Abstract
Genetic drift, which is particularly effective within small populations, can shape the size and complexity of genomes by affecting the fixation of deleterious mutations. In Bacteria, assessing the contribution of genetic drift to genome evolution is problematic because the usual methods, based on intraspecific polymorphisms, can be thwarted by difficulties in delineating species' boundaries. The increased availability of sequenced bacterial genomes allows application of an alternative estimator of drift, the genome-wide ratio of replacement to silent substitutions in protein-coding sequences. This ratio, which reflects the action of purifying selection across the entire genome, shows a strong inverse relationship with genome size, indicating that drift promotes genome reduction in bacteria.
Bacteria are the most ancient, abundant, and genetically diverse organisms on earth. The current repertoire of fully sequenced bacterial genomes spans a significant portion of this diversity; for example, sequenced representatives of 20 bacterial phyla are available, with genome sizes ranging from 0.16 to over 13 Mb (Nakabachi et al. 2006; Schneiker et al. 2007). The diversity observed among bacterial genomes results from the interplay among mutation, natural selection, and genetic drift. Although the effects of mutation and selection are relatively well understood, the importance of genetic drift in influencing the evolutionary trajectory of genome complexity has begun to be appreciated only recently (Lynch and Conery 2003; Charlesworth and Barton 2004; Daubin and Moran 2004; Lynch and Conery 2004; Lynch 2006; Hershberg et al. 2007).
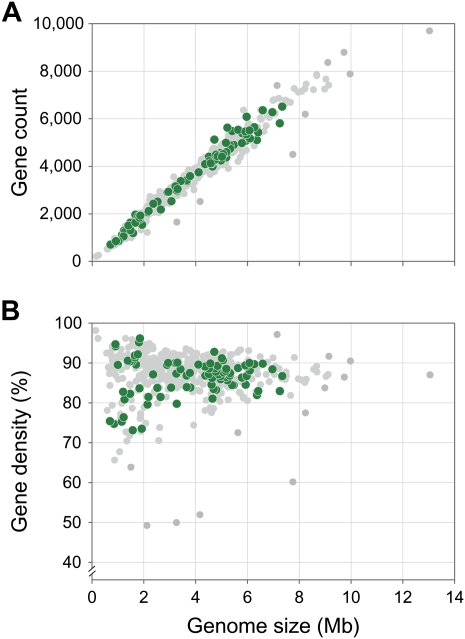
Unlike eukaryotes, in which there is wide variation in gene density and little association between genome size and gene number or organismal complexity (Gregory 2002; Lynch and Conery 2003), genome size in bacteria is tightly linked to gene number (Mira et al. 2001; Giovannoni et al. 2005) (Fig. 1A) (r = 0.98, P < 2.2 × 10−16). Consequently, evolutionary forces that act on individual genes have profound effects on the overall architecture of bacterial genomes. Due to the constant onslaught of new mutations, which are biased toward deletions in bacteria (Andersson and Andersson 2001; Mira et al. 2001; Nilsson et al. 2005; Hershberg et al. 2007), all genes will undergo inactivation and loss unless maintained by selection. At the extremes, those genes that are essential must, by definition, be preserved, whereas those that offer no beneficial effect will decay over time. However, most genes lie somewhere between these extremes, and the extent of genetic drift will govern how many such genes are maintained (Ochman and Davalos 2006; Khachane et al. 2007).
Figure 1.
Association between genome size and gene count (A) and gene density (B) for 488 bacterial species. Green points represent the 84 genomes considered in the present study; gray points are other published genomes.
To elucidate the role of genetic drift in bacterial genome evolution, we have investigated the relationship between the level of genetic drift and genome complexity, measured as genome size and gene density. Such an analysis requires examination of bacteria that display a wide range of population structures and genomic attributes. This has only recently become possible due to the increased availability of genome sequences that better represent the ecological and phylogenetic diversity of Bacteria.
