Abstract
We investigated functional epigenetic changes that occur in primary human T lymphocytes during entry into the cell cycle and mapped these at the single-nucleosome level by ChIP-chip on tiling arrays for chromosomes 1 and 6. We show that nucleosome loss and flanking active histone marks define active transcriptional start sites (TSSs). Moreover, these signatures are already set at many inducible genes in quiescent cells prior to cell stimulation. In contrast, there is a dearth of the inactive histone mark H3K9me3 at the TSS, and under-representation of H3K9me2 and H3K9me3 defines the body of active genes. At the DNA level, cytosine methylation (meC) is enriched for nucleosomes that remain at the TSS, whereas in general there is a dearth of meC at TSSs. Furthermore, a drop in meC also marks 3′ transcription termination, and a peak of meC occurs at stop codons. This mimics the 3′ nucleosomal distribution in yeast, which we show does not occur in human T cells.
Mature T lymphocytes can remain in a quiescent (G0) state in the peripheral blood for many years before entering the cell cycle and proliferating in response to an antigenic stimulus. Costimulation via CD3 and CD28 induces signal transduction pathways that result in commitment and entry into the cell cycle, the growth cycle, and expression of T cell effector (activation) functions (Acuto and Michel 2003). Many of the transcription factors that regulate expression of genes involved in these pathways have been characterized in some detail (Diehn et al. 2002). For example, RUNX3 is required for CD8 development and silencing of the CD4 locus (Woolf et al. 2003; Taniuchi and Littman 2004; Grueter et al. 2005), and IRF4 regulates T cell activation and cytokine production (Hu et al. 2002; Pernis 2002).
Alterations in chromatin compaction need to occur in order to allow access by proteins that regulate transcription or DNA replication (for reviews, see Mellor 2006; Rando and Ahmad 2007). Changes in chromatin structure can occur as a result of post-translational modifications of histones such as methylation, acetylation, sumoylation, phosphorylation, and ubiquitination (for reviews, see Bernstein et al. 2007; Kouzarides 2007). DNA cytosine methylation (meC) is also implicated in maintaining a generally inactive heterochromatic state and gene silencing (Jones and Baylin 2002). Recently, we demonstrated that CpG methylation experimentally targeted to a promoter inserted into the genome of mammalian cells directly caused gene repression and led to heritable changes in the histone code that persisted through many cell divisions (Smith et al. 2008). Under normal conditions, epigenetic changes are often associated with chromatin reprogramming that is laid down during DNA replication (Bird 2002). A handful of examples also exist for active demethylation/methylation in somatic cells, such as demethylation of specific CpGs in the promoter of the IL2 gene (Murayama et al. 2006) and strand-specific CpG demethylation and methylation (Kangaspeska et al. 2008; Metivier et al. 2008). Histone changes can also be replication-dependent (Sarma and Reinberg 2005); however, such epigenetic marks often change without cells entering S phase (Chow et al. 2005).
Genome-wide analyses of histone modifications by chromatin immunoprecipitation (ChIP) have resulted in a series of high-content publications from the ENCODE Consortium (Genome Res, June 2007) as well as from individual labs (Roh et al. 2004, 2005, 2006; Bernstein et al. 2006, 2007; Barski et al. 2007). Structural features of chromatin have also been classified by mapping DNase I hypersensitive sites and cytosine methylation (Crawford et al. 2006; Eckhardt et al. 2006). In genome-wide studies on T cells, active genes are shown to be associated with mono-, di-, and trimethylated histone H3 lysine 4 (H3K4me1, 2, and 3), H3K9me1, and acetylated H3 (H3Ac) near the transcriptional start site (TSS), and H2BK5me1, H3K9me2/3, H3K36me3, H3K27me1, and H4K20me1 throughout actively transcribed regions (Bernstein et al. 2005; Liu et al. 2005; Vakoc et al. 2005; Barski et al. 2007; Wang et al. 2008). In contrast, inactive genes are associated with high levels of H3K27me3, H3K79me3, and historically H3K9 mono- and dimethylation (Bannister et al. 2001; Roopra et al. 2004; Vakoc et al. 2006; Barski et al. 2007) and low levels of H3K27me1, H3K36me3, H3K9me1, H4K20me1, and H3K4 methylation (Barski et al. 2007). There are also genes in ES cells that carry marks associated with both activation and repression (“bivalent” marks), such as H3K4me3 and H3K27me3 (Azuara et al. 2006; Bernstein et al. 2006; Roh et al. 2006; Barski et al. 2007; Jorgensen et al. 2007; Cui et al. 2009).
Although a lot is known about the distribution of epigenetic marks throughout the genome, relatively little is known about how they change, for example, during entry into the cell cycle (G0→G1). We investigated active and repressive histone marks at the single-nucleosome level, as well as CpG DNA methylation, across two chromosomes (chr 1 and chr 6) in primary human T cells during the G0→G1 transition. Epigenetics was related to expression data derived from both conventional expression arrays and tiling arrays.
Results
Epigenetic analyses of human T cells during G0→G1
We wished to analyze nucleosome positioning and investigate epigenetic marks at single-nucleosome resolution in primary human T cells during entry into the first cell cycle from a quiescent (G0) state. Micrococcal nuclease (MNase) digests DNA that is not protected by tightly bound protein complexes. A genome-wide analysis of MNase and histone H3 ChIP samples showed that MNase predominantly releases nucleosomes rather than nonhistone complexes and is a more accurate way of determining nucleosome positioning (Schones et al. 2008). Thus, we isolated nuclei from T cells in G0 and 72 h post-stimulation with PMA/ionomycin. At this point a significant number of cells are in the first cell cycle (Supplemental Fig. S1A; Lea et al. 2003). Mainly mononucleosomes were then released by digestion with MNase (Supplmental Fig. S1B), and native ChIP was carried out with antibodies to activated (H3K4me3, H3Ac [H3K9/K14Ac]) and repressive (H3K9me2, H3K9me3) histone marks. Analysis of DNA from each ChIP showed that the predominant length of DNA is consistent with ChIP isolation from mononucleosomes for H3Ac and H3K4me3, whereas H3K9me2 and H3K9me3 isolated both mono- and dinucleosome-length DNA (Supplemental Fig. S1C). We also carried out immunoprecipitations for meC using purified nucleosomal DNA from the same samples as well as randomly sheared (sonicated) nucleosome-free DNA. We assessed nucleosomal occupancy at promoters directly by comparing input nucleosomal DNA with total sonicated DNA. The enrichment obtained in each ChIP experiment was validated by qPCR for known active and inactive genes, or known methylated and nonmethylated regions (Supplemental Fig. S2A–D). Enrichment was also confirmed for anti-meC immunoprecipitations using methylated oligodeoxynucleotides (Supplemental Fig. S2E). The selected DNA was then used to probe high-resolution tiling arrays that cover chromosomes 1 and 6. Enrichment data were visualized on the IGB software suite (Affymetrix, Inc.) and were based on statistically significant signal values processed using the MAT software package, which normalized for specific probe behavior based on sequence (Fig. 1A; Johnson et al. 2006). Where indicated, the Affymetrix TAS software package was used instead of MAT. Each ChIP experiment was normalized against “input” nucleosomal DNA by subtracting log input signal values from log ChIP signal values. P-value cut-offs with an enrichment value <10−5 or <10−4 were also applied (raw data files available on request). Gene-specific epigenetic patterns (P-value < 10−4 or where enrichment peaks identified manually were consistent in quiescent and stimulated cells) were subsequently verified by ChIP–qPCR or bisulfite sequencing to confirm the validity of the data (Supplemental Figs. S2F–I, S3). Files containing complete chromosome signal values were extracted from IGB in SGR format. These were used in global analyses where values were binned and averaged every 50 bp or 400 bp around fixed positions in all genes, such as the TSS (see below). Data are typically shown for chr 1, but similar analyses for chr 6 agree, and examples are shown in the Supplemental Results (Supplemental Fig. SR1).
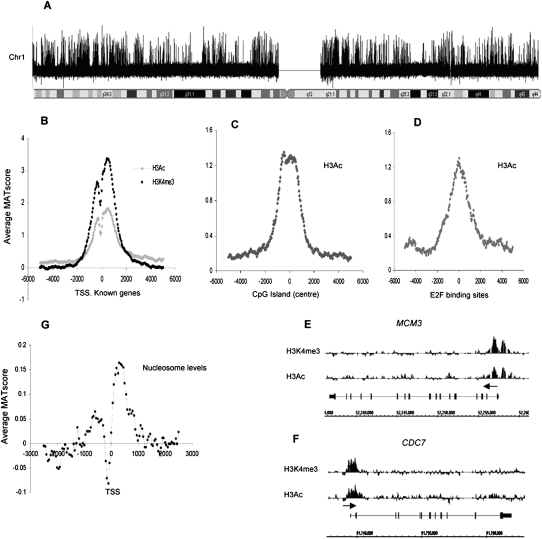
Figure 1.
