR44).
Our results showed that the GPR173–HUWE1 region was located very close to the distal end of the long arm of the tammar wallaby X chromosome. BAC clones containing GLA from human Xq22.1–22.2 have also been localized to this region of the tammar wallaby X chromosome (Deakin et al. 2008). We screened the tammar wallaby AGI BAC library with overgos for two other Xq22.1–22.2 genes, DRP2 and WDR44 (Supplemental Table S1). One BAC clone was isolated containing DRP2, and another was found to contain WDR44 (Table 1). The two BAC clones were localized to the distal end of the long arm of the tammar wallaby X chromosome by FISH (Fig. 2B). Two-color and sequential co-hybridization to the same metaphase chromosome preparations confirmed that the GPR173–HUWE1 region, DRP2 and WDR44 all co-localized to the distal end of the long arm of the tammar wallaby X chromosome, but the resolution was not sufficient to determine the order of these genes (Fig. 2B).
The co-localization of the GPR173–HUWE1 region (from stratum 2a at human Xp11.22–11.23) and GLA–DRP2 (from human Xq22.2) to the end of the marsupial X chromosome was unexpected, given that these regions are separated by around 100 Mb on the human X chromosome and are thought to belong to two separate evolutionary blocks. To determine which arrangement of genes was present in the therian common ancestor, the locations of the GPR173–HUWE1 region and GLA–DRP2 genes were examined in the platypus.
Platypus orthologs of stratum 2 genes (from human Xp11)
The location of the GPR173–HUWE1 region in the platypus was investigated by FISH. BAC clones that contained genes from this region were hybridized to platypus metaphase chromosomes. Using the Ensembl Biomart tool, we found that platypus GPR173 and KDM5C were contained in platypus ultracontig 295 (480 kb), RIBC1 was in platypus contig 3093 (70 kb) and HUWE1 was in platypus ultracontig 403 (1.3 Mb). Contig 3093 and ultracontig 403 were linked because each contains sequence from opposite ends of the same BAC clone (BAC clone ID 0398I14; platypus female genomic library ID KAAG; Washington University Genome Sequencing Center). This indicated that RIBC1 and HUWE1 are located together in the platypus, as they are in humans and marsupials.
None of these contigs had been previously physically anchored to platypus chromosomes. We identified BAC clones to be used to anchor these contigs (Table 1) by aligning the contigs to the platypus BAC end sequences trace archive using BLAST. FISH was used to map BAC clones that anchored GPR173–KDM5C (platypus ultracontig 295) and HUWE1 (platypus ultracontig 403) to the same position below the midpoint of the euchromatic long arm of platypus chromosome 6 (Fig. 2D). Platypus chromosome 6 is the sixth largest platypus chromosome with a distinctive nucleolus organizer region on the short arm (McMillan et al. 2007). This indicated that this group of genes (GPR173–KDM5C–RIBC1–HUWE1) co-localize in platypus as they do in humans and marsupials.
The GLA and DRP2 genes, which are located ∼200 kb apart in humans, are ∼100 kb apart in the middle of the large platypus ultracontig 519 (10 Mb). A BAC clone located ∼3 Mb from the GLA and DRP2 genes toward one end of ultracontig 519, was used to anchor this contig (Table 1) to the long arm of platypus chromosome 6 by FISH (Fig. 2D). Two-color and sequential co-hybridization using BAC clones representing GPR173, HUWE1, and ultracontig 519 (containing GLA and DRP2) to the same metaphase chromosome preparations showed that GPR173 and HUWE1 co-localize on platypus chromosome 6, whereas ultracontig 519 mapped to a separate location more distal than the other two genes (Fig. 2D).
Localization of these genes on platypus chromosome 6 suggests that the two regions that together make up the end of the marsupial X chromosome are both part of the original autosome that became the conserved region of the therian proto-X chromosome. As genes from these two regions (GPR173–HUWE1 and GLA–DRP2) lie on different scaffolds in the frog, and are separated in platypus and human, it is most likely that the two regions were separate in a vertebrate ancestor and fused early in marsupial evolution.
Identification of chicken GPR173 and KDM5C homologs as paralogs
To confirm the ancestral arrangement of these two regions, we also examined their locations in the chicken genome. Searches of Ensembl v50 found that genes from within the GLA–DRP2 region were annotated as located on chromosome 4 among other XCR genes, but our searches for genes within the GPR173–HUWE1 region yielded no clear orthologs in the chicken genome.
We used the Ensembl Biomart tool to examine orthologs of the GPR173–HUWE1 region of the opossum X chromosome in other vertebrates, expecting to find homologs mainly on chicken chromosomes 12 and 26 and zebrafish chromosome 8, as was reported previously (Kohn et al. 2004). We failed to find orthologs of these opossum genes on chicken chromosomes 12 and 26, although we could detect orthologs of these genes in other species. We therefore explored the possibility that the chicken orthologs are missing from the chicken assembly, and the genes detected are paralogs.