In contrast to either genome size or gene density (i.e., the proportion of a genome that is composed of genes) that can each be calculated directly from a complete genome sequence, quantifying the level of genetic drift affecting a lineage is less straightforward. One commonly used index is based on the level of polymorphism within a species (Tajima 1983). Although widely applied to animals and plants, this measure is difficult to apply to microbes due to uncertainties regarding species boundaries and other factors (Daubin and Moran 2004; Katz et al. 2006; Snoke et al. 2006). An alternative index of the degree of genetic drift can be based on the efficacy of purifying selection in protein-coding sequences (Yang and Bielawski 2000; Daubin and Moran 2004; Novichkov et al. 2009). Because point mutations causing amino acid replacements are often deleterious, the rate of nonsynonymous substitution per site (Ka) is usually much less than the rate of synonymous substitution per site (Ks) in functional genes. An increased level of genetic drift, resulting from either reduced effective population size (Ne), genome-wide relaxation of selection, or some combination, can result in increased incidence of slightly deleterious amino acid replacements and an increase in the genome-wide Ka/Ks ratio. Although Ka can also increase as a result of positive selection favoring certain amino acid changes, such positive selection will be focused on particular genes and sites and is not expected to drive changes throughout the genome (Novichkov et al. 2009).
Results
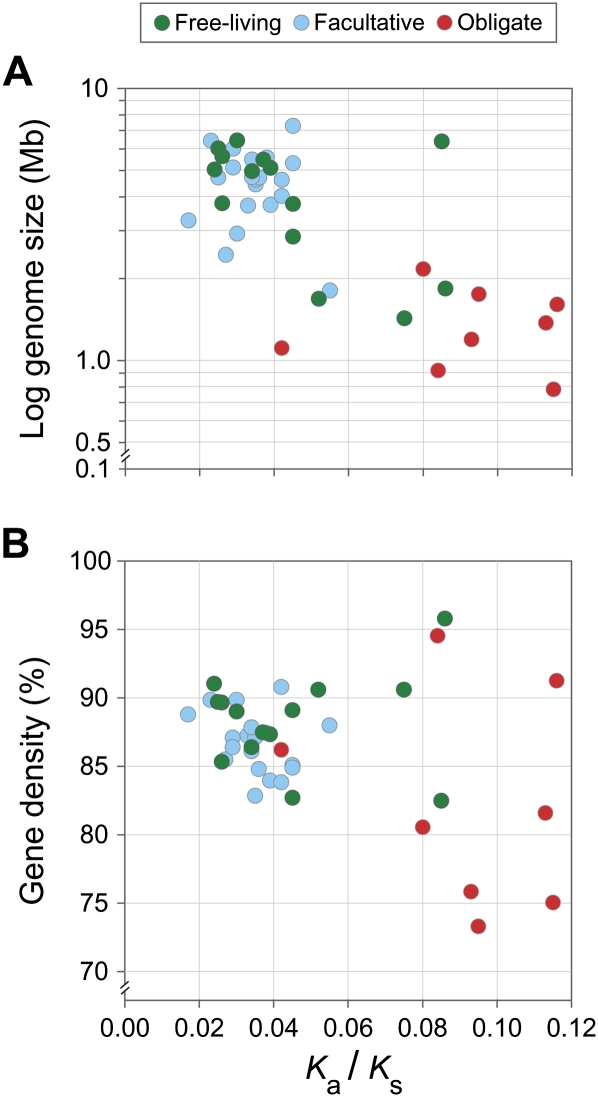
We utilized genome-wide Ka/Ks ratios as a proxy for the level of genetic drift experienced by 42 species-pairs of bacteria representing varied lifestyles and eight phyla. By limiting our analysis to pairs consisting of closely related species, we were able to obtain robust estimates of Ka and Ks. Genome size, which in bacteria is a close correlate of metabolic capabilities and organismal complexity, exhibits a strong negative correlation with the level of genetic drift (Fig. 2A) (r = –0.72, P = 6.3 × 10 −8). Although the overall relationship might appear to rely strongly on the inclusion of obligate symbionts and pathogens, which almost universally have small genome sizes and high levels of drift, a significant negative correlation is also apparent when only free-living bacteria are considered (r = –0.86, P = 0.0018; see Supplemental Table 1 for the 13 species-pairs included in this analysis; one anomalous pair of free-living cyanobacteria is excluded, see explanation below).
