Abstract
To address the role of small regulatory RNAs in rice development, we generated a large data set of small RNAs from mature leaves and developing roots, shoots, and inflorescences. Using a spatial clustering algorithm, we identified 36,780 genomic groups of small RNAs. Most consisted of 24-nt RNAs that are expressed in all four tissues and enriched in repeat regions of the genome; 1029 clusters were composed primarily of 21-nt small RNAs and, strikingly, 831 of these contained phased RNAs and were preferentially expressed in developing inflorescences. Thirty-eight of the 24-mer clusters were also phased and preferentially expressed in inflorescences. The phased 21-mer clusters derive from nonprotein coding, nonrepeat regions of the genome and are grouped together into superclusters containing 10–46 clusters. The majority of these 21-mer clusters (705/831) are flanked by a degenerate 22-nt motif that is offset by 12 nt from the main phase of the cluster. Small RNAs complementary to these flanking 22-nt motifs define a new miRNA family, which is conserved in maize and expressed in developing reproductive tissues in both plants. These results suggest that the biogenesis of phased inflorescence RNAs resembles that of tasiRNAs and raise the possibility that these novel small RNAs function in early reproductive development in rice and other monocots.
Endogenous small regulatory RNA pathways play diverse and fundamental roles in the regulation of gene expression in a broad range of eukaryotes, including plants. In these diverse organisms a common set of genes mediates small RNA biogenesis and function: DICER ribonucleases generate small RNAs from double-stranded RNA (dsRNA), while ARGONAUTE proteins form the core of effector complexes that mediate their function. In addition, plants and worms encode RNA-dependent RNA polymerases (RDRs) that can produce dsRNA substrates for DICER or facilitate DICER-independent small RNA biogenesis. Several distinct classes of small RNAs have been identified to date, and these play important roles in a variety of processes, including development, genome stability, and response to both biotic and abiotic stress. However, the full spectrum of small RNA pathways and their functions is not yet known, and the identification of novel small RNA generating loci remains an area of intense investigation (for recent reviews of small RNA pathways, see Ambros and Chen 2007; Eamens et al. 2008; Farazi et al. 2008; Girard and Hannon 2008; Ramachandran and Chen 2008).
Many types of small RNAs have been identified in plants, the best characterized of which are microRNAs (miRNAs), trans-acting siRNAs (tasiRNAs), and small-interfering RNAs (siRNAs). The plant miRNAs play a major role in many developmental processes, often targeting the transcription factors that mediate transition from one developmental stage to the next (Jones-Rhoades and Bartel 2004). The genes that encode miRNAs are transcribed to produce a precursor RNA with the mature miRNA located within a stem–loop structure that is processed by DICER-LIKE1 (DCL1) or in some cases by other DCLs to give a single small RNA duplex with two nucleotide 3′ overhangs (Rajagopalan et al. 2006; Vazquez et al. 2008). One strand of the small RNA duplex is the mature miRNA, which binds to an ARGONAUTE protein to form an effector complex that directs cleavage or translational repression of target mRNAs (Mallory et al. 2008). The opposite strand of the duplex, termed miRNA*, is rapidly degraded and normally does not accumulate. DCL1 knockout plants are embryo lethal, pointing to the importance of this small RNA pathway in development (Schauer et al. 2002).
Trans-acting siRNAs (tasiRNAs) constitute a small class of phased small RNAs that play a role in phase transition during Arabidopsis leaf development (Peragine et al. 2004; Vazquez et al. 2004; Yoshikawa et al. 2005). In tasiRNA biogenesis, the tasiRNA locus is transcribed into a single-stranded RNA that is cleaved by a particular miRNA. The miRNA cleavage product serves as a substrate for RDR6, which generates a dsRNA with a discrete end defined by the miRNA cleavage site. The dsRNA is then processively cleaved by DCL4 to produce the phased 21-nt tasiRNAs. Most of the genomic loci that produce phased siRNAs are flanked by two miRNA/tasiRNA binding sites, both of which, for reasons that are not yet fully clear, are important for proper biogenesis of the tasiRNAs (Axtell et al. 2006). The different phased tasiRNAs from a single tasiRNA locus typically accumulate to different levels, and some form complexes with ARGONAUTE proteins to direct cleavage of specific target mRNAs (Allen et al. 2005). In Arabidopsis, tasiRNA function appears to be confined to vegetative phase change. However, tasiRNAs or other DCL4-dependent small RNAs might play a broader role in rice because the developmental phenotype of dcl4 loss-of-function mutants is much more severe in rice than in Arabidopsis (Liu et al. 2007).