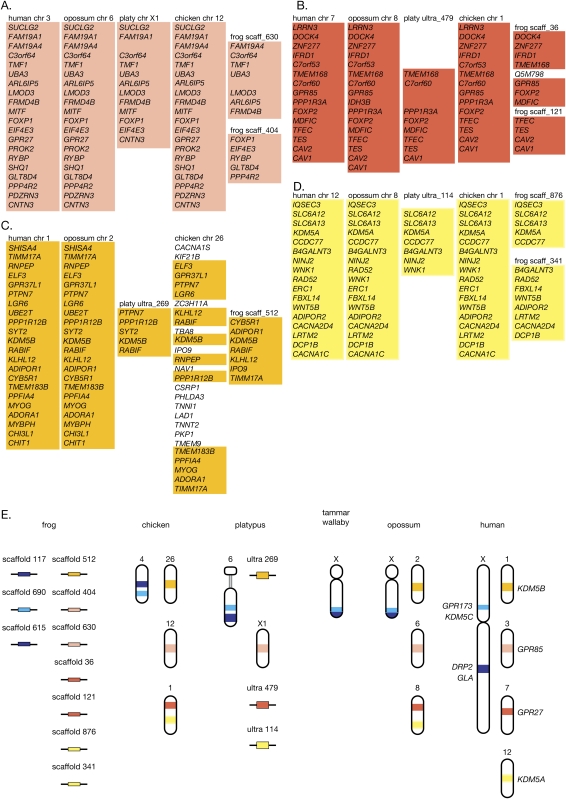
The resolution of this inconsistency can be illustrated by examining the location of GPR173 and KDM5C paralogs in more detail in different species. There are two genes with homology with GPR173 in the human genome; paralogous genes GPR85 and GPR27 lying on human chromosomes 7 and 3, respectively. The same two genes lay on opossum chromosomes 8 and 6, platypus ultracontig 479 and chromosome X1, and chicken chromosomes 1 (outside the region containing XAR genes) and 12, respectively. Two GPR173 homologous regions were also located on separate scaffolds in a frog (Fig. 3). As the surrounding gene content and order is also consistent and conserved in all species including chicken, we concluded that these homologous genes represent paralogs rather than orthologs of GPR173. Thus, there is no true ortholog of GPR173 in the chicken assembly.
Figure 3.
Conservation of the genomic context surrounding GPR173 and KDM5C indicates that their chicken homologs are paralogs and not orthologs. The gene arrangements surrounding the GPR173 and KDM5C paralogs are well conserved: (A) GPR27 (light red), (B) GPR85 (dark red), (C) KDM5B (orange), and (D) KDM5A (yellow). (E) Schematic representation of the gene arrangements of paralogous and orthologous regions to Xp11 stratum 2 genes across the vertebrates. Genes from the proposed Xp11 stratum 2a region (light blue) and the human Xq22 region (dark blue) containing GLA, DRP2, and WDR44 is indicated.
The same logic showed that the chicken homolog of KDM5C in the assembly is also a paralog rather than an ortholog. In addition to its homolog on the Y chromosome, there are two other regions in the human genome with homology with KDM5C. Paralogs KDM5A and KDM5B lie on human chromosomes 12 and 1, respectively. In other species KDM5A and KDM5B lay on opossum chromosomes 8 and 2, platypus contigs 114 and 269, and chicken chromosomes 1 (outside the region containing XAR genes) and 26, respectively. KDM5A and KDM5B also lie on separate scaffolds in the frog (Fig. 3). Again, as the surrounding gene content and order is also consistent and conserved in all species in these regions, we concluded that these genes are likely to represent paralogs rather than orthologs of KDM5C. Thus, there is no true ortholog of KDM5C in the chicken assembly.
The identification of genes on chicken chromosomes 12 and 26 as paralogs rather than orthologs of human X chromosome genes is crucial for establishing the origin of the stratum 2 genes. Therefore, we examined in greater detail all the genes from stratum 2, located at both human Xp11 and Xq28.
Paralogs of stratum 2 genes in chicken and other vertebrates
To investigate further the proposed origin of stratum 2 genes, we analyzed genes from both human Xp11 and Xq28. To address the hypothesis that homologs of stratum 2 genes on chicken chromosomes 12 and 26 were paralogs, rather than orthologs of stratum 2 genes, we looked at all paralogs of human stratum 2 genes found on chromosomes other than the human X, and examined the gene content of the 500 kb region on each side of each gene in different species.
There are 307 protein-coding genes within the Xp11 and Xq28 regions of the human X chromosome: 186 genes in Xp11 and 121 genes in Xq28 (Supplemental Table S2). Over half (187 genes) of the genes in these regions have paralogs on chromosomes other than the human X chromosome. We found that the content and order of genes flanking the paralogs is extremely well conserved in the genomes of human, rat, opossum, and chicken.
Of particular interest to us was the genomic context of the human paralogs that were conserved on chicken chromosomes 1, 12, and 26, because the homology between these chicken genes and human X genes had previously been used to define a new stratum of the human X chromosome (Kohn et al. 2004). We used comparisons of flanking genes to unambiguously identify homologous genes as either orthologs or paralogs of stratum 2 genes. We found that most genes on chicken chromosome 12 with homology with the human X genes have true orthologs on human chromosome 3 (Fig. 4). Similarly, most chicken chromosome 26 genes with homology with human X genes are orthologous to genes on human chromosome 1 (Fig. 4), with a few on human chromosomes 6 and 22 (data not shown). Chicken chromosome 1 homologs (outside the region containing XAR orthologs) of human X genes have orthologs largely on human chromosomes 7, 10, and 12 (Fig. 4), with a smaller number also found on human chromosomes 11 and 22, as well as other human chromosomes (data not shown).
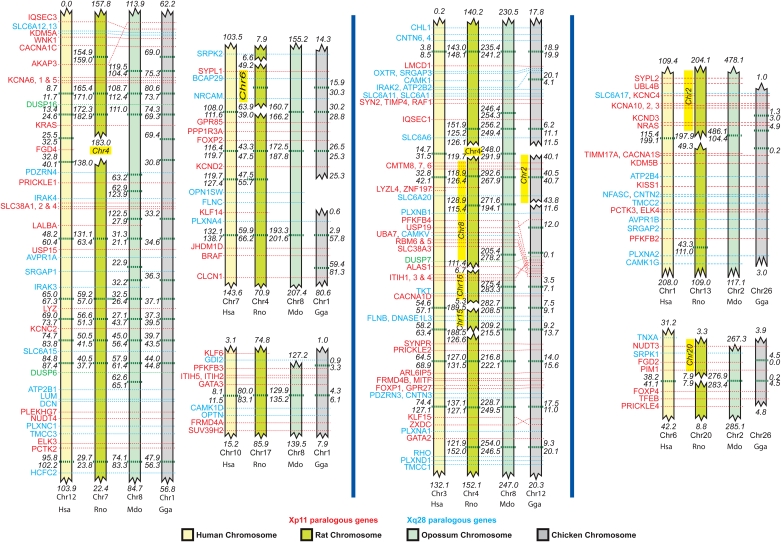
Figure 4.