Figure 2.
Association between level of genetic drift and genome size (A) and gene density (B) for the 42 pairs of bacterial genomes. The level of genetic drift exhibits a strong negative correlation with genome size (r = –0.72, P = 6.3 × 10−8). When only strictly free-living bacteria are considered, the correlation remains statistically significant (r = –0.55, P = 0.039), particularly when the anomalous pair of cyanobacteria is excluded (r = –0.86, P = 0.0018; see Results for explanation).
Because each of the species-pairs harbors a unique set of orthologs that might collectively be subject to different selection constraints, the average Ka/Ks ratios included in Figure 2 are calculated using only those genes that are shared by at least 30 of the 42 selected pairs to ensure that we compared a similar set of genes across different taxa. The strong correlation (r = 0.99, P < 2.2 × 10−16; see Supplemental Fig. 1) between the average Ka/Ks ratios based on broadly distributed genes (average = 322 genes/genome pair; most of these genes are involved in central cellular processes, such as replication and translation, and the identity and annotation of these 13,557 genes are listed in Supplemental Table 2) and those based on the orthologs common to the two genomes in a pair (average = 1326 genes/genome-pair) confirms that the criteria applied to selecting shared genes do not bias the result. Moreover, the observed pattern is not attributable to artifacts associated with variation in pairwise divergence levels: A similar pattern is observed when we restrict our analysis to genome-pairs that have low or intermediate levels of divergence (i.e., average Ks < 0.4 or Ks = 0.4–0.6, see Supplemental Fig. 2).
Although measuring the Ne of any species is difficult (and perhaps even more so in bacteria), the ecological niches occupied by an organism often provide some clues to the relative magnitude of Ne. In this regard, genome-wide Ka/Ks ratio appears to be a reliable predictor of the expected Ne. For example, all but one of the 10 species-pairs displaying relatively high levels of genetic drift (i.e., average Ka/Ks > 0.06) might be expected to have reduced Ne based on their lifestyles; these include insect endosymbionts (Wolbachia pipentis), extremophiles (Dehalococcoides ethenogenes and Thermotoga spp.), vector-borne pathogens (Bartonella spp., Rickettsia spp., Borrelia spp., and phytoplasmas), and human pathogens with limited transmission routes (Neisseria spp. and Helicobacter spp.). Thus, lifestyles expected to result in an increase in the level of genetic drift appear to be associated with genome reduction. The only exception is a pair of cyanobacteria in the order Nostocales (average Ka/Ks = 0.09 and average genome size =6.4 Mb). In contrast to most other species-pairs in our analysis, these two species display different ecological niches and potentially differ in typical values of Ne. As a result, the average Ka/Ks ratio may not, in the case of this particular pair, be an appropriate measure of drift. It is noteworthy that several obligate endosymbionts have also been reported to display small genome sizes as a consequence of high levels of genetic drift; however, these cases, including Buchnera, Blochmannia, and Wigglesworthia were not included because available genome sequences do not have a suitable relative to meet the specifications for our analyses (see Methods).
The majority of the bacterial lineages that we examined (32 of the 42 genome-pairs) appear to have experienced low levels of genetic drift (average Ka/Ks < 0.06). This observation is consistent with the commonly held view that most bacterial species have a large Ne and experience effective purifying selection (Lynch and Conery 2003; Lynch 2006). Almost all of these organisms (including all members of Actinobacteria, Firmicutes, and most Proteobacteria that we examined) have intermediate-to-large genomes (i.e., 2–7 Mb) that are typical sizes for known bacterial lineages (Fig. 1A). Only three pairs possess genomes of <2 Mb, including Campylobacter jejuni (a leading cause of bacterial food poisoning), Mycoplasma spp. (mammalian pathogens with multiple host species), and Prochlorococcus marinus (phytoplanktonic marine cyanobacteria). The numerous cyanobacterial species designated as Prochlorococcus marinus together comprise some of the most abundant photosynthetic organisms on earth (Partensky et al. 1999), and, along with two other broadly distributed marine microbes (Pelagibacter ubique [Giovannoni et al. 2005] and a group of methylotrophs from the Betaproteobacteria [Giovannoni et al. 2008]), are the only documented examples of free-living bacterial lineages with highly reduced genomes. Although the exact explanation remains unclear, natural selection for decreased cell volumes and nutritional loads has been hypothesized to have caused genome reduction in such lineages (Dufresne et al. 2005; Giovannoni et al. 2005).