Activated histone marks and nucleosome positioning in quiescent T cells. (A) Relative enrichment of H3K4me3 across chr 1 following chromatin immunoprecipitation experiments. All analyses of microarray signal intensity were performed using the MAT program (output: log signal values). (B) Average relative enrichment based on average MAT scores (y-axis) of active histone marks H3Ac (H3K9/14Ac) and H3K4me3 across a 10-kb region centered on the TSS (x-axis) for 2104 unique TSSs, corresponding to 1935 known chr 1 genes. Values were binned at 50-bp intervals for all analyses unless stated otherwise. (C) Average MAT score for H3Ac for 2463 CpG islands on chr 1 aligned to the center of each CpG island. (D) Average MAT score for H3Ac centered on a set of 1000 E2F binding sites on chr 1 identified by The ENCODE Project Consortium (2007). (E,F) Relative enrichment (MAT score) of H3Ac and H3K4me3 for the genes encoding DNA replication and cell cycle proteins MCM3 and CDC7. (x-axis) Genomic structure and relative size of these genes, (y-axis) normalized signal values of the modifications present. (Horizontal arrow) Direction of transcription. (G) Average relative nucleosome levels at the TSS for 1935 chr 1 genes.
Nucleosome deposition and active histone modifications mark the TSS of T cell genes
The most striking enrichment associated with the TSS was for active histone marks. A strong enrichment of H3Ac (H3AcK9/K14) and H3K4me3 occurs from −2000 to +2000 bp across the TSS (Fig. 1B). A slight drop in enrichment 150–300 bp 5′ of the TSS also occurs, which could be due to “bimodal” deposition of H3Ac and H3K4me3 or as an indirect consequence of lower nucleosome levels in this region (see below). A similar distribution of these marks was observed across CpG islands and E2F1 binding sites (Fig. 1C,D), as would be expected since both are associated with large numbers of TSSs. A bimodal distribution was not seen on these occasions, as the database coordinates of such elements do not specify the direction of transcription. Specific examples of genes with active histone marks are shown in Figure 1, E and F.
We assessed histone occupancy across the TSS directly by comparing the single-nucleosome input with total sonicated DNA. There is a relative drop in nucleosomal levels 150–300 bp 5′ of the TSS, bisecting peaks of enrichment ∼500 bp up- and downstream (Fig. 1G). Nucleosomal enrichment is thus highly correlated with the position of active histone marks in T cell promoters.
Repressive histone marks in T cell genes
The “repressive” histone modification H3K9me2 was significantly underrepresented through the body of genes but not at promoters, although enrichment levels of H3K9me2 were generally low on average in these regions (Fig. 2A). A second “repressive” mark, H3K9me3, was also underrepresented through the body of genes. However, unlike H3K9me2 deposition, H3K9me3 was significantly underrepresented at the TSS (Fig. 2B). Furthermore, H3K9me2 but not H3K9me3 exhibits a small relative peak of enrichment −800 to −1200 bp upstream of the TSS, which partially overlaps with the active histone signature. The complete H3K9me2 TSS profile is unchanged when mapped against the top 50% of genes enriched for H3Ac or H3K4me3, indicating that these opposing histone marks are indeed found at the same genes in many instances (Supplemental Fig. S4A). When the top 100 genes enriched for H3K9me2 were assessed on an individual basis, significant overlap of active and inactive marks were seen in 30% of cases. In contrast, overlap of the two active histone marks occurred in all cases when the top 100 genes enriched for each active histone mark were assessed (Supplemental Table S1). Gene-specific examples are shown in Figure 2, E–H, which highlights the global averages while also demonstrating the presence of high H3K9me2 and H3K9me3 outside known genes.
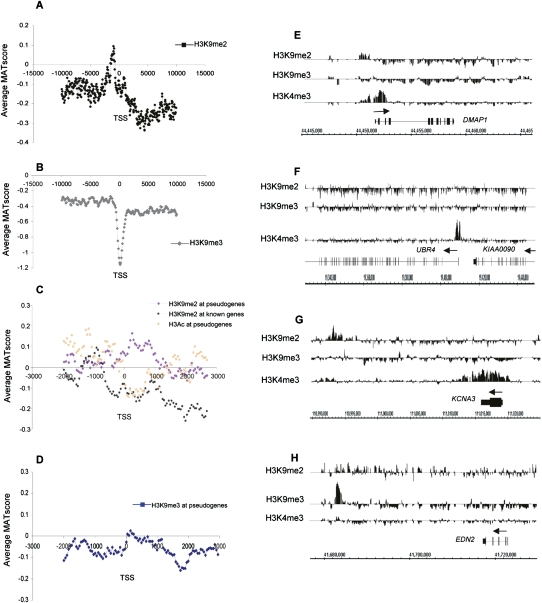
Figure 2.
H3K9me2 and H3K9me3 patterns in quiescent T cells. (A,B) Average MAT score for the repressive histone marks H3K9me2 (A) and H3K9me3 (B) (y-axis) across a 20-kb region centered on the TSS of 1935 chr 1 genes (x-axis). (C) Average MAT score for the H3K9me2 modification in all known and predicted pseudogenes (pink line; 2229 loci) and known protein-encoding genes (black line) on chr 1. (Orange) H3Ac enrichment for the same group of pseudogenes. (D) Average MAT score for H3K9me3 modification in all known and predicted chr 1 pseudogenes (blue line; 2229 loci). (E–H) Examples of relative enrichment of H3K9me2, HeK9me3, and H3K4me3 modifications for the genes DMAP1 (E), UBR4 and KIAA0090 (F), KCNA3 (G), and EDN2 (H). Peaks of enrichment for H3K9me2 and H3K4me3 modifications around the TSS are seen for DMAP1. For UBR4 and KIAA0090, under-representation of the H3K9me2 and HeK9me3 marks occur throughout the gene body. Under-representation of H3K9me2 and contrasting high levels of H3K4me3 occur throughout the body of KCNA3. (G,H) An example of an intergenic H3K9me2 peak 25 kb downstream of KCNA3 (G) and an intergenic H3K9me3 peak 20 kb downstream of EDN2 (H).
The significance of the underrepresentation of these marks throughout the gene body is further highlighted when the average distribution of H3K9me2 across known genes is compared with that of pseudogenes. In pseudogenes, relatively strong enrichment of H3K9me2 occurs throughout the element body that is reciprocal to the deposition of the active histone mark H3Ac (Fig. 2C). However, unlike H3K9me2, H3K9me3 is not enriched in pseudogenes (Fig. 2D). The relationship between H3K9me2 (Fig. 2A) and nucleosomal occupancy (Fig. 1G) around the TSS is less obvious than for active histone marks. There is a partial overlap of the H3K9me2 upstream peak and nucleosome occupancy, but the dearth of H3K9me3 at the TSS maps one nucleosome downstream of the position of nucleosome loss. In addition, underrepresentation of H3K9me2 and H3K9me3 at the start of the gene body are reciprocal to nucleosomal occupancy adjacent to the TSS. Thus, underrepresentation of H3K9me3 at the TSS and both inactive marks 3′ of the TSS are active events and not just a passive result of nucleosomal loss.