Conservation of Xp11 and Xq28 paralogs and their genomic contexts in different species. Schematic representation of the location of Xp11 paralogs (red) and Xq28 paralogs (blue), including 1 Mb of genomic context surrounding each, on chicken (Gga, gray) chromosomes 1, 12, and 26. Conservation of the positions of these genes is indicated by the red and blue dotted lines across human (Hsa, yellow), rat (Rno, yellow-green), and opossum (Mdo, blue-green) chromosomes. Chromosome (Chr) numbers are indicated either below or at the side of each conserved segment, and the start and end points of the conserved sections along the chromosomes are indicated in megabases from the terminus of the short arm. Small intervals between the conserved blocks on a single chromosome are indicated by green horizontal dotted lines, and the start and end points of the intervals are also indicated in megabases from the tip of the short arm. Members of the DUSP gene family are found in both Xp11 and Xq28, and their paralogs are indicated in green.
This conservation of genes and their surrounding gene context can also be seen in other mammalian species. The region conserved between chicken chromosome 12 and human chromosome 3 can also be seen on opossum chromosome 8, which also contains blocks of conserved genomic context between chicken chromosome 1 and human chromosomes 7, 10, and 12. The conserved genomic context between chicken chromosome 26 and human chromosome 1 can also be seen on opossum chromosome 2. All these regions can also be found in the rat genome, although they are more rearranged (Fig. 4).
The analysis of conservation of genomic context included the paralogs of GPR173 and KDM5C that had previously been identified (Fig. 3). We conclude that homologous genes on chicken chromosomes 1, 12, and 26 are not true orthologs of human Xp11 and Xq28 genes, but are paralogs.
Identification of orthologs of human stratum 2 genes from chicken EST/cDNA sequences
If the chicken homologs of stratum 2 genes are paralogs rather than orthologs as we have determined, then what and where are the true chicken orthologs of human stratum 2 genes? There are 307 known protein-coding genes in the Xp11 (186 genes) and Xq28 (121 genes) regions of the human X chromosome (Ensembl v50). Previous work has defined the origin of many of these genes within the conserved (XCR) or the recently added (XAR) regions of the human X chromosome. The XCR and XAR are also represented as orthologous blocks at 4p11–p14 and 1q13–q31 respectively in the chicken (Schmid et al. 2000; Ross et al. 2005). In an attempt to identify true orthologs of stratum 2 genes from human Xp11 and Xq28 that were not present in the current annotated chicken genome, we searched for orthologs of Xp11 and Xq28 genes in a phylogenetic tree database and the less well-annotated chicken cDNA and EST databases.
For these analyses we removed the cancer-testis (CT) antigen genes from our data set of Xp11 and Xq28 genes, as members of these gene families (CSAG, CTAG, MAGE, SSX, PAGE, XAGE, and GAGE) in Xp11 and Xq28 are the result of recent amplifications in the primate lineage (Ross et al. 2005; Delbridge and Graves 2007). Therefore, our final data set of human stratum 2 genes include 106 genes from Xp11 (46.8–57.9 Mb, stratum 2a) and 45 genes from Xq28 between BGN and IKBKG (152.4–153.4 Mb, stratum 2b) (Supplemental Table S2).
The TreeFam (Li et al. 2006) database contains gene family annotations for all animal genes calculated using non-animal (plant and yeast) genes as outgroups. We chose this database to check for the presence of one-to-one orthologs of human Xp11 and Xq28 genes in chicken genome. There are 37 XAR genes in human Xp11. We first examined the phylogenetic relationships of these 37 defined XAR genes to determine the completeness and accuracy of the TreeFam database. Our analysis of the TreeFam data showed that 27 of the 37 XAR genes (73%) have orthologs in the chicken genome; 26 of which mapped within the XAR region of chicken chromosome 1 (1q13–q31) (Schmid et al. 2000; Kohn et al. 2004; Ross et al. 2005) and one (ZNF673), which was located on an unanchored contig (chrUn). Similarly, when we analyzed XCR genes from Xq28 that flank stratum 2b, 32 of the 55 genes (58%) within this region had orthologs on chicken chromosome 4 (4p11–p14) as expected (Kohn et al. 2004).
We then analyzed the TreeFam database for chicken orthologs of the remaining 106 Xp11 and 45 Xq28 genes from stratum 2. The TreeFam database contains chicken orthologs for only seven genes of stratum 2a and no orthologs of stratum 2b genes. Four of these Xp11 stratum 2a orthologs (RRAGB, GLOD5, CLCN5, and BMP15) mapped to the chicken chromosome 4 (4p11–p14), and the other three (KLF8, PFKFB1, and WASF4) were on an unanchored contig (chrUn). We also noted that Xp11 and Xq28 stratum 2 genes had paralogs on chicken chromosomes 1, 12, 26, and other microchromosomes (data not shown). Our mapping of four orthologs of stratum 2a genes to chicken chromosome 4p11–p14 suggests that the stratum 2 at Xp11 is a part of the XCR.
As the TreeFam database is generated from computer analysis of genomic assembly data present in Ensembl, it is not surprising that the gene content of the TreeFam database correlates closely to that of the chicken genome assembly, in that there is little additional information for most of the stratum 2 genes. We therefore sought to check an independent source of sequences for orthologs of stratum 2 genes, such as the EST/cDNA sequence data sets of chicken and zebra finch (Taeniopygia guttata). We employed two independent strategies to identify orthologous EST/cDNA sequences for the human X chromosome stratum 2 genes in the avian lineage.
The first strategy involved identifying orthologous sequences using the “reciprocal best hit match” criterion, which means that orthologous sequences are defined when a sequence from the first genome identifies a sequence from a second genome as its homolog, and the homologous sequence from the second genome reciprocally identifies the original sequence from the first genome as its best match. Using this criterion, we identified orthologous EST/cDNA sequences for 26 of the 106 Xp11 stratum 2a genes and 10 of the 45 Xq28 stratum 2b genes. We mapped these EST/cDNA sequences on to the chicken genome by using a BLAST search to identify the location of homologous genomic sequences. Of the 36 novel stratum 2 orthologs identified, six orthologs mapped to chicken chromosome 4p, 10 orthologs were located on unanchored contigs (chrUn) and 20 orthologs could not be localized to a chromosome as a search did not detect any regions of significant homology with the current chicken genome assembly.