In addition to size, gene density (i.e., the proportion of a genome that is composed of annotated genes) is also associated with levels of genetic drift (Fig. 2B). The 32 genome-pairs with low levels of drift (average Ka/Ks < 0.06) display a relatively narrow range of gene densities, ranging from 83% in Psychrobacter spp. (cold-adapted bacteria isolated from permafrost) to 91% in Anaeromyxobacter spp. (spore-forming soil bacteria). In sharp contrast, the 10 genome-pairs subject to higher levels of drift (average Ka/Ks ratio > 0.06) span a much wider range, from 73% in Bartonella spp. (insect-borne human pathogens) to 96% in Thermotoga spp. (anaerobic thermophiles). Moreover, most of the lineages that experience high levels of drift lie outside of the 85%–90% gene density that is typical of bacterial genomes (Fig. 1B).
Discussion
Our results indicate that the variation in level of genetic drift coupled with the inherent bias toward deletions in bacterial genomes (Andersson and Andersson 2001; Mira et al. 2001; Nilsson et al. 2005; Hershberg et al. 2007) are the key forces that govern the evolution of genome complexity in bacteria. The increased values of Ka/Ks are probably a consequence of reductions in Ne (i.e., increases in drift), though an alternative hypothesis might be that low Ka/Ks reflects relaxed selection, for example, due to constant environments within host cells. We note that a substantial fraction of the genes showing elevated Ka/Ks underlie central cellular processes such as translation and replication (e.g., 43/80 in the phytoplasma species-pair; see Supplemental Table 2 for a complete list), and are thus essential regardless of life style. This observation suggests that reduced population size substantially outweighs relaxed selection as a force affecting both Ka/Ks and gene retention.
When a bacterial lineage adapts to a lifestyle that reduces its long-term Ne, such as obligate symbiosis (e.g., Wolbachia and Rickettsia) or limited habitat range (e.g., Dehalococcoides and Thermotoga), new mutations (with a propensity toward deletions) become more likely to be fixed in the population due to an elevated level of drift. In addition to reducing overall genome size (Fig. 2A), the random fixation of mutations will also increase the variation in coding densities (Figs. 1B, 2B). This wide range of coding densities likely represents various stages in the process of genome reduction. Because coding regions constitute the largest mutational target in a typical bacterial genome, the fixation of mildly deleterious mutations often reduces the gene density through the creation of pseudogenes. Bacterial lineages that only recently became host restricted, such as Mycobacterium leprae (Cole et al. 2001) and Sodalis glossinidius (Toh et al. 2006), illustrate this initial stage of genome reduction and have the lowest coding densities (at about 50%) among sequenced bacterial genomes. Even more recent is the host-restricted lifestyle of the human pathogen Mycobacterium tuberculosis, which has been found to show both elevated polymorphism and elevated ratios of nonsynonymous to synonymous changes (Hershberg et al. 2008), but which retains relatively large genome size. As random deletions remove the pseudogenes, essential genes will be retained, ultimately resulting in tight gene packing in the most highly reduced genomes. In the extreme examples of genome reduction, such as Carsonella ruddii (Nakabachi et al. 2006) and Sulcia muelleri (McCutcheon and Moran 2007), the genomes are <0.25 Mb and contain <4% of noncoding DNA.