meC at promoters is low on average, but nucleosomes that remain at the TSS are associated with relatively high meC levels
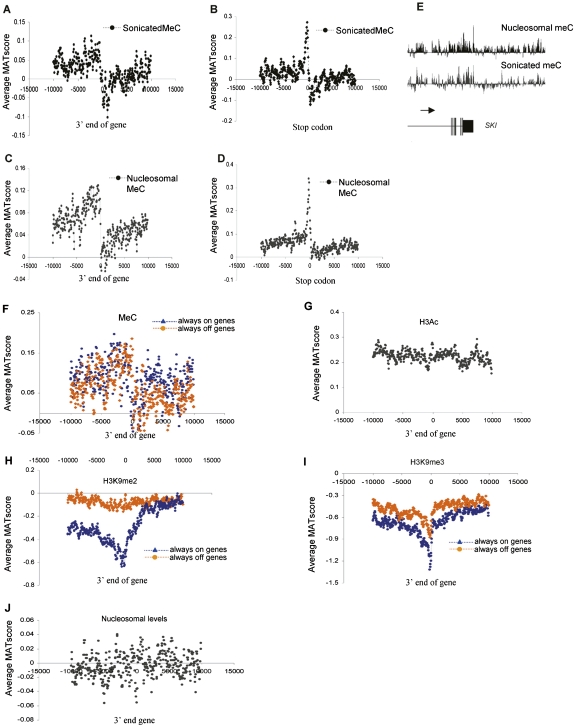
Next, we determined the distribution of cytosine methylation across TSS. Analysis of meC using sonicated DNA showed a dearth of meC at the TSS (Fig. 3A), in agreement with previously published bisulfite sequencing data (Eckhardt et al. 2006). However, a peak of meC enrichment was observed at the TSS when DNA from nucleosomal preparations was analyzed (Fig. 3B). These data suggest that there is a subpopulation of genes or cells where a nucleosome remains at the TSS, which contains DNA with a relatively high level of meC. Hypermethylation of CpG dinucleotides at the TSS was confirmed by bisulfite sequencing of enriched promoters, and hypomethylation was shown in underrepresented promoters (Fig. 3D–F; Supplemental Fig. S3). We did not observe evidence of allelic imbalance in methylation for any of the genes tested, which would manifest as a 1:1 hyper:hypo-methylated ratio of bisulfite-sequenced clones. The false-positive rate was one in 15 for regions apparently enriched for meC on the array that were subsequently shown to be hypomethylated by bisulfite sequencing.
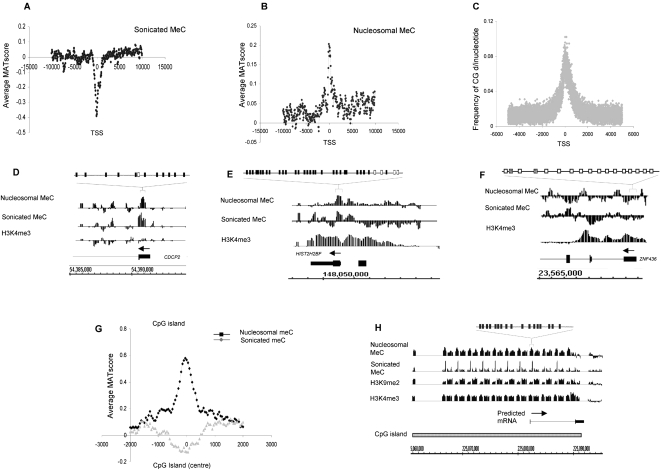
Figure 3.
Cytosine methylation in quiescent T cells. (A,B) Average MAT score for meC in randomly sonicated DNA (A) and nucleosomal DNA (B) (y-axis) across a 20-kb region centered on the TSS (x-axis), corresponding to 1935 chr 1 genes. (C) Frequency of the CG dinucleotide (y-axis) relative to the TSS for 615 sense-strand genes. (D–F) Relative enrichment (MAT score) of meC in sonicated and nucleosomal DNA samples, as well as H3K4me3 for the genes CDCP2, HIST2H2BF, and ZNF436. The presence of DNA methylation at these loci was verified by bisulfite genomic sequencing. (Black boxes) Methylated CpG; (white boxes) unmethylated CpG; (gray boxes) low level CpG methylation (<15%). (G) Average MAT score for nucleosomal meC for 2463 CpG islands on chr 1 (black line) and for meC in randomly sonicated DNA (gray line) aligned to the center of each CpG island. (H) An example of a putative CpG island on chr 1 (shaded box) with high levels of meC and H3K9me2. The presence of methylation at the peaks identified was verified by bisulfite genomic sequencing. (Black boxes) Methylated CpG; (white boxes) unmethylated CpG.
CG content at the TSS is high relative to flanking regions (Fig. 3C), further highlighting how such low meC levels must be actively maintained by the cell. The drop in average meC enrichment at CpG islands was not as pronounced as at the TSS. Moreover, meC enrichment at CpG island nucleosomal DNA was significantly higher than at the TSS. As CpG islands contain promoters as well as other CG-rich sequences, our data therefore concur with there being higher methylation levels at non-promoter CpG islands (Fig. 3G,H).
Active histone marks in inducible genes are laid down in G0 prior to their induction
On average, no significant differences in the deposition of histone marks were observed when comparing T cells in G0 with cells stimulated to enter the cell cycle (Fig. 4A; data not shown). This was unexpected, as many genes are induced and some are repressed in T cells in response to stimulation with PMA/ionomycin (NC Lea, SJ Orr, and NSB Thomas, unpubl.; Diehn et al. 2002). Moreover, when looked at individually, it was clear that epigenetic changes occurred at a number of the induced genes (Fig. 4B). Therefore, we subdivided our analyses of genes on the basis of their expression profiles during the G0→G1 transition (see Methods).
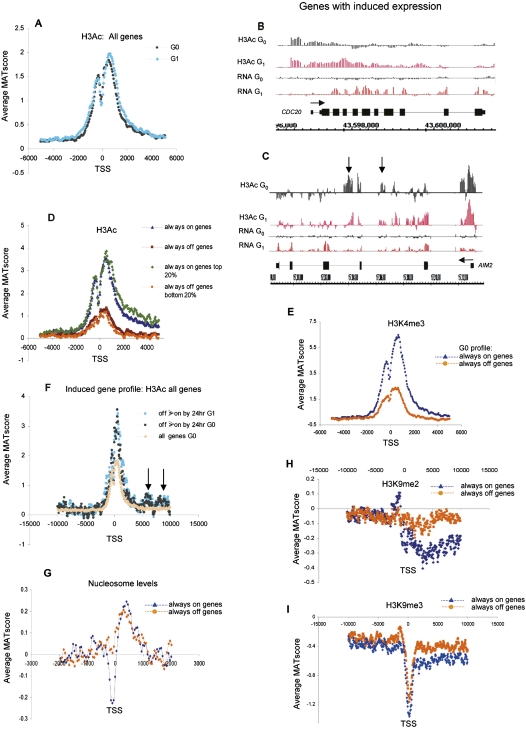
Figure 4.
Epigenetic state and nucleosome positioning in G0 and G1 for active vs. inactive genes. (A) Average MAT score (y-axis) for H3Ac across a 10-kb region relative to the TSS (x-axis) for 1935 chr 1 genes in quiescent T lymphocytes (G0; black line) and after stimulation for 72 h with PMA/ionomycin (G1; blue line). (B,C) H3Ac modification of genes with induced expression in quiescent (G0; black bars) and stimulated cells (G1; red bars). Although both AIM2 (cell cycle and tumorigenesis control regulator) and CDC20 (anaphase protein complex co-factor) become induced upon G0→G1 progression, H3Ac distribution changes only for CDC20. (Vertical arrows) Examples of downstream H3Ac peaks in introns 2 and 3 of the induced AIM gene. (D) Average MAT score for H3Ac in quiescent T cells (G0) plotted according to expression. (Blue) Average values of active genes (“always on”), (brown) inactive genes (“always off”), (green) top 20% most active genes, (orange) bottom 20% most inactive genes. (E) Average relative enrichment of H3K4me3 for “always on” (blue line) and “always off” (orange line) genes of chr 1. (F) Average MAT score for H3Ac for genes that become induced by 24 h post PMA/ionomycin stimulation. Analyses were carried out on quiescent (black line) and PMA/ionomycin-stimulated T cells (blue line). Enrichment peaks 5–10 kb downstream of the TSS are present only in induced genes (arrows). (Orange) Pattern for all 1935 chr 1 genes in G0. (G) Average relative nucleosome levels (MAT score) aligned to the TSS for “always on” (blue line) vs. “always off” (orange) genes on chr 1. Analyses were carried out on quiescent T cells. (H,I) Average MAT score for H3K9me2 (H) and H3K9me3 (I) aligned to the TSS for all “always on” (blue line) and “always off” (orange line) genes of chr 1. Analyses were carried out on quiescent T cells.
Analyses of the epigenetic state of genes that are on or off in G0 and at serial time-points during entry into G1 show that the active histone marks H3Ac and H3K4me3 display the same overall pattern, with a bimodal enrichment peak over the TSS (Fig. 4D,E). However, expressed genes (always on) are significantly more enriched (two to three times MAT score) for these histone modifications on average than nonexpressed (always off) genes. Significantly, the enrichment of both modifications corresponding to the top 20% most highly expressed “always on” genes and bottom 20% least expressed “always off” genes was the same as that for all the “always on” and “always off” genes, respectively (Fig. 4D). This again points to a less fluid and more set chromatin structure.