Several chicken orthologs were identified on chromosomes other than the XAR and XCR regions of the chicken chromosomes, 1q13–q31 and 4p11–p14, respectively. We identified orthologous chicken EST/cDNA sequences for six stratum 2 genes that map to chicken chromosome 12 (GPR173, ZXDB, PFKFB1, ATP2B3, BGN, and SLC6A8) one gene that maps to chicken chromosome 14 (GSPT2), and one gene that maps to the chicken chromosome 1 outside the 1q13–q31 region (GDI1). However, we already showed that these chicken genes are paralogs rather than orthologs of human X chromosome stratum 2 genes (Figs. 3, 4). A similar strategy was used in previous studies that resulted in the misidentification of paralogs as true orthologs, and led to the definition of an independent stratum 2 (Kohn et al. 2004). The reason this has occurred is that the reciprocal best hit match strategy is inherently flawed whenever there is under-representation of a part of the genome. If a true ortholog is missing from the searched database, then closely related paralogous genes are erroneously identified as the reciprocal best hit match homologs.
Our second strategy was to use neighbor-joining phylogenetic trees to establish the orthology of chicken EST/cDNA sequences. To ascertain the true homology status of the chicken/zebra finch EST/cDNA sequences, these sequences were combined with the TreeFam derived data set, and phylogenetic trees were reconstructed for all of the stratum 2 genes, which then included translated EST/cDNA sequence data. We identified one-to-one orthology for 28 Xp11 stratum 2a genes; four orthologs map to chicken chromosome 4p, eight genes are located on unanchored contigs (chrUn), and 16 genes do not map to any part of the current chicken genome assembly. Similarly we identified eight chicken orthologs for Xq28 stratum 2b genes; one gene, which is located on an unanchored contig (chrUn), and seven genes that do not map to any part of the current chicken genome assembly. An interesting result of this analysis was that orthologs that had previously been assigned to chromosome 12 and others by the reciprocal best hit match criterion were correctly placed in the phylogenetic trees as paralogs rather than orthologs.
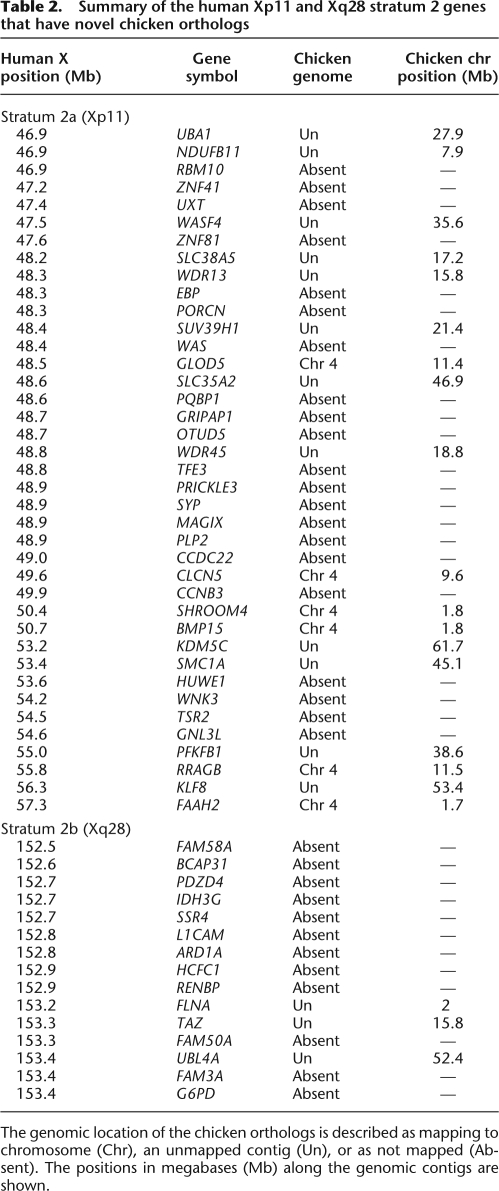
In summary, our search for novel chicken orthologs for 106 genes from human Xp11 stratum 2a genes and 45 genes from human Xq28 stratum 2b genes resulted in the detection of orthologs for 39 of the 106 Xp11 stratum 2a genes and 15 of the 45 Xq28 stratum 2b genes. Six chicken orthologs from Xp11 map to the chicken chromosome 4p suggesting that this region is not independent, but is part of the XCR of the human X chromosome. We have found evidence for the existence of an additional 48 chicken orthologs of human stratum 2 genes. Fifteen of these orthologs map to unanchored contigs that have not yet been assigned to chromosomes in the current chicken genome assembly (chrUn), and 33 chicken orthologs are not represented in the current chicken genome assembly (Table 2).
Table 2.
Summary of the human Xp11 and Xq28 stratum 2 genes that have novel chicken orthologs
The genomic location of the chicken orthologs is described as mapping to chromosome (Chr), an unmapped contig (Un), or as not mapped (Absent). The positions in megabases (Mb) along the genomic contigs are shown.
Steps were taken to try to locate the unanchored contigs (chrUn) containing the chicken orthologs in the chicken genome. EST/cDNA sequences are too small to be reliably mapped to chromosomes using FISH; therefore, larger genomic clones were sought. However, there were no BAC clones associated with the unanchored contigs, which could be used for physical mapping of these contigs. All sequences in the current chicken assembly are derived from the available chicken genomic BAC library, either unanchored or as anchored to chromosomes, and another chicken genomic library was not available to screen. These novel orthologs of stratum 2 genes from Xp11 and Xq28 could therefore not be physically mapped.