A commonly held view of bacteria genome size evolution argues that selection for rapid and efficient replication is a major force that drives the streamlining of bacterial genomes (Maniloff 1996). However, two factors argue against this view: The first is the overall lack of an association between genome size and doubling time either within or among bacterial species (Bergthorsson and Ochman 1998; Mira et al. 2001; Couturier and Rocha 2006; Froula and Francino 2007), and the second is that the bacteria harboring the smallest genomes are often obligate endosymbionts (e.g., Buchnera and Carsonella), whose lifestyle does not promote selection for rapid cell division. Our results provide additional evidence that selection favoring deletions that remove excess DNA is not a major determinant of genome size in bacteria, although marine bacterioplanktons may be exceptions (Dufresne et al. 2005; Giovannoni et al. 2005, 2008). The strong association between elevated levels of drift and small genome sizes (Fig. 2A) suggests that genome reduction in bacteria is predominately a nonadaptive process. On a larger scale, the lack of Bacteria with very large genomes may reflect selection that imposes constraints on the upper limits of bacterial genome size. Such constraints might result from limitations on bacterial cell volume and on chromosome structure (e.g., bacterial chromosomes only have a single origin of replication). Such selection could drive the genome-wide deletional bias that appears to be pervasive in bacterial genomes (Mira et al. 2001; Nilsson et al. 2005; Hershberg et al. 2007), even though there is little evidence that selection for smaller genome size underlies observed variation among bacterial lineages.
Two earlier studies investigated the role of genetic drift in bacterial genome size evolution. The first, by Daubin and Moran (2004), failed to detect a significant correlation between the level of genetic drift and genome size. However, their analysis was limited to the relatively few species-pairs available at the time, including some that were likely too divergent for accurate estimates of Ka/Ks, and used computational methods that were susceptible to biases caused by variation in base composition and codon usage patterns. A study examining a more extensive set of bacterial genomes (Novichkov et al. 2009) detected a relationship between genome size and the impact of purifying selection on coding sequences, but ascribed it to the inclusion of obligate parasites, which were concluded to experience weak purifying selection due to small Ne “despite the sometimes dramatic shrinkage caused by gene loss.” We show that this relationship is not simply an effect of including host-restricted bacteria and have established the generality of the effect of genetic drift on genome complexity across all, even free-living, Bacteria.
Although our finding of a major effect of drift on genome complexity is in accord with a major thesis of Lynch and Conery (2003), the direction of the relationship is with the opposite of their prediction, suggesting a fundamental difference between the genome biology of bacteria and eukaryotes. Whereas gene duplication and the proliferation of mobile elements are widespread in eukaryotic genome evolution, these processes are less prevalent in bacterial genomes. Although the proliferation of mobile elements has been observed in bacteria that experienced recent reduction in Ne, these mobile elements (along with nonessential genes) are eventually removed from the genomes through deletions in the process of genome reduction (Moran and Plague 2004). Because the model of genome evolution proposed by Lynch and Conery (2003) assumes that most DNA in a genome is of no benefit to organismal fitness, the model may be appropriate for higher eukaryotes, but it does not adequately explain genome evolution in Bacteria.
Methods
Data source and genome-pair selection
We obtained the 703 fully sequenced bacteria genomes available from NCBI GenBank (Benson et al. 2008) on October 1, 2008. The level of divergence between each possible pair within the same order was estimated by calculating the average Ks value for five conserved single-copy genes, dnaE, polA, rpoB, argS, and metG (see below for details). Genome-pairs with an average Ks between 0.2 and 1.2 were identified, and after removing redundant pairs (e.g., there were 121 possible Escherichia coli–Salmonella sp. pairs), we selected 42 genome-pairs from 24 orders for further analysis. Genome project id (GPID) and species name of each of these 84 genomes are listed in Supplemental Table 1. The genome size and gene density of each genome were calculated based on the GenBank file of all chromosomes (excluding plasmids) using a custom Perl script written with Bioperl modules (Stajich et al. 2002). The genome size expressed for each of these 42 pairs was calculated as the average of the two genomes forming a given pair. In cases where multiple genome pairs displayed appropriate levels of divergence, we selected the two genomes that were most similar in size. Of the 42 pairs analyzed, 31 deviate from the average by <5% and only two (Brucella ovis_Ochrobactrum anthropi and Rhodobacter sphaeroides 17029_Rhodobacter sphaeroides 17025) deviate from the average by >10%. Since the total range in genome size across the pairs analyzed is more than 30-fold, the variation within pairs constitutes only a small fraction of the variation.