Endogeneous siRNAs derive from two separate pathways and may be associated with either transcriptional or post-transcriptional gene silencing. The post-transcriptional silencing pathway serves primarily defensive purposes and is induced by invading nucleic acids such as transgenes or viruses; it produces 21- or 22-nt siRNAs via the activity of DCL4 and DCL2, respectively (Deleris et al. 2006; Fusaro et al. 2006; Mlotshwa et al. 2008). The transcriptional pathway is involved in heterochromatin formation in repeated regions of the genome and the silencing of other regions of the genome as well. This pathway involves the activity of RNA polymerase IV/V, RDR2 and DCL3, producing 24-nt small RNAs referred to as Pol IV/V siRNAs (Henderson and Jacobsen 2007; Huettel et al. 2007). Whereas individual Pol IV/V siRNAs do not accumulate to high levels like miRNAs, as a size class they are usually the most abundant small RNAs in the cell. Because these small RNAs are involved in controlling transposons and other repeated regions of the genome, they could potentially play a bigger role in plants such as rice and maize that have large genomes with a high proportion of repeated sequences. Furthermore, Arabidopsis plants having a mutation in RDR2 display virtually no developmental defects, whereas mutation of the maize homolog of RDR2 (MOP1) results in a number of developmental anomalies (Dorweiler et al. 2000; Alleman et al. 2006; Woodhouse et al. 2006). These observations raise the possibility that some aspects of Pol IV/V siRNA-mediated transcriptional silencing have a greater role in regulating development in cereals than in Arabidopsis.
Here we report the results of high-throughput sequencing of small RNA populations from four different rice tissues: root apices, shoot apices, developing inflorescence, and mature leaves. Our bioinformatic analysis has identified previously unreported groups or clusters of phased 21-nt and 24-nt small RNAs that are preferentially expressed in the inflorescence of rice plants.
Results
Rice small RNAs from root apices, shoot apices, developing inflorescence, and mature leaves were sequenced using the 454 Life Sciences (Roche) pyrosequencing technology (Margulies et al. 2005) to identify developmentally regulated small RNAs. These tissues were chosen because they represent a comprehensive range of different developmental stages and cell types, and might be expected to display different patterns of small RNA expression. The root apices, shoot apices, and inflorescence samples are expected to be enriched for young differentiating cells and meristematic tissues each giving rise to cells having different fates. In contrast, the leaf tissues examined in this study were terminally differentiated.
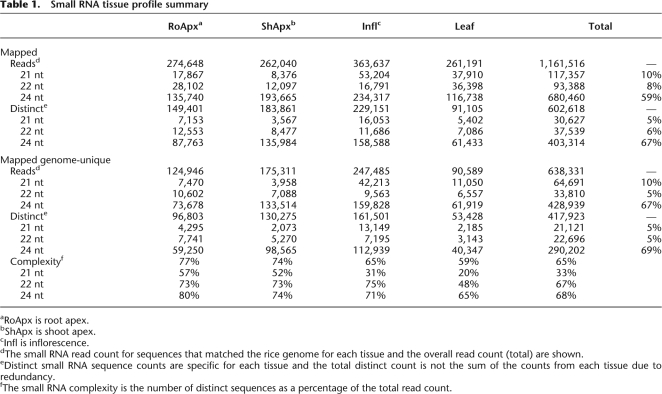
Pyrosequencing generated a total of 1,409,217 small RNA sequence reads representing 781,885 different (distinct) sequences. The vast majority of these distinct sequences (77% or 602,618) could be mapped to the ∼400-Mbp rice genome (Table 1; Goff et al. 2002). Little overlap of the mapped distinct sequences was observed from one tissue to the next; for each tissue, ∼90% of the distinct sequences were detected only within that tissue. Over half of the mapped distinct small RNAs (69% or 417,923) mapped to only one genome location and are referred to hereafter as genome-unique small RNAs. The remainder of the small RNAs mapped to two to a few thousand different genomic locations (Supplemental Fig. 1), and this distribution was generally similar in all four tissues. The majority of the small RNAs were 24-nt in length in all tissues (Table 1). Because 24-nt small RNAs have been implicated in transcriptional gene silencing, this result suggests that most of the small RNAs in all four tissues are involved in suppression of transcription. Consistent with this idea, 48% of the 24-nt small RNAs in all our data sets overlapped TIGR annotated repeats, most of which are known transposons or other mobile elements (data not shown). Interestingly, the ratio of the distinct sequences to the total reads expressed as a percentage, i.e., the relative complexity, was lowest in the terminally differentiated leaf tissue. This characteristic was especially true for the 21-nt small RNAs, which had a relative complexity of only 20% in leaf as compared to 31%–57% in developing shoot, root, and inflorescence tissues (Table 1).