We then analyzed genes that are induced during cell cycle entry. The fraction of genes that were either induced or repressed was 17% (greater than a twofold change); these data suggested that corresponding epigenetic changes may indeed be hidden within the general trend. An example of epigenetic induction is shown in Figure 4B, but a number of transcriptionally induced genes remained unchanged at the epigenetic level after cell stimulation (e.g., Fig. 4C). Moreover, analyses of all genes that are off in G0 and that are then induced by 24 h or 48 h post-stimulation (induced gene sets 1 and 2) have a signature of H3Ac and H3K4me3 across the TSS in G0 that is the same as that for genes that are on at all time points (Fig. 4C,F; Supplemental Fig. S4B). These signatures were the same in quiescent cells, where these mRNAs where not significantly expressed, as in proliferating cells, when these mRNA are expressed, again suggesting that such marks may “prime” genes in quiescent cells in preparation for induction by new transcription factor input. This profile was similar for genes that displayed any pattern of induction (Supplemental Fig. S4B; data not shown). Additionally, for induced genes, H3Ac deposition was not confined to the TSS region, but rather was also found much further downstream of the promoter, forming a novel motif of smaller peaks of enrichment at around +6000 and +8000 bp from the TSS (Fig. 4C,F; Supplemental Fig. S4B). Enrichment of active histone marks at such downstream positions is also seen for H3K4me3 on an individual gene basis, but not when averaged across all genes (data not shown). This H3Ac distal motif is again the same in quiescent and proliferating cells and is therefore also established in G0 before these genes are induced.
Nucleosome loss 5′ of the TSS defines gene activity
Next, we determined whether nucleosome positioning across the TSS was dependent on gene activity. Absence of nucleosomes adjacent to the TSS (−150 bp) is specific to active “always on” genes (Fig. 4G). In contrast, the nucleosomal profile for inactive “always off” genes shows a general spread of enrichment through promoters with little nucleosomal loss immediately 5′ of the TSS. Surprisingly, this was in direct contrast to the pattern seen for active histone marks, where a drop in enrichment levels adjacent to the TSS occurs in both active and inactive genes (cf. Fig. 4D,E with 4G). These data suggest that a lower deposition of H3Ac and H3K4me3 at this position is an active event and not just the passive result of nucleosomal loss. A relative absence of nucleosomes at active genes is the same for cells in G0 and cells stimulated to enter the cell cycle (Supplemental Fig. S4C). Furthermore, relative nucleosomal loss is seen for induced genes prior to induction, providing further evidence that induction of expression is a response to a primed active epigenetic state (Supplemental Fig. S4D).
A dearth of inactive histone marks in the body of active genes
In contrast to active histone marks, the inactive chromatin marks H3K9me2 and H3K9me3 distinguish active from inactive genes through an underrepresentation in the body of expressed genes specifically (Fig. 4H). This signature is maintained as cells progress from G0 to G1 (Supplemental Fig. S4E). Moreover, active, rather than inactive, genes are associated with a small peak of H3K9me2 upstream of the TSS (Fig. 4H), suggesting further levels of control for active chromatin. In contrast, H3K9me3 has a different pattern (Fig. 4I), with a dearth at the TSS of both active and inactive genes.
Gene-specific epigenetic patterns
In addition to global averaging across all relevant genes on chr 1 and chr 6, we took a systematic approach to identify epigenetic changes associated with induction of specific genes encoding T cell proteins of functional importance (SJ Orr, R Wang, C Chronis, NC Lea, GJ Mufti, EM Marcotte, and NSB Thomas, in prep.). These data agreed with the epigenetic patterns observed on average, but there were also gene-specific epigenetic events not represented by the average profiles. These include the induction and spread of active histone marks throughout the body of the IRF4 gene accompanying induction of gene expression and are detailed in the Supplemental Results (Supplemental Fig. SR2).
meC at the TSS shows no bias toward active or inactive genes
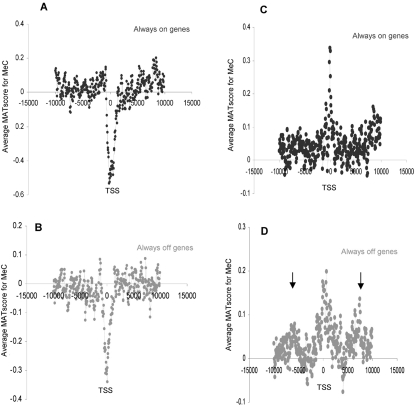
Next, we determined whether meCG-rich gene promoters were associated with any particular gene transcriptional profile. Both active (always on) and inactive (always off) genes had a dearth of meC across the TSS when total genomic sonicated DNA was studied (Fig. 5A,B). However, when residual nucleosomal DNA was analyzed, a peak of meC was seen at the TSSs of both active and inactive genes (Fig. 5C,D). However, the peak of meC enrichment was spread more evenly across the TSS for inactive genes (approximately −2000 to +2000), while expressed genes displayed an increased level of nucleosome-associated cytosine methylation precisely at the TSS (−100 to +50 bp). This apparent focusing of meC at the core TSS nucleosomes for active genes was also seen when methylation maps were made for the top 50% of H3Ac-modified promoters (Supplemental Fig. S5A). Moreover, when the top 200 meC-enriched TSSs in either nucleosomal or sonicated DNA were categorized, no bias was found in favor of nonexpressed genes. These results strongly indicate that when hypermethylation is found at a TSS, it has no definitive association with gene inactivation (Supplemental Table S3). Furthermore, it is not just a passive result of differences in CG or cytosine content (Fig. 5B), which is similar in both expressed and nonexpressed genes.
Figure 5.
Cytosine methylation status of genes according to their expression profiles. (A,B) Average MAT scores (y-axis) of meC in sonicated DNA across a 20-kb region centered on the TSS (x-axis) for “always on” (black line) and “always off” (gray line) chr 1 genes. (C,D) As for A and B, but for nucleosomal DNA.
Interestingly, nucleosomal DNA methylation at promoters is associated with small secondary peaks of enrichment at 6000–7000 bp up- and downstream of the TSS, and this is seen particularly for inactive genes (Fig. 5D; Supplemental Fig. S5C). This periodicity is not driven by CG distribution and was not observed when sonicated DNA was analyzed (Fig. 5A,B), indicating that such patterns again represent nucleosomes containing meC.
Cytosine methylation and H3K9 histone methylation patterning denote the 3′ end of genes
Since the pattern of meC peaks precisely at the TSS where hnRNA transcription initiates, we investigated whether DNA methylation also defines the 3′ terminus of genes. The levels of meC reduce significantly over a 2000-bp window immediately 5′ of 3′ hnRNA termini. There is also a peak of enrichment at stop codons (Fig. 6A–D). A gene-specific example is shown in Figure 6E. The 3′-end pattern weakly correlated with CG content, and a small increase in cytosine and CG levels occurs at the stop codon (Supplemental Fig. S6). The 3′ pattern of meC was not dependent on gene activation (Fig. 6F). As expected, the 3′ pattern of meC was different from that of the active histone mark, H3Ac, which has no 3′-end pattern (Fig. 6G). At first glance, the meC pattern seemed related to the distribution of H3K9me2 at the 3′ end of inactive genes and H3K9me3 at inactive and to a lesser extent active genes (Fig. 6H,I). These H3Ac and H3K9me3 patterns are likely to represent to some extent a dearth of these modifications in the body of genes, as seen when analyses were performed from the TSS (Fig. 4H,I). The significance of this is strengthened by the acute drop of H3K9me3 levels precisely at the 3′ end of the gene.
Figure 6.
Epigenetic patterns at the 3′ end of genes. (A,B) Average MAT score (y-axis) for meC in sonicated DNA, mapped across a 20-kb region centered on the 3′end (A) and stop codon (B) (x-axis) of 1935 chr 1 genes. (C,D) As for A and B, but for nucleosomal DNA. (E) An example of 3′ end meC (nucleosomal and sonicated) pattern for the SKI gene. (Horizontal arrow) direction of transcription. (F) Average MAT score for meC at the 3′ end of active “always on” (blue) and inactive “always off” (orange) genes on chr 1. (G) Average MAT score (y-axis) for H3Ac across a 20-kb region centered on the 3′ end of the gene (x-axis) for 1935 chr 1 genes in quiescent T cells. (H) Average MAT score for the inactive histone mark H3K9me2 in quiescent T cells plotted according to expression and centered around the 3′ end of chr 1 genes. (Blue) Average values of active “always on” genes, (orange) inactive “always off” genes. (I) As for H, but for the H3K9me3 modification. (J) Average relative nucleosome levels (MAT score) aligned with respect to the 3′ end of 1935 chr 1 genes over 20 kb in quiescent T cells.