Ortholog identification
To identify genome-pairs that are sufficiently, but not excessively diverged so that robust estimates of substitution rates can be obtained, we used the protein sequences of the five conserved genes from E. coli MG1655 as the queries (GenBank accession nos. NP_414726, NP_418300, NP_418414, NP_416390, and NP_416617) to find the best BLASTP (Altschul et al. 1990) hit from each of the other 702 genomes for substitution rate calculations. After the 42 candidate genome-pairs were selected, a set of more stringent criteria was applied for defining orthologs within a pair. A pair of genes were defined as orthologs between the two closely related genomes if: (1) the protein sequences were reciprocal best-hits, (2) the BLASTP E-value was less than or equal to 1 × 10−15, (3) the difference in length was no more than 20% of the shorter sequence, (4) the high-scoring pair (HSP) accounted for at least 80% of the shorter gene, and (5) the amino acid sequence similarity was at least 90% within the HSP. The close relationship of the genomes within each of the 42 selected pairs coupled with the high stringency of our ortholog selection minimized, if not entirely eliminated, the presence of paralogs in our comparisons.
Substitution rate calculations
To calculate the nonsynonymous and synonymous substitution rates between a pair of orthologs, we aligned amino acid sequences in MUSCLE (Edgar 2004) with the default settings. The resulting protein alignments were converted to nucleotide alignments with PAL2NAL (Suyama et al. 2006). Because many highly reduced genomes have a strong base compositional bias, we applied the YN00 method (Yang and Nielsen 2000) implemented in the PAML package (Yang 2007) to calculate the Ka/Ks ratios. In addition to base composition and codon usage biases, the mutation model used in the YN00 method accounts for transition/transversion rate bias. To avoid the problem of biased Ka/Ks ratios due to insufficient sequence divergence or saturation, genes having an estimated Ks of <0.1 or >1.5 were excluded from subsequent analyses. In addition to improving the inference of Ka/Ks ratios, the exclusion of genes with high Ks also helped our resolution of true orthologs, because Ks values are expected to be elevated between paralogs.
Defining sets of shared genes
To compare average Ka/Ks ratios across taxa, we confirmed the presence and absence of each ortholog-pair in every genome-pair by BLASTP with an E-value cutoff of 1 × 10−5. For inclusion in our final calculations of the average Ka/Ks ratio for a given genome-pair, we required a gene to be present in at least 30 genome-pairs (i.e., shared by at least 71% of the genome-pairs).
Statistical test
We used the linear model function implemented in the R package (http://www.R-project.org/) to perform linear regressions. Genome sizes were log-transformed prior to regression analysis to improve the goodness of fit.
Acknowledgments
This work is funded by NIH grants GM56120 and GM74738 to H.O. We thank Becky Nankivell for preparing the figures.
Footnotes
[Supplemental material is available online at www.genome.org.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.091785.109.