Our analysis indicated that there are still many genes from the Xp11 and Xq28 regions of the human X chromosome without identified orthologs in the chicken genome. The absence of these orthologs may mean either that they are simply missing from an incomplete chicken genome assembly, or that they have been deleted from the chicken genome. Our EST/cDNA analysis indicated that there are sequences representing 33 expressed chicken orthologs that do not have a corresponding genomic location. This implies that the assembly of the chicken genome is still incomplete.
Discussion
Our results challenge the hypothesis that there is a third, independent evolutionary block of genes on the human X chromosome. This block is conserved in all vertebrates, but forms a part of the ancestral therian X chromosome.
We show here that four genes from the putative human Xp11 stratum 2 (GPR173, KDM5C, RIBC1, and HUWE1) lie together near the telomere of Xq in two distantly related marsupials, the tammar wallaby and the American opossum. The GPR173, KDM5C, and HUWE1 genes lie close together on the X in all therian mammals including human. GPR173 and KDM5C are only 12 kb apart on the opossum X chromosome. The localization of these three genes in the tammar wallaby within two overlapping BACs, each ∼250 kb in size, implies that all three genes lie within a 500 kb region of the X chromosome in both these distantly related marsupials.
It is not clear whether these genes maintain the same arrangement in other vertebrates. We show that the GPR173 and KDM5C homologs previously detected in the chicken genome assembly (Kohn et al. 2004) are not, after all, orthologous to the GPR173 and KDM5C genes on the X chromosome of all therian mammals, and chromosome 6 in platypus. Our identification of the previously identified chicken GPR173 and KDM5C homologs as paralogs in well-conserved regions on other chromosomes now means that the locations, and even the existence, of the true chicken orthologs of the GPR173, KDM5C, RIBC1, and HUWE1 genes are unknown.
These four genes are part of a larger region on the marsupial X chromosome, which previously comprised part of the proposed stratum 2 at Xp11 (from 46.8 to 57.9 Mb) on the human X chromosome. Genes from this region of the human X chromosome are therefore located on the X chromosome of all therian mammals, implying that they were present on the therian proto-X chromosome. Their presence also on platypus chromosome 6 implies that they were present on the ancestral mammal autosome that became the proto-X when its partner acquired a sex-determining gene.
We have shown that six of these genes are located on chromosome 4 in the chicken, suggesting that they are part of the ancient conserved region of the therian X chromosome. The location of the proposed Xp11 stratum 2 genes on the therian X chromosome, platypus chromosome 6, and chicken chromosome 4 supports the hypothesis that this region was part of an ancestral mammalian autosome, which eventually became the therian X chromosome, after monotremes diverged from therian mammals 166 Mya and prior to divergence of marsupials and eutherians 145 Mya (Kohn et al. 2004, 2006).
We also investigated more widely the ortholog and paralog status of human Xp11 and Xq28 genes that have been proposed to make up stratum 2 of the human X chromosome. These two regions have been suggested to have evolutionary origins (Kohn et al. 2004) that are different from the previously defined recently added and conserved regions of the human X chromosome (Graves 1995). Our analysis of Xp11 and Xq28 homologs and their genomic context showed that chicken homologs of human Xp11 and Xq28 genes located on chicken chromosomes 1, 12, and 26 were not orthologs of the X chromosome genes. We showed that these chicken genes were paralogs of the X chromosome genes. These paralogous genes were conserved within the same genomic context and located on homologous autosomal regions in three mammalian species, including human, rodent, and a distantly related marsupial species.
Interestingly, the paralogs of Xp11 and Xq28 genes, and their counterparts in other species, are not scattered randomly throughout the genomes of either human, rat, opossum, or chicken as might be expected. Instead they tend to be located on the same chromosomes in all species, although they are not clustered together, spanning over 100 Mb in the case of paralogs located on human chromosomes 3 and 12 (Fig. 4). Moreover, paralogs from both the Xp11 and Xq28 regions are intermingled on the same chromosomes in all the species examined (Fig. 4). This suggests that the Xp11 and Xq28 regions were originally located together in mammals and birds, and may have arisen from an ancient genome duplication or segmental duplication. Rearrangements and a dramatic loss of genes in this duplicated region on the human X chromosome have resulted in the current gene content of the Xp11 and Xq28 regions.
Our phylogenetic analysis of combined TreeFam and cDNA/EST sequence data has identified many more chicken orthologs of Xp11 and Xq28 genes than was previously reported using a reciprocal best match BLAST strategy (Kohn et al. 2004; Ross et al. 2005). We have reassigned the chicken orthologs of Xp11 and Xq28 genes and shown that several of these genes map to chicken chromosomes 4p11–p14 and 1q13–q31, consistent with their human counterparts having an origin in the conserved and recently added regions of the human X chromosome, respectively. The location of 48 of 151 (31%) chicken orthologs of single copy human Xp11 and Xq28 genes is still unknown, because they map to an unanchored contig that has not yet been assigned to a chromosome. The presence of these chicken orthologs in the cDNA/EST database, but not the chicken genomic assembly indicates that these genes are not missing from the chicken genome, but that the genome assembly is incomplete in this region.
What could have driven the rearrangement and gene loss from the Xp11 and Xq28 regions of the human X chromosome? The region around Xp11 on the short arm, and Xq28 on the long arm of the human X chromosome contains several large inverted repeats, which contain concentrations of the expanded gene families (e.g., MAGE, SSX) with testis-specific expression patterns. It has been proposed that the formation of cruciform structures by the inverted repeat sequences might permit escape of crucial genes involved in late spermatogenesis from meiotic sex chromosome inactivation (Skaletsky et al. 2003; Warburton et al. 2004). Alternatively, it has been suggested that the formation of the inverted repeat secondary structures might suppress expression of genes within them (Wu and Xu 2003). It is therefore possible that the formation of inverted repeat secondary structures contributed to the rearrangement and gene loss from these regions of the human X chromosome.