Table 1.
Small RNA tissue profile summary
aRoApx is root apex.
bShApx is shoot apex.
cInfl is inflorescence.
dThe small RNA read count for sequences that matched the rice genome for each tissue and the overall read count (total) are shown.
eDistinct small RNA sequence counts are specific for each tissue and the total distinct count is not the sum of the counts from each tissue due to redundancy.
fThe small RNA complexity is the number of distinct sequences as a percentage of the total read count.
Clusters of small RNAs
An analysis of genomic clustering was used to examine the distribution of small RNA generating loci in the rice genome and to determine if small RNAs from these loci were differentially expressed in the sampled tissues. Only the 417,923 genome-unique small RNAs were utilized for this analysis, because these small RNAs could be attributed with certainty to a particular chromosome locus. One approach to defining clusters of small RNAs is to assign them to discrete sized bins. However, such groups are arbitrary, and small RNAs that arise from different transcripts might frequently be inappropriately assigned to the same group. Similarly, small RNAs that arise from the same transcript might be assigned to different groups. Although there is no way to completely avoid these problems, we took the approach of defining a cluster as a group of small RNAs in which each small RNA is ≤100 nt from its nearest neighbor. Thus, small RNAs at the ends of a cluster are >100 nt away from the next nearest small RNA outside the cluster. This analysis identified 73,983 small RNA clusters containing at least two distinct genome-unique small RNAs, and this number was reduced to 36,780 by eliminating clusters that did not contain small RNAs from at least three of 12 samples (see Methods). Therefore, the minimum number of small RNA reads in a cluster is three. This filtering process removed clusters that did not have much support and allowed a reduced data set that permitted the use of more computationally demanding algorithms for the analysis. The clusters and included small RNAs can be viewed using a genome browser at http://sundarlab.ucdavis.edu/cgi-bin/smrna_browse/ (Johnson et al. 2007).
Of the 36,780 clusters examined in our analysis, the cluster size ranged up to 2037 nt, with 59% spanning at least 100 nt (Supplemental Fig. 2). The vast majority of the clusters (29,765 clusters, 81% of the total) contained primarily 24-nt small RNAs (24-mer clusters) and included 167,146 distinct 24-nt small RNAs. In contrast, only a small number of clusters (1029, 2.8% of the total) were found to be composed primarily of 21-nt small RNAs (21-mer clusters), and from these 21-mer clusters, 6356 distinct 21-nt small RNAs were detected (Supplemental Table 1). In 16.2% of the clusters (5986), neither 21- nor 24-nt small RNAs were in the majority.