References
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Andersson JO, Andersson SGE. Pseudogenes, junk DNA, and the dynamics of Rickettsia genomes. Mol Biol Evol. 2001;18:829–839. doi: 10.1093/oxfordjournals.molbev.a003864. [DOI] [PubMed] [Google Scholar]
- Benson DA, Karsch-Mizrachi I, Lipman DJ, Ostell J, Wheeler DL. GenBank. Nucleic Acids Res. 2008;36:D25–D30. doi: 10.1093/nar/gkm929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergthorsson U, Ochman H. Distribution of chromosome length variation in natural isolates of Escherichia coli. Mol Biol Evol. 1998;15:6–16. doi: 10.1093/oxfordjournals.molbev.a025847. [DOI] [PubMed] [Google Scholar]
- Charlesworth B, Barton N. Genome size: Does bigger mean worse? Curr Biol. 2004;14:R233–R235. doi: 10.1016/j.cub.2004.02.054. [DOI] [PubMed] [Google Scholar]
- Cole ST, Eiglmeier K, Parkhill J, James KD, Thomson NR, Wheeler PR, Honore N, Garnier T, Churcher C, Harris D, et al. Massive gene decay in the leprosy bacillus. Nature. 2001;409:1007–1011. doi: 10.1038/35059006. [DOI] [PubMed] [Google Scholar]
- Couturier E, Rocha EPC. Replication-associated gene dosage effects shape the genomes of fast-growing bacteria but only for transcription and translation genes. Mol Microbiol. 2006;59:1506–1518. doi: 10.1111/j.1365-2958.2006.05046.x. [DOI] [PubMed] [Google Scholar]
- Daubin V, Moran NA. Comment on “The origins of genome complexity.”. Science. 2004;306:978a. doi: 10.1126/science.1098469. [DOI] [PubMed] [Google Scholar]
- Dufresne A, Garczarek L, Partensky F. Accelerated evolution associated with genome reduction in a free-living prokaryote. Genome Biol. 2005;6:R14. doi: 10.1186/gb-2005-6-2-r14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Froula JL, Francino MP. Selection against spurious promoter motifs correlates with translational efficiency across bacteria. PLoS One. 2007;2:e745. doi: 10.1371/journal.pone.0000745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giovannoni SJ, Tripp HJ, Givan S, Podar M, Vergin KL, Baptista D, Bibbs L, Eads J, Richardson TH, Noordewier M, et al. Genome streamlining in a cosmopolitan oceanic bacterium. Science. 2005;309:1242–1245. doi: 10.1126/science.1114057. [DOI] [PubMed] [Google Scholar]
- Giovannoni SJ, Hayakawa DH, Tripp HJ, Stingl U, Givan SA, Cho JC, Oh HM, Kiter JB, Vergin KL, Rappe MS. The small genome of an abundant coastal ocean methylotroph. Environ Microbiol. 2008;10:1771–1782. doi: 10.1111/j.1462-2920.2008.01598.x. [DOI] [PubMed] [Google Scholar]
- Gregory TR. Genome size and developmental complexity. Genetica. 2002;115:131–146. doi: 10.1023/a:1016032400147. [DOI] [PubMed] [Google Scholar]
- Hershberg R, Tang H, Petrov DA. Reduced selection leads to accelerated gene loss in Shigella. Genome Biol. 2007;8:R164. doi: 10.1186/gb-2007-8-8-r164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hershberg R, Lipatov M, Small PM, Sheffer H, Niemann S, Homolka S, Roach JC, Kremer K, Petrov DA, Feldman MW, et al. High functional diversity in M. tuberculosis driven by genetic drift and human demography. PLoS Biol. 2008;6:e311. doi: 10.1371/journal.pbio.0060311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz LA, Snoeyenbos-West O, Doerder FP. Patterns of protein evolution in Tetrahymena thermophila: Implications for estimates of effective population size. Mol Biol Evol. 2006;23:608–614. doi: 10.1093/molbev/msj067. [DOI] [PubMed] [Google Scholar]
- Khachane AN, Timmis KN, Martins dos Santos VAP. Dynamics of reductive genome evolution in mitochondria and obligate intracellular microbes. Mol Biol Evol. 2007;24:449–456. doi: 10.1093/molbev/msl174. [DOI] [PubMed] [Google Scholar]
- Lynch M. Streamlining and simplification of microbial genome architecture. Annu Rev Microbiol. 2006;60:327–349. doi: 10.1146/annurev.micro.60.080805.142300. [DOI] [PubMed] [Google Scholar]
- Lynch M, Conery JS. The origins of genome complexity. Science. 2003;302:1401–1404. doi: 10.1126/science.1089370. [DOI] [PubMed] [Google Scholar]