We then determined whether the meC pattern at the 3′ end of genes is due to nucleosome positioning. The patterns of meC at the 3′ termini and stop codons were observed when either sonicated or nucleosomal DNA was analyzed, indicating that they were not due to nucleosomal positioning (Fig. 6A,C). Furthermore, nucleosome positioning did not exhibit any clear distribution patterns at either 3′ position (Fig. 6J; data not shown). This is different from yeast, which have a 3′ nucleosome pattern as well as a nucleosome positioned at the stop codon (Shivaswamy et al. 2008).
Discussion
There are three main conclusions from our work. First, an active histone code occurs around the TSS of inducible genes in quiescent human T cells prior to their induction, including a loss of core TSS nuclesomes. Secondly, although in general there is a dearth of meC at TSSs, meC is present at residual nucleosomal DNA at the TSS. Thirdly, a drop in meC as well as H3K9me3 and H3K9me2 rather than nucleosome positioning demarcates the 3′ end of genes.
An active histone code marks inducible genes prior to induction
Our work verifies previous T cell analyses, indicating a high correlation between H3Ac and H3K4me3 signals in active gene promoters (Roh et al. 2005, 2006). However, by correlating epigenetics with detailed gene expression data, we find that active histone modifications are often already set in quiescent T cells prior to stimulation and induction of gene expression. Relatively low nucleosome levels are also observed at the TSS (−150 to −300 bp) before cell stimulation and up-regulation of gene expression. This is suggestive of a more open chromatin conformation that is maintained in readiness for transcription factor (TF) regulation and not simply due to an induction of TF binding leading to a cycle of nucleosome remodeling, histone modification, and increased transcription.
Our analyses show that marking a gene promoter with active histone modifications before gene induction is a more general phenomenon in T cells that may prime genes in G0 for induction in G1. The T cells that we analyzed were predominantly memory T cells (Lea et al. 2003), and such genes may have been active in these T cells when they proliferated in the body during prior exposure to antigen. Thus, T cells that can remain quiescent in the peripheral blood for months or years may maintain the epigenetic state of sets of genes that were previously induced. Additionally, genes involved in both Th1 and Th2 responses are thought to be available for induction and only become epigenetically fixed once Th1/Th2 switching has occurred (Martins et al. 2005).
For the subset of genes in T cells that are induced at the level of transcription and epigenetic modification, such as CDC20 and IRF4, we observed an increase in the levels of active histone marks at the TSS and sometimes throughout the body of the gene. This spread of active histone marks also occurs in genes that were classified here as always on and adds to previous data that indicate clusters of active histone marks toward the 3′ end of genes (Roh et al. 2005). Perhaps such a trend favors more efficient transcriptional elongation across a whole gene or region, as has been observed previously in yeast (Kristjuhan and Svejstrup 2004). This may be through a more open chromatin structure associated with H4Ac and correlates with predicted charged protein effects (Wade et al. 1997; Shogren-Knaak et al. 2006). Interestingly, the deposition of H3Ac in the body of genes is thought to be destabilizing and leads to internal aberrant transcription events (Kaplan et al. 2003; Carrozza et al. 2005). In addition, short, capped transcripts adjacent to TSSs within annotated genes and elsewhere in the genome are very common, and some have been shown to be regulatory (Affymetrix/Cold Spring Harbor Laboratory ENCODE Transcriptome Project 2009). It is possible that some genes require an unstable local chromatin structure for appropriate transcription to override “repressive” local conditions, or that certain transcription complexes favor a more open structure, and aberrant internal starts may be suppressed by properly coordinated 5′ transcription events.
Additionally, our data indicate that the distribution of active marks downstream of the TSS is more specific and localizes to peaks of enrichment occurring +6000 and +8000 bp that are particular to H3Ac. These secondary peaks were again present both in quiescent cells, where genes associated with this motif were off, and in cycling cells in which transcription of this gene set is induced. This has not been reported previously and may correspond to downstream enhancer regions for induced genes. Active histone marks were also enriched in a number of specific genes that are “always off” in T cells (Supplemental Fig. S7), and significant but relatively low levels were noted when values for “always off” genes were averaged. This raises the question, “What is on or off?” A more robust model for inactive genes or regions is perhaps that for pseudogenes, where underrepresentation of active histone marks occurs at the 5′ end and throughout the body of such “inactive” elements.
In contrast to active histone marks at the TSS, the pattern of nucleosome positioning for active “always on” and inactive “always off” genes is distinct. Active but not inactive genes are associated with a clear dearth of nucleosomes immediately upstream of the TSS (−150 bp), which is consistent with previous findings in yeast and for studies of DNase I hypersensitive sites and histone occupancy in humans (Han and Grunstein 1988; Bernstein et al. 2004; Yuan et al. 2005; Dion et al. 2007; Boyle et al. 2008; Shivaswamy et al. 2008). Importantly, a dip in enrichment levels of active histone marks adjacent to the TSS is seen in both active and inactive genes to a similar extent, whereas nucleosomal loss is much higher for active than inactive genes. This is suggestive of an active process that regulates the placement of histone modifications at the TSS and indicates that a dip in enrichment levels of such marks is not just the passive result of nucleosomal loss.
A dearth of inactive histone marks in the body of genes
There is a relative dearth of the inactive histone marks H3K9me2 and H3K9me3 throughout the body of active “always on” genes, and a relative enrichment of the H3K9me2 mark specifically in “inactive” pseudogenes. This is consistent with previous findings for enrichment of H3K9me2 at inactive genes and heterochromatic regions but not at active genes (Bannister et al. 2001; Heard et al. 2001; Fuks 2005). This clearly develops the idea that epigenetics of the gene body are of fundamental importance, such as that for H3K36me3 (Barski et al. 2007; Bell et al. 2007; Mikkelsen et al. 2007). The trend toward underrepresentation of H3K9me2 in the body of genes is seen in supplemental data presented elsewhere, but the significance of this is unclear (Barski et al. 2007). We also found that H3K9me2 but not H3K9me3 was relatively enriched upstream of the TSS specifically for active genes, thus again highlighting a different functional repertoire for different H3K9 methylation marks. Previous findings show that the mammalian EHMT2 (also known as G9a) H3K9 mono- and dimethyltransferase mainly localizes at (active) euchromatin and has been shown to be enriched 2000 bp away from the TSS (Tachibana et al. 2001, 2002; Peters et al. 2003; Rice et al. 2003). In our study, colocalization of H3K9me2 with active histone marks on the same nucleosome occurred in 30% of cases; thus, this “inactive” mark is not always placed in an inactive context.
meC is present in nucleosomal DNA at the TSS
We first analyzed DNA cytosine methylation patterns in the same nucleosomal samples that we used to investigate histone marks. There was a peak of meC at the TSS, although “hypermethylation” (very high cytosine methylation levels that often mean most CG sites are methylated) occurs only at a minority of promoters, and relatively low meC:CG levels were largely responsible for the TSS peak observed. In contrast to this, an underrepresentation of meC was observed when randomly sonicated DNA was used rather than nucleosomal DNA. As there is a drop in nucleosomal levels at the TSS, this represents an analysis of the residual nucleosomes left at this position. These data therefore indicate that meC is present in residual nucleosomal DNA at the TSS and is likely to represent a subpopulation of cells or certain genes where nucleosomes are not lost at the TSS. As we show “always on” genes exhibiting at least as much TSS nucleosomal meC as “always off” genes, this clearly is not the consequence of simply analyzing inactive genes.
meC does not correlate with gene activity
Although inactive genes are historically associated with higher meC levels, we show here that this is not the case. We also present evidence that when DNA methylation is present at the TSS it has little correlation with transcriptional activity. However, individual CpG islands at promoters and elsewhere were often associated with consistently low cytosine methylation levels for both nucleosomal and sonicated DNA, which were reciprocal to levels of active histone marks and vice versa (Supplemental Fig. S8), thus highlighting the importance of studying individual examples as well as general trends.
It is widely held that CpGs found in promoter CpG islands are generally unmethylated, while CpG island promoter methylation correlates with transcriptional inactivation. This occurs in normal cells, and aberrant promoter hypermethylation correlates with gene silencing in cancers (Walsh and Bestor 1999; Bird 2002; Jones and Baylin 2007). Furthermore, we have shown that targeting cytosine methylation to specific genomic loci can induce gene silencing in mammalian cells (Smith et al. 2008). However, data exist that complicate this paradigm. For instance, in studies of chromosomes 6, 20, and 22 (Eckhardt et al. 2006), only 37% of methylated 5′ untranslated regions inversely correlated with mRNA expression levels for 43 genes studied. In previous studies from the same group, the data were consistent with DNA methylation being indicative of lower expression levels; however, this was not the case for one of the tissue types tested, and the number of promoter regions studied was low. Moreover, others indicate that DNA methylation flanking active histone marks is not necessarily repressive (Brinkman et al. 2007), and a more recent study indicates that the methyl cytosine binding protein MECP2 is required for promoter activity in some cases (Chahrour et al. 2008). We therefore add to the growing body of evidence for significant methylation at a subset of promoters and indicate that where methylation exists at the TSS there is no bias toward transcriptional inactivity or activity. Furthermore, we suggest that any reciprocal relationship between cytosine methylation and active histone marks is highly gene- or region-specific. Thus, much is still left to be resolved as to the extent and functional relevance of meC at gene promoters.