21-mer but not 24-mer clusters are arranged in superclusters
Visual inspection of the distribution of 21-mer and 24-mer clusters across the rice genome using a genome browser (Johnson et al. 2007) indicated that there were marked differences in their global distribution. Whereas the 24-mer clusters were relatively evenly spread across the genome (data not shown), many of the 21-mer clusters tended to be grouped near each other. The region occupied by a group of 21-mer clusters was defined using a method similar to that used to define individual clusters. In this case, a distance of 100 kbp was chosen as the minimum separation distance between regions of clusters. In other words, clusters that are closer than this were considered part of the same region. This was the optimal margin as determined by the ratio of simulated to real cluster regions (see Methods; Supplemental Fig. 3). Using this criterion, we identified 168 cluster regions that contained one or more clusters, ranging in size up to as large as 558 kb, with a median length of 0.98 kb. Strikingly, the majority of the 21-mer clusters (689 of 1029) were contained in just 31 larger regions containing 10 or more clusters (superclusters), with the largest supercluster containing 46 clusters (Fig. 1; Supplemental Table 2). These superclusters range in size from 31 to 546 kb with a median of 215 kb and are scattered more or less evenly across the genome, with the exception of the bottom half of chromosome 12, which appears to have a higher concentration of superclusters (Fig. 1).
Figure 1.
Distribution of 21-mer superclusters and phased 24-mer clusters in the rice genome. Regions of 21-mer clusters are represented as black lines (<10 clusters) or if they are superclusters as red lines (≥10 clusters). Phased 24-mer clusters are represented as cyan lines. The number of individual clusters within a region is indicated next to the line.
The 21-mer clusters tend not to overlap TIGR annotated repeats or gene loci, and supercluster regions have properties distinct from other parts of the genome
In order to identify any general associations of 21-mer and 24-mer clusters with either repetitive sequences or likely protein coding genes, the numbers of individual small RNA clusters that overlap TIGR annotated repeats or annotated gene loci were counted (Table 2). Simulations of the placement of the 24-mer and 21-mer clusters onto the genome were used to evaluate the possibility that any overlap would occur by chance (see Methods). This analysis showed that 48% of 24-mer clusters overlapped TIGR annotated repeats, ∼1.2-fold more than expected by chance (P = 2.16 × 10−211). In contrast, only 8% of the 21-mer clusters overlapped annotated repeats, about one fifth that expected by chance (P = 9 × 10−89). However, ∼20% of the 21-mer clusters can be unambiguously aligned with another such cluster when using the cluster sequence plus an additional 200 nt on either side. By definition there were no 21-nt small RNAs that were found in more than one cluster. However, 54 of the 6356 distinct small RNAs that comprise the 21-mer clusters are similar to another 21-nt small RNA in the set when up to three mismatches are allowed.
Table 2.
Features of clusters with dominant source tissue
aStatistically significantly differentially-expressed clusters with P-values less than or equal to alpha cutoff of 0.001.
bNumber of clusters that are considered in phase with FDRs of 1.2% and 10% for the 21-mer and 24-mer clusters, respectively.
cNumber of clusters overlapping at least one annotated protein coding gene locus.
dNumber of clusters overlapping at least one repeat annotation.
eTissue dominance at >50% of total normalized read count.
fClusters in which no source tissue constituted >50% of normalized reads.
The 24-mer clusters were relatively devoid of TIGR annotated gene loci (Table 2), most of which are expected to code for proteins that include transposon genes, hypothetical genes, and unknown expressed genes as well as characterized protein coding genes. Only 18% of the 24-mer clusters overlap gene loci, about one half to one third that expected by chance (P = 0). The 21-mer clusters showed the same tendency, with only 9.5% overlapped by likely protein coding genes, about one-fifth that expected by chance (P = 2.9 × 10−147) (Table 2). There was virtually no overlap of 21-mer or 24-mer clusters with tRNA or noncoding RNA loci (data not shown). This analysis indicates that the 21-mer clusters are significantly devoid of both annotated genes and repeats. The 24-mer clusters are relatively devoid of annotated gene loci and, although they are slightly enriched for repeats, 52% of the 24-mer clusters do not overlap repeats.