- Lynch M, Conery JS. Response to comment on “The origins of genome complexity.”. Science. 2004;306:978b. doi: 10.1126/science.1100559. [DOI] [PubMed] [Google Scholar]
- Maniloff J. The minimal cell genome: “On being the right size.”. Proc Natl Acad Sci. 1996;93:10004–10006. doi: 10.1073/pnas.93.19.10004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCutcheon JP, Moran NA. Parallel genomic evolution and metabolic interdependence in an ancient symbiosis. Proc Natl Acad Sci. 2007;104:19392–19397. doi: 10.1073/pnas.0708855104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mira A, Ochman H, Moran NA. Deletional bias and the evolution of bacterial genomes. Trends Genet. 2001;17:589–596. doi: 10.1016/s0168-9525(01)02447-7. [DOI] [PubMed] [Google Scholar]
- Moran NA, Plague GR. Genomic changes following host restriction in bacteria. Curr Opin Genet Dev. 2004;14:627–633. doi: 10.1016/j.gde.2004.09.003. [DOI] [PubMed] [Google Scholar]
- Nakabachi A, Yamashita A, Toh H, Ishikawa H, Dunbar HE, Moran NA, Hattori M. The 160-kilobase genome of the bacterial endosymbiont Carsonella. Science. 2006;314:267. doi: 10.1126/science.1134196. [DOI] [PubMed] [Google Scholar]
- Nilsson AI, Koskiniemi S, Eriksson S, Kugelberg E, Hinton JCD, Andersson DI. Bacterial genome size reduction by experimental evolution. Proc Natl Acad Sci. 2005;102:12112–12116. doi: 10.1073/pnas.0503654102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novichkov PS, Wolf YI, Dubchak I, Koonin EV. Trends in prokaryotic evolution revealed by comparison of closely related bacterial and archaeal genomes. J Bacteriol. 2009;191:65–73. doi: 10.1128/JB.01237-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochman H, Davalos LM. The nature and dynamics of bacterial genomes. Science. 2006;311:1730–1733. doi: 10.1126/science.1119966. [DOI] [PubMed] [Google Scholar]
- Partensky F, Hess WR, Vaulot D. Prochlorococcus, a marine photosynthetic prokaryote of global significance. Microbiol Mol Biol Rev. 1999;63:106–127. doi: 10.1128/mmbr.63.1.106-127.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneiker S, Perlova O, Kaiser O, Gerth K, Alici A, Altmeyer MO, Bartels D, Bekel T, Beyer S, Bode E, et al. Complete genome sequence of the myxobacterium Sorangium cellulosum. Nat Biotechnol. 2007;25:1281–1289. doi: 10.1038/nbt1354. [DOI] [PubMed] [Google Scholar]
- Snoke MS, Berendonk TU, Barth D, Lynch M. Large global effective population sizes in Paramecium. Mol Biol Evol. 2006;23:2474–2479. doi: 10.1093/molbev/msl128. [DOI] [PubMed] [Google Scholar]
- Stajich JE, Block D, Boulez K, Brenner SE, Chervitz SA, Dagdigian C, Fuellen G, Gilbert JGR, Korf I, Lapp H, et al. The Bioperl toolkit: Perl modules for the life sciences. Genome Res. 2002;12:1611–1618. doi: 10.1101/gr.361602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suyama M, Torrents D, Bork P. PAL2NAL: Robust conversion of protein sequence alignments into the corresponding codon alignments. Nucleic Acids Res. 2006;34:W609–W612. doi: 10.1093/nar/gkl315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajima F. Evolutionary relationship of DNA sequences in finite populations. Genetics. 1983;105:437–460. doi: 10.1093/genetics/105.2.437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toh H, Weiss BL, Perkin SAH, Yamashita A, Oshima K, Hattori M, Aksoy S. Massive genome erosion and functional adaptations provide insights into the symbiotic lifestyle of Sodalis glossinidius in the tsetse host. Genome Res. 2006;16:149–156. doi: 10.1101/gr.4106106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z. PAML 4: Phylogenetic analysis by maximum likelihood. Mol Biol Evol. 2007;27:1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- Yang Z, Bielawski JP. Statistical methods for detecting molecular adaptation. Trends Ecol Evol. 2000;15:496–503. doi: 10.1016/S0169-5347(00)01994-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Nielsen R. Estimating synonymous and nonsynonymous substitution rates under realistic evolutionary models. Mol Biol Evol. 2000;17:32–43. doi: 10.1093/oxfordjournals.molbev.a026236. [DOI] [PubMed] [Google Scholar]