Our results indicate that neither human Xp11 nor Xq28 have an independent evolutionary origin. Human Xp11 contains genes from both the XAR and XCR of the human X chromosome. The XAR (Xpter-Xp11.3) was added to the human X chromosome after marsupials diverged from eutherians 148 Mya and before the eutherian radiation 105 Mya. The remainder of Xp11 and Xq28 were part of the original autosomal pair that became the therian sex chromosome pair, following the divergence of the monotremes from the therians 166 Mya. Thus, there is no stratum 2 on the human X chromosome that arose from an independent genome block. This result, as well as the demonstration that the conserved region of the human X is autosomal in monotremes, requires a rethinking of how and when the mammal X and Y chromosomes became different.
The dates estimated from sequence divergence of genes with partners on the Y chromosome (Lahn and Page 1999) cannot be supported. The claims that initiation of differentiation of the oldest stratum (Stratum 1) of X and Y occurred 320–240 Mya are inconsistent with incontrovertible new evidence (Veyrunes et al. 2008) that this region was autosomal until 166–145 Mya. Although the claim for an independent stratum 2 (Lahn and Page 1999) is not supported, the dating of divergence of the two genes in this region (170–130 Mya) is consistent with an origin 166–145 Mya. The dates estimated for stratum 3 as 75–130 Mya and stratum 4 (30–50 Mya) (Lahn and Page 1999) are each compatible with the dating of fusion with an autosomal region (to both the X and Y) after the divergence of marsupials 145 Mya and the eutherian radiation 105 Mya. This new understanding of how and when the human sex chromosomes evolved is summarized in Figure 5.
Figure 5.
Summary of the current understandings of how and when the human sex chromosomes evolved. The XCR and XAR of the human sex chromosomes are shown and their corresponding regions of the tammar wallaby, platypus, and chicken. The proposed stratum 2 is now known to be a part of the XCR of the human X chromosome and does not have a separate evolutionary origin. Our localization of four Xp11 orthologs to chicken 4p suggests that stratum 2 genes are located on chicken chromosome 4p, although the majority of these genes were unable to be mapped. Divergence times given between each of the groups are millions of years ago.
Methods
Polymerase chain reaction (PCR)
PCR amplifications were carried out using 15 pmol of each primer (Geneworks), 2.0 mM each of dATP, dCTP, dGTP, dTTP (Roche), and 0.625U Taq polymerase in the recommended buffer containing 1.5 mM MgCl2 (Promega). Following an initial denaturation at 94°C for 2 min, cycling conditions were 35 cycles of 94°C for 30 sec; 50–59°C for 30 sec; 72°C for 1 min; with a final extension of 10 min at 72°C. Primer pairs and annealing temperatures are listed in Supplemental Table S1. All PCR products were cloned into the TA TOPO Cloning Kit for sequencing and verification of their identity. Sequencing was done at the Australian Genomic Research Facility (AGRF), Brisbane, Australia.
BAC library screening
PCR generated DNA probes were radioactively labeled using 32P-dCTP using the Megaprime DNA labeling system. Alternatively, 40 bp overgo probes (Supplemental Table S1) were labeled with [32P]dCTP and [32P]dATP (Ross et al. 1999). DNA probes were hybridized to BAC library filters for 16 h at 60°C in Church's buffer (Church and Gilbert 1984) and washed twice in 2× SSC/0.1% SDS at 60°C. Overgo probes were washed further, twice more in 0.1× SSC/0.1% SDS at 60°C (Ross et al. 1999). Hybridization was detected by exposure to X-ray film for up to 7 d.
Positive BAC clones were isolated from the commercially available female M. eugenii (tammar wallaby) BAC library (Arizona Genomics Institute [AGI]). M. domestica (opossum) and O. anatinus (platypus) BAC clones were obtained from the BACPAC Resources Center at the Children's Hospital Oakland Research Institute (CHORI) (http://bacpac.chori.org/home.htm).
Fluorescence in situ hybridization (FISH)
Tammar wallaby, opossum, and platypus male fibroblast cells were cultured and metaphase chromosome spreads prepared on glass slides, as previously described (Koina et al. 2005). BAC DNA was labeled with Spectrum Orange or Spectrum Green (Abbott Molecular Inc.) by nick translation and hybridized to the chromosome preparations, as previously described (Alsop et al. 2005). Multiple BACs were hybridized sequentially to the same slide, as previously described (McMillan et al. 2007). Fluorescence was visualized with a Zeiss Axioplan epifluorescence microscope fitted with a 100-W mercury lamp and a SPOT RT Monochrome CCD camera (Diagnostic Instruments Inc.). IPLab imaging software (Scanalytics Inc.) was used to capture and enhance images.
Multispecies comparison of Xp11 and Xq28 homologs
Paralogs of human Xp11 and Xq28 genes, and their genomic locations were obtained from Ensembl v50 (July 2008) (http://www.ensembl.org/index.html) using the BioMart tool (Vilella et al. 2009). We also obtained the protein-coding genes and their genomic location data from the 500-kb flanking region on either side of these paralogs. Orthologous regions of these 1-Mb genomic regions in rat, opossum, chicken, and frog, each containing a paralog of a human Xp11 or Xq28 gene, were identified using BioMart tool of Ensembl v50 and compared manually (Figs. 3, 4).
TreeFam database analysis
The TreeFam (Li et al. 2006) database contains phylogenetic gene trees for all animal genes. The gene trees are calculated using nonanimal (plant and yeast) genes as outgroups. The TreeFam database can also compute the topology of the phylogenetic trees and assign within species paralogs and interspecies orthologs. The TreeFam data are submitted in MySQL data dumps at ftp://ftp.sanger.ac.uk/pub/treefam/. The TreeFam database records were processed using in-house Perl scripts to search for orthologs of human X genes in the chicken genome.