This analysis also shows that the fraction of 21-mer clusters that overlap annotated gene loci differs among the four tissues sampled. Table 2 shows that the percentage of 21-mer clusters that overlap annotated gene loci is much higher for clusters dominated by the leaf small RNAs than those dominated by inflorescence small RNAs, 42.5% versus 7.7%. In contrast, the percentage of the 24-mer clusters that overlap annotated gene loci is similar in the four tissue types examined. Furthermore, of the 75 inflorescence clusters found to overlap annotated gene loci, only five overlap characterized protein coding genes, whereas the others overlap hypothetical genes, unknown expressed genes, or transposons (data not shown). In contrast, of the 17 leaf derived clusters that overlap annotated gene loci, 11 overlap characterized protein coding genes (data not shown). Interestingly, in virtually all the cases where a 21-mer cluster from either inflorescence or leaves overlaps a characterized protein-coding locus, the overlap occurs within an intron rather than an exon.
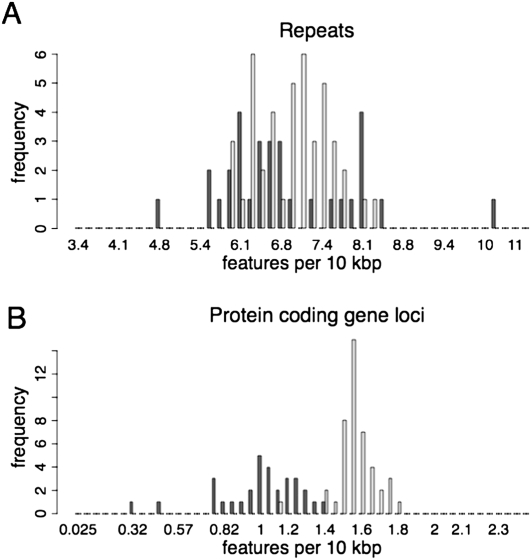
To determine if the 21-mer superclusters are similarly devoid of protein coding and repeat loci, the frequency of such loci within superclusters was compared to that in the sequences between superclusters. For each region, the number of annotated gene loci and the number of repeats were tallied up and the density of these features per 10 kbp was plotted on two overlapping histograms. The frequency distribution of protein coding gene loci in the superclusters was lower than that for the intervening regions, centering at ∼1.1–1.2 loci per 10 kbp as compared with ∼1.5 loci per 10 kbp (Fig. 2B), a statistically significant difference (Mann-Whitney, P < 2.2 × 10−16; see Methods). In contrast, no significant difference in the frequency of repeat loci between supercluster regions and intervening regions was observed (Mann-Whitney, P = 0.29; Fig. 2A). However, this analysis did not rule out the possibility that a particular type of repeat was enriched within the superclusters. To address this possibility, the superclusters and the intervening regions were scored for the presence or absence of 30 types of repeats, including individual members of retrotransposons and DNA transposons. The “CACTA” and “MITE-adh-10-like” repeats (Ouyang and Buell 2004) were the most significantly positively associated with the cluster regions, and were increased 2.2- and 5.1-fold, respectively, in the 21-mer cluster regions as compared to intervening regions (Supplemental Table 3; see Methods). The observation that supercluster regions were biased against characterized gene loci, yet have a higher frequency of some repeats but not repeats in general, indicates that these regions have properties distinct from other parts of the genome. This result raises the possibility that some “CACTA” and/or “MITE-adh-10-like” repeats may play a role in the biogenesis of the 21-mer supercluster small RNAs.
Figure 2.
Density of TIGR annotated features in 21-mer superclusters versus sequences between superclusters. The frequency of annotated repeats (A) or annotated protein coding genes (B) per 10 kbp is plotted for 21-mer superclusters (dark gray) versus the sequences between the 21-mer superclusters (light gray). The difference in the frequency is statistically significant for protein coding gene loci (P < 2.2 × 10−16), but not statistically significant for the repeats (P = 0.29).