Cytosine methylation and H3K9 histone methylation marks the 3′ end of genes
A more surprising finding was a drop in cytosine methylation levels at the 3′ end of genes and a peak of meC enrichment at stop codons. These patterns were observed when we analyzed sonicated DNA as well as nucleosomal DNA, indicating that the patterns are not dependent on nucleosome positioning. In yeast, a “fixed” nucleosome denotes the 3′ end of genes at the stop codon (Shivaswamy et al. 2008). Our analyses and those of others (Boyle et al. 2008) indicate that humans and yeast exhibit the same general pattern of nucleosome structure at the TSS. However, we found that no definite nucleosome patterning occurs at the 3′ end of genes in human T cells, either centered around the 3′ end or at the stop codon. The pattern of meC enrichment at the 3′ end of genes was instead similar to the yeast nucleosomal patterning, being associated with a drop in levels around the point of transcription termination and a peak of enrichment at the stop codon. Such meC patterning was not dependent on gene expression and was not just a passive by-product of higher CG content. Relatively high levels of meC at the 3′ end of genes has previously been observed in plants (Zhang et al. 2006), but the landmark 3′ deposition of meC shown in our studies has not been reported previously. A recent study of imprinting in mice shows that cytosine methylation can orchestrate allele-specific transcription termination at the HM13 gene by regulating the activity of an internal promoter and second polyadenlyation site (Wood et al. 2008). In addition, previous studies have shown that splicing of the final 3′ exon and polyadenylation are coordinated and dependent reactions (Berget 1995; Cooke et al. 1999). We suggest that 3′ cytosine methylation is a relatively common phenomenon that demarcates the 3′ end of genes in human cells and may be required for normal transcription termination. We also speculate that 3′ cytosine methylation substitutes for the 3′ fixed nucleosome observed in yeast.
Significantly, H3K9 methylation also dropped to its lowest levels at the gene terminus, albeit in a more gradual manner. For H3K9me2, this pattern was specific for active genes. In contrast, for H3K9me3 the same pattern occurred in both active and inactive genes, albeit with a less pronounced 3′ dip for the inactive gene set. These findings are likely to represent a dearth of H3K9 methylation in the body of genes, agreeing with data for these marks when mapped from the TSS, but the data also indicate that the methylation state of H3K9 at the 3′ end of genes is tightly regulated.
Gene-specific examples
Examination of individual genes verified the average epigenetic profiles at the TSS and 3′ end of genes described above, as well as providing information on possible new exons and use of alternative promoters. Importantly, additional novel epigenetic motifs were also observed on a gene-by-gene basis and highlight the variety of strategies employed by the cell to regulate chromatin structure and gene expression at the DNA level.
In conclusion, by categorizing epigenetic changes according to detailed gene expression data and combining empirical global analyses with more qualitative gene-specific assessments, we have generated novel data on gene regulation as T cells are stimulated to come out of quiescence and enter the first cell cycle. We suggest that many genes are “primed” with an active chromatin structure in quiescent T cells in readiness for regulatory transcription factor activation following stimulation to enter the cell cycle. In other cases, gene induction also results in epigenetic changes and fits the model whereby transcription factor input drives chromatin remodeling and allows transcription to commence. The “inactive” chromatin mark H3K9me2 is also found upstream in gene promoters, but in this case it is specific for active genes and points to a hitherto unknown level of regulation. Instead of an expected absence of this mark consistently in gene promoters, a dearth of H3K9me2 in the gene body defines active genes. We propose that cytosine methylation is a landmark defining the 3′ terminus of genes and stop codons. We show also that meC is often present where nucleosomes remain at the TSS, and this is the case for both active and inactive genes. Moreover, the meC profile at the TSS, stop codon, and 3′ end of genes is not affected by their expression status, adding to the growing body of evidence that indicates that meC is not necessarily a repressive mark. We suggest that the meC pattern at the 3′ end of genes is an epigenetic mark defining a genetic boundary for transcriptional termination.
Methods
Routine methods
Human primary T cells were isolated from buffy coats by negative selection and stimulated with PMA/ionomycin or CD3/CD28 as we described previously (Lea et al. 2003). Preparation of nuclei and ChIP were carried out essentially according to Wagschal et al. (2007) with anti-H3Ac (H3K9/K14Ac) (Upstate Inc), anti-H3K4me3 (b8580), anti-H3K9me2 (ab7312 or 1220-25, Abcam), and anti-H3K9me3 (Upstate Inc). Extraction of DNA and RNA and bisulfite sequencing were as described previously (Smith et al. 2008), and MeDIP was carried out essentially according to Weber et al. (2005). Gene expression (U133A) and tiling (GeneChip Human Tiling 2.0R A) arrays were probed as per the manufacturer's protocols (Affymetrix). Methods are described in more detail in the Supplemental Methods.
Analysis of ChIP and RNA expression and global averaging of tiling array data
Raw array data were analyzed using the Model-based Analysis of Tiling arrays (MAT) tool (Johnson et al. 2006) and/or the Tiling Array analysis Software (TAS; Affymetrix). The MAT score results were visualized in the Integrated Genome Browser (IGB; Affymetrix). Perl scripts were used to perform global averaging of the data with respect to genome features such as TSS, 3′ termini, and CpG islands. See Supplemental Methods for full details.
Acknowledgments
We thank Kevin Ford (King's College London) and Vishwanath Iyer (University of Texas at Austin) for critical comments on the manuscript; Shirley Liu and Wei Li (Harvard University) for help and advice on analyzing tiling data using MAT; and Paul Lavender and Audrey Kelly (King's College London) for advice on nucleosome purification. This work was supported by grants from the Leukaemia Research Fund and the Department of Trade and Industry (DTI UK-Texas Bioscience Collaboration).
Footnotes
[Supplemental material is available online at www.genome.org. The array data from this study have been submitted to ArrayExpress (http://www.ebi.ac.uk/microarray-as/ae/) under accession no. E-MEXP-2188.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.085530.108.