Reciprocal best hit match search of EST/cDNA databases
Chicken EST/cDNA sequences were downloaded from the BBSRC ChickEST database at http://www.chick.manchester.ac.uk/ (Boardman et al. 2002; Hubbard et al. 2005). Zebra finch EST and cDNA sequences were downloaded from the Songbird Neurogenomics (SoNG) Initiative website http://titan.biotec.uiuc.edu/songbird/ (Replogle et al. 2008). Human cDNA sequences (including alternatively spliced transcripts) were downloaded from Ensembl v50. Human cDNA sequences of stratum 2 genes were used as query sequences for reciprocal best hit BLAST search (Altschul and Lipman 1990) against the chicken/zebra finch EST/cDNA database. Default nucleotide BLAST search parameters were used, except e-value cutoff was set to 1.0 and the minimum alignment length was set to be more than 30 bp. BLAST search results were parsed and full-length EST/cDNA sequences that passed the above filter criteria were aligned against the repeat masked human genome to identify reciprocal best hit matches. All reciprocal best hit match sequences were then mapped to the chicken genome using the megablast tool of the BLAST suite.
Addition of EST/cDNA sequences to TreeFam neighbor-joining phylogenetic trees
Chicken/zebra finch EST/cDNA sequences were translated in all six reading frames to make a translated protein database. All protein sequences that make up a gene family in the TreeFam database were extracted using in-house Perl scripts. These protein sequences were used to isolate homologous chicken/zebra finch EST/cDNA sequences using default BLASTP search parameters. A subdata set of the homologous chicken/zebra finch EST/cDNA six-frame translated sequences was created for each gene family after parsing of the BLASTP results. This subdata set was then filtered for true homologous sequences using the hidden-Markov model search using the HMMER program (Eddy 1998) (E-value cutoff 0.1) and gene family HMM models obtained from TreeFam. All positive homologous sequences for a gene family were merged with protein sequences of the gene family (obtained from TreeFam) and multiple sequence alignments were performed using ClustalW (Higgins et al. 1994). ClustalW parameters were set to obtain accurate pairwise alignments for multiple sequence alignments (parameters: PWGAPOPEN = 50, PWGAPEXT = 2, GAPOPEN = 3, GAPEXT = 1, NUMITER = 25, MAXDIV = 30, and the GONNET series matrix). TreeBeST (Li et al. 2006) was then used to filter and retain columns with a score greater than 15, and construct neighbor-joining trees. The tree-building algorithm parameters were set to “t = kimura” along with other default parameters. The resulting neighbor-joining phylogenetic trees were than read for topology using “ortho” module of the TreeBest program. The results were then processed using in-house Perl scripts. The chicken/zebra finch EST/cDNA sequences that were orthologous to human stratum 2 genes were then mapped to the chicken genome using the megablast tool of the BLAST suite.
Acknowledgments
We thank R. Doherty for her expert technical assistance. This work was supported by ARC grants to M.L.D. and J.A.M.G., and the ARC Centre of Excellence in Kangaroo Genomics. Thank you to the anonymous reviewers for their comments.
Author contributions: M.L.D. carried out the library screening and identification of BAC clones of human Xp11.22–11.23 genes and was responsible for the design of the study and preparation of the manuscript. H.R.P. carried out the bioinformatics analysis of the Xp11 and Xq28 regions. P.D.W. identified BAC clones of the human Xq22.1–22.2 genes. DAM and PDW carried out the localization experiments. J.A.M.G. contributed to the design of the study, and preparation of the manuscript.
Footnotes
[Supplemental material is available online at www.genome.org.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.088625.108.
References
- Alsop AE, Miethke P, Rofe R, Koina E, Sankovic N, Deakin JE, Haines H, Rapkins RW, Graves JAM. Characterizing the chromosomes of the Australian model marsupial Macropus eugenii (tammar wallaby) Chromosome Res. 2005;13:627–636. doi: 10.1007/s10577-005-0989-2. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Lipman DJ. Protein database searches for multiple alignments. Proc Natl Acad Sci. 1990;87:5509–5513. doi: 10.1073/pnas.87.14.5509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bininda-Emonds ORP, Cardillo M, Jones KE, MacPhee RDE, Beck RMD, Grenyer R, Price SA, Vos RA, Gittleman JL, Purvis A. The delayed rise of present-day mammals. Nature. 2007;446:507–512. doi: 10.1038/nature05634. [DOI] [PubMed] [Google Scholar]
- Boardman PE, Sanz-Ezquerro J, Overton IM, Burt DW, Bosch E, Fong WT, Tickle C, Brown WR, Wilson SA, Hubbard SJ. A comprehensive collection of chicken cDNAs. Curr Biol. 2002;12:1965–1969. doi: 10.1016/s0960-9822(02)01296-4. [DOI] [PubMed] [Google Scholar]
- Charlesworth D, Charlesworth B, Marais G. Steps in the evolution of heteromorphic sex chromosomes. Heredity. 2005;95:118–128. doi: 10.1038/sj.hdy.6800697. [DOI] [PubMed] [Google Scholar]
- Church GM, Gilbert W. Genomic sequencing. Proc Natl Acad Sci. 1984;81:1991–1995. doi: 10.1073/pnas.81.7.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deakin JE, Koina E, Waters PD, Doherty R, Patel VS, Delbridge ML, Dobson B, Fong J, Hu Y, van den Hurk C, et al. Physical map of two tammar wallaby chromosomes: A strategy for mapping in non-model mammals. Chromosome Res. 2008;16:1159–1175. doi: 10.1007/s10577-008-1266-y. [DOI] [PubMed] [Google Scholar]
- Delbridge ML, Graves JAM. Origin and evolution of spermatogenesis genes on the human sex chromosomes. Soc Reprod Fertil Suppl. 2007;65:1–17. [PubMed] [Google Scholar]