References
- Acuto O, Michel F. CD28-mediated co-stimulation: A quantitative support for TCR signalling. Nat Rev Immunol. 2003;3:939–951. doi: 10.1038/nri1248. [DOI] [PubMed] [Google Scholar]
- Affymetrix/Cold Spring Harbor Laboratory ENCODE Transcriptome Project. Post-transcriptional processing generates a diversity of 5′-modified long and short RNAs. Nature. 2009;457:1028–1032. doi: 10.1038/nature07759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azuara V, Perry P, Sauer S, Spivakov M, Jorgensen HF, John RM, Gouti M, Casanova M, Warnes G, Merkenschlager M, et al. Chromatin signatures of pluripotent cell lines. Nat Cell Biol. 2006;8:532–538. doi: 10.1038/ncb1403. [DOI] [PubMed] [Google Scholar]
- Bannister AJ, Zegerman P, Partridge JF, Miska EA, Thomas JO, Allshire RC, Kouzarides T. Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature. 2001;410:120–124. doi: 10.1038/35065138. [DOI] [PubMed] [Google Scholar]
- Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- Bell O, Wirbelauer C, Hild M, Scharf AN, Schwaiger M, MacAlpine DM, Zilbermann F, van Leeuwen F, Bell SP, Imhof A, et al. Localized H3K36 methylation states define histone H4K16 acetylation during transcriptional elongation in Drosophila. EMBO J. 2007;26:4974–4984. doi: 10.1038/sj.emboj.7601926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berget SM. Exon recognition in vertebrate splicing. J Biol Chem. 1995;270:2411–2414. doi: 10.1074/jbc.270.6.2411. [DOI] [PubMed] [Google Scholar]
- Bernstein BE, Liu CL, Humphrey EL, Perlstein EO, Schreiber SL. Global nucleosome occupancy in yeast. Genome Biol. 2004;5:R62. doi: 10.1186/gb-2004-5-9-r62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein BE, Kamal M, Lindblad-Toh K, Bekiranov S, Bailey DK, Huebert DJ, McMahon S, Karlsson EK, Kulbokas EJ, III, Gingeras TR, et al. Genomic maps and comparative analysis of histone modifications in human and mouse. Cell. 2005;120:169–181. doi: 10.1016/j.cell.2005.01.001. [DOI] [PubMed] [Google Scholar]
- Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- Bernstein BE, Meissner A, Lander ES. The mammalian epigenome. Cell. 2007;128:669–681. doi: 10.1016/j.cell.2007.01.033. [DOI] [PubMed] [Google Scholar]
- Bird A. DNA methylation patterns and epigenetic memory. Genes & Dev. 2002;16:6–21. doi: 10.1101/gad.947102. [DOI] [PubMed] [Google Scholar]
- Boyle AP, Davis S, Shulha HP, Meltzer P, Margulies EH, Weng Z, Furey TS, Crawford GE. High-resolution mapping and characterization of open chromatin across the genome. Cell. 2008;132:311–322. doi: 10.1016/j.cell.2007.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brinkman AB, Pennings SW, Braliou GG, Rietveld LE, Stunnenberg HG. DNA methylation immediately adjacent to active histone marking does not silence transcription. Nucleic Acids Res. 2007;35:801–811. doi: 10.1093/nar/gkl1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrozza MJ, Li B, Florens L, Suganuma T, Swanson SK, Lee KK, Shia WJ, Anderson S, Yates J, Washburn MP, et al. Histone H3 methylation by Set2 directs deacetylation of coding regions by Rpd3S to suppress spurious intragenic transcription. Cell. 2005;123:581–592. doi: 10.1016/j.cell.2005.10.023. [DOI] [PubMed] [Google Scholar]
- Chahrour M, Jung SY, Shaw C, Zhou X, Wong ST, Qin J, Zoghbi HY. MeCP2, a key contributor to neurological disease, activates and represses transcription. Science. 2008;320:1224–1229. doi: 10.1126/science.1153252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow CM, Georgiou A, Szutorisz H, Maia e Silva A, Pombo A, Barahona I, Dargelos E, Canzonetta C, Dillon N. Variant histone H3.3 marks promoters of transcriptionally active genes during mammalian cell division. EMBO Rep. 2005;6:354–360. doi: 10.1038/sj.embor.7400366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooke C, Hans H, Alwine JC. Utilization of splicing elements and polyadenylation signal elements in the coupling of polyadenylation and last-intron removal. Mol Cell Biol. 1999;19:4971–4979. doi: 10.1128/mcb.19.7.4971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford GE, Holt IE, Whittle J, Webb BD, Tai D, Davis S, Margulies EH, Chen Y, Bernat JA, Ginsburg D, et al. Genome-wide mapping of DNase hypersensitive sites using massively parallel signature sequencing (MPSS) Genome Res. 2006;16:123–131. doi: 10.1101/gr.4074106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui K, Zang C, Roh TY, Schones DE, Childs RW, Peng W, Zhao K. Chromatin signatures in multipotent human hematopoietic stem cells indicate the fate of bivalent genes during differentiation. Cell Stem Cell. 2009;4:80–93. doi: 10.1016/j.stem.2008.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diehn M, Alizadeh AA, Rando OJ, Liu CL, Stankunas K, Botstein D, Crabtree GR, Brown PO. Genomic expression programs and the integration of the CD28 costimulatory signal in T cell activation. Proc Natl Acad Sci. 2002;99:11796–11801. doi: 10.1073/pnas.092284399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dion MF, Kaplan T, Kim M, Buratowski S, Friedman N, Rando OJ. Dynamics of replication-independent histone turnover in budding yeast. Science. 2007;315:1405–1408. doi: 10.1126/science.1134053. [DOI] [PubMed] [Google Scholar]
- Eckhardt F, Lewin J, Cortese R, Rakyan VK, Attwood J, Burger M, Burton J, Cox TV, Davies R, Down TA, et al. DNA methylation profiling of human chromosomes 6, 20 and 22. Nat Genet. 2006;38:1378–1385. doi: 10.1038/ng1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The ENCODE Project Consortium. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuks F. DNA methylation and histone modifications: Teaming up to silence genes. Curr Opin Genet Dev. 2005;15:490–495. doi: 10.1016/j.gde.2005.08.002. [DOI] [PubMed] [Google Scholar]
- Grueter B, Petter M, Egawa T, Laule-Kilian K, Aldrian CJ, Wuerch A, Ludwig Y, Fukuyama H, Wardemann H, Waldschuetz R, et al. Runx3 regulates integrin alpha E/CD103 and CD4 expression during development of CD4−/CD8+ T cells. J Immunol. 2005;175:1694–1705. doi: 10.4049/jimmunol.175.3.1694. [DOI] [PubMed] [Google Scholar]
- Han M, Grunstein M. Nucleosome loss activates yeast downstream promoters in vivo. Cell. 1988;55:1137–1145. doi: 10.1016/0092-8674(88)90258-9. [DOI] [PubMed] [Google Scholar]
- Heard E, Rougeulle C, Arnaud D, Avner P, Allis CD, Spector DL. Methylation of histone H3 at Lys-9 is an early mark on the X chromosome during X inactivation. Cell. 2001;107:727–738. doi: 10.1016/s0092-8674(01)00598-0. [DOI] [PubMed] [Google Scholar]
- Hu CM, Jang SY, Fanzo JC, Pernis AB. Modulation of T cell cytokine production by interferon regulatory factor-4. J Biol Chem. 2002;277:49238–49246. doi: 10.1074/jbc.M205895200. [DOI] [PubMed] [Google Scholar]
- Johnson WE, Li W, Meyer CA, Gottardo R, Carroll JS, Brown M, Liu XS. Model-based analysis of tiling-arrays for ChIP-chip. Proc Natl Acad Sci. 2006;103:12457–12462. doi: 10.1073/pnas.0601180103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones PA, Baylin SB. The fundamental role of epigenetic events in cancer. Nat Rev Genet. 2002;3:415–428. doi: 10.1038/nrg816. [DOI] [PubMed] [Google Scholar]
- Jones PA, Baylin SB. The epigenomics of cancer. Cell. 2007;128:683–692. doi: 10.1016/j.cell.2007.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgensen HF, Azuara V, Amoils S, Spivakov M, Terry A, Nesterova T, Cobb BS, Ramsahoye B, Merkenschlager M, Fisher AG. The impact of chromatin modifiers on the timing of locus replication in mouse embryonic stem cells. Genome Biol. 2007;8:R169. doi: 10.1186/gb-2007-8-8-r169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kangaspeska S, Stride B, Metivier R, Polycarpou-Schwarz M, Ibberson D, Carmouche RP, Benes V, Gannon F, Reid G. Transient cyclical methylation of promoter DNA. Nature. 2008;452:112–115. doi: 10.1038/nature06640. [DOI] [PubMed] [Google Scholar]
- Kaplan CD, Laprade L, Winston F. Transcription elongation factors repress transcription initiation from cryptic sites. Science. 2003;301:1096–1099. doi: 10.1126/science.1087374. [DOI] [PubMed] [Google Scholar]
- Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- Kristjuhan A, Svejstrup JQ. Evidence for distinct mechanisms facilitating transcript elongation through chromatin in vivo. EMBO J. 2004;23:4243–4252. doi: 10.1038/sj.emboj.7600433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lea NC, Orr SJ, Stoeber K, Williams GH, Lam EW, Ibrahim MA, Mufti GJ, Thomas NS. Commitment point during G0→G1 that controls entry into the cell cycle. Mol Cell Biol. 2003;23:2351–2361. doi: 10.1128/MCB.23.7.2351-2361.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu CL, Kaplan T, Kim M, Buratowski S, Schreiber SL, Friedman N, Rando OJ. Single-nucleosome mapping of histone modifications in S. cerevisiae. PLoS Biol. 2005;3:e328. doi: 10.1371/journal.pbio.0030328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martins GA, Hutchins AS, Reiner SL. Transcriptional activators of helper T cell fate are required for establishment but not maintenance of signature cytokine expression. J Immunol. 2005;175:5981–5985. doi: 10.4049/jimmunol.175.9.5981. [DOI] [PubMed] [Google Scholar]
- Mellor J. Dynamic nucleosomes and gene transcription. Trends Genet. 2006;22:320–329. doi: 10.1016/j.tig.2006.03.008. [DOI] [PubMed] [Google Scholar]
- Metivier R, Gallais R, Tiffoche C, Le Peron C, Jurkowska RZ, Carmouche RP, Ibberson D, Barath P, Demay F, Reid G, et al. Cyclical DNA methylation of a transcriptionally active promoter. Nature. 2008;452:45–50. doi: 10.1038/nature06544. [DOI] [PubMed] [Google Scholar]
- Mikkelsen TS, Ku M, Jaffe DB, Issac B, Lieberman E, Giannoukos G, Alvarez P, Brockman W, Kim TK, Koche RP, et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448:553–560. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murayama A, Sakura K, Nakama M, Yasuzawa-Tanaka K, Fujita E, Tateishi Y, Wang Y, Ushijima T, Baba T, Shibuya K, et al. A specific CpG site demethylation in the human interleukin 2 gene promoter is an epigenetic memory. EMBO J. 2006;25:1081–1092. doi: 10.1038/sj.emboj.7601012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pernis AB. The role of IRF-4 in B and T cell activation and differentiation. J Interferon Cytokine Res. 2002;22:111–120. doi: 10.1089/107999002753452728. [DOI] [PubMed] [Google Scholar]
- Peters AH, Kubicek S, Mechtler K, O'Sullivan RJ, Derijck AA, Perez-Burgos L, Kohlmaier A, Opravil S, Tachibana M, Shinkai Y, et al. Partitioning and plasticity of repressive histone methylation states in mammalian chromatin. Mol Cell. 2003;12:1577–1589. doi: 10.1016/s1097-2765(03)00477-5. [DOI] [PubMed] [Google Scholar]
- Rando OJ, Ahmad K. Rules and regulation in the primary structure of chromatin. Curr Opin Cell Biol. 2007;19:250–256. doi: 10.1016/j.ceb.2007.04.006. [DOI] [PubMed] [Google Scholar]
- Rice JC, Briggs SD, Ueberheide B, Barber CM, Shabanowitz J, Hunt DF, Shinkai Y, Allis CD. Histone methyltransferases direct different degrees of methylation to define distinct chromatin domains. Mol Cell. 2003;12:1591–1598. doi: 10.1016/s1097-2765(03)00479-9. [DOI] [PubMed] [Google Scholar]
- Roh TY, Ngau WC, Cui K, Landsman D, Zhao K. High-resolution genome-wide mapping of histone modifications. Nat Biotechnol. 2004;22:1013–1016. doi: 10.1038/nbt990. [DOI] [PubMed] [Google Scholar]
- Roh TY, Cuddapah S, Zhao K. Active chromatin domains are defined by acetylation islands revealed by genome-wide mapping. Genes & Dev. 2005;19:542–552. doi: 10.1101/gad.1272505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roh TY, Cuddapah S, Cui K, Zhao K. The genomic landscape of histone modifications in human T cells. Proc Natl Acad Sci. 2006;103:15782–15787. doi: 10.1073/pnas.0607617103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roopra A, Qazi R, Schoenike B, Daley TJ, Morrison JF. Localized domains of G9a-mediated histone methylation are required for silencing of neuronal genes. Mol Cell. 2004;14:727–738. doi: 10.1016/j.molcel.2004.05.026. [DOI] [PubMed] [Google Scholar]
- Sarma K, Reinberg D. Histone variants meet their match. Nat Rev Mol Cell Biol. 2005;6:139–149. doi: 10.1038/nrm1567. [DOI] [PubMed] [Google Scholar]
- Schones DE, Cui K, Cuddapah S, Roh TY, Barski A, Wang Z, Wei G, Zhao K. Dynamic regulation of nucleosome positioning in the human genome. Cell. 2008;132:887–898. doi: 10.1016/j.cell.2008.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shivaswamy S, Bhinge A, Zhao Y, Jones S, Hirst M, Iyer VR. Dynamic remodeling of individual nucleosomes across a eukaryotic genome in response to transcriptional perturbation. PLoS Biol. 2008;6:e65. doi: 10.1371/journal.pbio.0060065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shogren-Knaak M, Ishii H, Sun JM, Pazin MJ, Davie JR, Peterson CL. Histone H4-K16 acetylation controls chromatin structure and protein interactions. Science. 2006;311:844–847. doi: 10.1126/science.1124000. [DOI] [PubMed] [Google Scholar]
- Smith AE, Hurd PJ, Bannister AJ, Kouzarides T, Ford KG. Heritable gene repression through the action of a directed DNA methyltransferase at a chromosomal locus. J Biol Chem. 2008;283:9878–9885. doi: 10.1074/jbc.M710393200. [DOI] [PubMed] [Google Scholar]
- Tachibana M, Sugimoto K, Fukushima T, Shinkai Y. Set domain-containing protein, G9a, is a novel lysine-preferring mammalian histone methyltransferase with hyperactivity and specific selectivity to lysines 9 and 27 of histone H3. J Biol Chem. 2001;276:25309–25317. doi: 10.1074/jbc.M101914200. [DOI] [PubMed] [Google Scholar]
- Tachibana M, Sugimoto K, Nozaki M, Ueda J, Ohta T, Ohki M, Fukuda M, Takeda N, Niida H, Kato H, et al. G9a histone methyltransferase plays a dominant role in euchromatic histone H3 lysine 9 methylation and is essential for early embryogenesis. Genes & Dev. 2002;16:1779–1791. doi: 10.1101/gad.989402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taniuchi I, Littman DR. Epigenetic gene silencing by Runx proteins. Oncogene. 2004;23:4341–4345. doi: 10.1038/sj.onc.1207671. [DOI] [PubMed] [Google Scholar]
- Vakoc CR, Mandat SA, Olenchock BA, Blobel GA. Histone H3 lysine 9 methylation and HP1gamma are associated with transcription elongation through mammalian chromatin. Mol Cell. 2005;19:381–391. doi: 10.1016/j.molcel.2005.06.011. [DOI] [PubMed] [Google Scholar]
- Vakoc CR, Sachdeva MM, Wang H, Blobel GA. Profile of histone lysine methylation across transcribed mammalian chromatin. Mol Cell Biol. 2006;26:9185–9195. doi: 10.1128/MCB.01529-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wade PA, Pruss D, Wolffe AP. Histone acetylation: Chromatin in action. Trends Biochem Sci. 1997;22:128–132. doi: 10.1016/s0968-0004(97)01016-5. [DOI] [PubMed] [Google Scholar]
- Wagschal A, Delaval K, Pannetier M, Arnaud P, Feil R. Chromatin immunoprecipitation (ChIP) on unfixed chromatin from cells and tissues to analyze histone modifications. Cold Spring Harbor Press; New York: 2007. [DOI] [PubMed] [Google Scholar]
- Walsh CP, Bestor TH. Cytosine methylation and mammalian development. Genes & Dev. 1999;13:26–34. doi: 10.1101/gad.13.1.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Zang C, Rosenfeld JA, Schones DE, Barski A, Cuddapah S, Cui K, Roh TY, Peng W, Zhang MQ, et al. Combinatorial patterns of histone acetylations and methylations in the human genome. Nat Genet. 2008;40:897–903. doi: 10.1038/ng.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber M, Davies JJ, Wittig D, Oakeley EJ, Haase M, Lam WL, Schubeler D. Chromosome-wide and promoter-specific analyses identify sites of differential DNA methylation in normal and transformed human cells. Nat Genet. 2005;37:853–862. doi: 10.1038/ng1598. [DOI] [PubMed] [Google Scholar]
- Wood AJ, Schulz R, Woodfine K, Koltowska K, Beechey CV, Peters J, Bourc'his D, Oakey RJ. Regulation of alternative polyadenylation by genomic imprinting. Genes & Dev. 2008;22:1141–1146. doi: 10.1101/gad.473408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woolf E, Xiao C, Fainaru O, Lotem J, Rosen D, Negreanu V, Bernstein Y, Goldenberg D, Brenner O, Berke G, et al. Runx3 and Runx1 are required for CD8 T cell development during thymopoiesis. Proc Natl Acad Sci. 2003;100:7731–7736. doi: 10.1073/pnas.1232420100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan GC, Liu YJ, Dion MF, Slack MD, Wu LF, Altschuler SJ, Rando OJ. Genome-scale identification of nucleosome positions in S. cerevisiae. Science. 2005;309:626–630. doi: 10.1126/science.1112178. [DOI] [PubMed] [Google Scholar]
- Zhang X, Yazaki J, Sundaresan A, Cokus S, Chan SW, Chen H, Henderson IR, Shinn P, Pellegrini M, Jacobsen SE, et al. Genome-wide high-resolution mapping and functional analysis of DNA methylation in Arabidopsis. Cell. 2006;126:1189–1201. doi: 10.1016/j.cell.2006.08.003. [DOI] [PubMed] [Google Scholar]