- Eddy SR. Profile hidden Markov models. Bioinformatics. 1998;14:755–763. doi: 10.1093/bioinformatics/14.9.755. [DOI] [PubMed] [Google Scholar]
- Graves JAM. The origin and function of the mammalian Y chromosome and Y-borne genes—An evolving understanding. Bioessays. 1995;17:311–320. doi: 10.1002/bies.950170407. [DOI] [PubMed] [Google Scholar]
- Graves JAM. Sex chromosome specialization and degeneration in mammals. Cell. 2006;124:901–914. doi: 10.1016/j.cell.2006.02.024. [DOI] [PubMed] [Google Scholar]
- Graves JAM. Weird animal genomes and the evolution of vertebrate sex and sex chromosomes. Annu Rev Genet. 2008;42:565–586. doi: 10.1146/annurev.genet.42.110807.091714. [DOI] [PubMed] [Google Scholar]
- Graves JAM, Shetty S. Sex from W to Z: Evolution of vertebrate sex chromosomes and sex determining genes. J Exp Zool. 2001;281:472–481. doi: 10.1002/jez.1088. [DOI] [PubMed] [Google Scholar]
- Higgins D, Thompson J, Gibson T, Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubbard SJ, Grafham DV, Beattie KJ, Overton IM, McLaren SR, Croning MD, Boardman PE, Bonfield JK, Burnside J, Davies RM, et al. Transcriptome analysis for the chicken based on 19,626 finished cDNA sequences and 485,337 expressed sequence tags. Genome Res. 2005;15:174–183. doi: 10.1101/gr.3011405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohn M, Kehrer-Sawatzki H, Vogel W, Graves JAM, Hameister H. Wide genome comparisons reveal the origins of the human X chromosome. Trends Genet. 2004;20:598–603. doi: 10.1016/j.tig.2004.09.008. [DOI] [PubMed] [Google Scholar]
- Kohn M, Hogel J, Vogel W, Minich P, Kehrer-Sawatzki H, Graves JAM, Hameister H. Reconstruction of a 450-My-old ancestral vertebrate protokaryotype. Trends Genet. 2006;22:203–210. doi: 10.1016/j.tig.2006.02.008. [DOI] [PubMed] [Google Scholar]
- Koina E, Wakefield MJ, Walcher C, Disteche CM, Whitehead S, Ross M, Graves JAM. Isolation, X location and activity of the marsupial homologue of SLC16A2, an XIST-flanking gene in eutherian mammals. Chromosome Res. 2005;13:687–698. doi: 10.1007/s10577-005-1006-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahn BT, Page DC. Four evolutionary strata on the human X chromosome. Science. 1999;286:964–967. doi: 10.1126/science.286.5441.964. [DOI] [PubMed] [Google Scholar]
- Li H, Coghlan A, Ruan J, Coin LJ, Heriche JK, Osmotherly L, Li R, Liu T, Zhang Z, Bolund L, et al. TreeFam: A curated database of phylogenetic trees of animal gene families. Nucleic Acids Res. 2006;34:D572–D580. doi: 10.1093/nar/gkj118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMillan D, Miethke P, Alsop AE, Rens W, O'Brien P, Trifonov V, Veyrunes F, Schatzkamer K, Kremitzki CL, Graves T, et al. Characterizing the chromosomes of the platypus (Ornithorhynchus anatinus) Chromosome Res. 2007;15:961–974. doi: 10.1007/s10577-007-1186-2. [DOI] [PubMed] [Google Scholar]
- Nanda I, Zend-Ajusch E, Shan Z, Grutzner F, Schartl M, Burt DW, Koehler M, Fowler VM, Goodwin G, Schneider WJ, et al. Conserved synteny between the chicken Z sex chromosome and human chromosome 9 includes the male regulatory gene DMRT1: A comparative (re)view on avian sex determination. Cytogenet Cell Genet. 2000;89:67–78. doi: 10.1159/000015567. [DOI] [PubMed] [Google Scholar]
- Replogle K, Arnold AP, Ball GF, Band M, Bensch S, Brenowitz EA, Dong S, Drnevich J, Ferris M, George JM, et al. The Songbird Neurogenomics (SoNG) Initiative: Community-based tools and strategies for study of brain gene function and evolution. BMC Genomics. 2008;18:131. doi: 10.1186/1471-2164-9-131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross MT, LaBrie S, McPherson J, Stanton VP., Jr . Screening large insert libraries by hybridisation. In: Dracopoli ST, et al., editors. Current protocols in human genetics. Wiley; New York: 1999. pp. 5.6.1–5.6.52. [Google Scholar]
- Ross MT, Grafham DV, Coffey AJ, Scherer S, McLay K, Muzny D, Platzer M, Howell GR, Burrows C, Bird CP, et al. The DNA sequence of the human X chromosome. Nature. 2005;434:325–337. doi: 10.1038/nature03440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid M, Nanda I, Guttenbach M, Steinlein C, Hoehn M, Schartl M, Haaf T, Weigend S, Fries R, Buerstedde JM, et al. First report on chicken genes and chromosomes 2000. Cytogenet Cell Genet. 2000;90:169–218. doi: 10.1159/000056772. [DOI] [PubMed] [Google Scholar]
- Skaletsky H, Kuroda-Kawaguchi T, Minx PJ, Cordum HS, Hillier L, Brown LG, Repping S, Pyntikova T, Ali J, Bieri T, et al. The male-specific region of the human Y chromosome is a mosaic of discrete sequence classes. Nature. 2003;423:825–837. doi: 10.1038/nature01722. [DOI] [PubMed] [Google Scholar]
- Veyrunes F, Waters PD, Miethke P, Rens W, McMillan D, Alsop AE, Grutzner F, Deakin JE, Whittington CM, Schatzkamer K, et al. Bird-like sex chromosomes of platypus imply recent origin of mammal sex chromosomes. Genome Res. 2008;18:965–973. doi: 10.1101/gr.7101908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilella AJ, Severin J, Ureta-Vidal A, Heng L, Durbin R, Birney E. EnsemblCompara GeneTrees: Complete, duplication-aware phylogenetic trees in vertebrates. Genome Res. 2009;19:327–335. doi: 10.1101/gr.073585.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warburton PE, Giordano J, Cheung F, Gelfand Y, Benson G. Inverted repeat structure of the human genome: The X-chromosome contains a preponderance of large, highly homologous inverted repeats that contain testes genes. Genome Res. 2004;14:1861–1869. doi: 10.1101/gr.2542904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu CI, Xu EY. Sexual antagonism and X inactivation—the SAXI hypothesis. Trends Genet. 2003;19:243–247. doi: 10.1016/s0168-9525(03)00058-1. [DOI] [PubMed] [Google Scholar]