Characterization of the 21- and 24-nt clusters with respect to strandedness, differential expression, and phasing
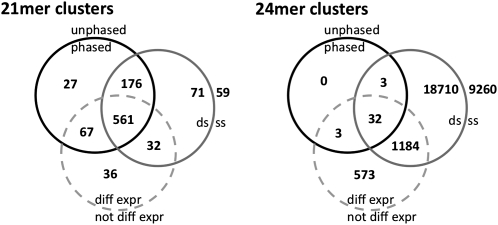
The 21-mer and 24-mer clusters were characterized with respect to their strandedness, differential expression, and phasing, and the results of this analysis are displayed on two Venn diagrams, one for 21-mer and the other for 24-mer clusters (Fig. 3). Clusters in which the polarities of the small RNA population were relatively balanced (having not more than 80% of the small RNAs in the same polarity) were classified as having been derived from double-stranded RNA (dsRNA), whereas clusters displaying more than 80% of the small RNAs in the same polarity were classified as having been derived from single-stranded RNA (ssRNA). Using this classification, 81.6% (or 840 of 1029) of the 21-mer clusters and 67.0% (or 19,929 of 29,765) of the 24-mer clusters were of dsRNA derivation. It is likely that the dsRNA-derived cluster RNAs arise primarily via DCL processing of dsRNA generated either from the transcription of overlapping transcription units or by the activity of an RNA-dependent RNA polymerase. The cluster RNAs derived from ssRNA likely arise via DCL cleavage of regions of high secondary structure within the RNA.
Figure 3.
Classification of clusters with regard to strandedness, differential expression, and phasing. Venn diagrams indicating the classification of 21-mer clusters (left) and 24-mer clusters (right) according to three binary criteria: origin from dsRNA (ds) versus origin from ssRNA (ss) (solid gray circle), differentially expressed (diff expr) versus not differentially expressed (not diff expr) (dashed gray circle), and phased versus unphased (black circle). Small RNAs were classified as having originated from ssRNA if >80% of the small RNAs in the cluster were of the same polarity. Classification as differentially expressed was at the 99.9% confidence level and classification as phased was with a FDR of 1.2% for 21-mer clusters and 10% for 24-mer clusters.
The differential expression of small RNA clusters in the various tissues was evaluated using a combination of χ2 and multinomial tests (see Methods) using a confidence limit of 99.9% (i.e., alpha of 0.001). The majority of the 21-mer clusters (68% of 1029) are significantly differentially expressed. Almost all of these (96%) are expressed specifically in the developing inflorescence, whereas 3% are preferentially expressed in the mature leaf tissue (Fig. 3; Table 2). A much lower fraction of the 24-mer clusters (6% of 29,765) were differentially expressed, and these are more evenly distributed among the four sampled tissues (Table 2).
The 21-mer and 24-mer clusters were evaluated to determine if the small RNAs within each were in phase. Phasing refers to the accumulation of small RNAs at regular intervals along the genome in a frame that matches the length of the small RNAs, as would occur by the processive cleavage of dsRNAs that consistently have the same unique end. To assess the level of small RNA phasing, the probability that the observed degree of phasing would occur by chance alone was estimated assuming a random placement of the small RNAs in the region occupied by the cluster (see Methods). This method easily detected the three known rice tasiRNA loci, TAS3a, TAS3b, and TAS3c, with in-phase P-values of 4.2 × 10−6, 2.2 × 10−6, and 3.4 × 10−6, respectively, validating its use for this analysis. Using a P-value of ≤0.01, 80.8% of the 21-mer clusters (831 of 1029) were classified as phased, and these were the source of 5684 of the 6356 21-nt small RNAs produced by the 21-mer clusters. The median P-value for these 21-mer clusters is ∼4 × 10−6 (Supplemental Fig. 4). Because the number of phased clusters expected by chance is ∼10, the false discovery rate (FDR) for the phased 21-mer clusters is 1.2% at this P-value cutoff. However, using the same maximum P-value of 0.01 to identify phased 24-mer clusters generated an unacceptably high FDR. We therefore chose a lower P-value cutoff (P = 0.00012766), which resulted in a FDR of 10%, and this allowed the identification of 38 phased 24-mer clusters that include 760 distinct 24-nt small RNAs (Table 2; Fig. 3). Thus, in contrast to the 21-mer clusters, the vast majority of which are phased, only a small percentage (0.13%,) of the 24-mer clusters were phased, and the phased 24-mer clusters, in contrast to the bulk of the 24-mer clusters, appear to display a nonrandom distribution in the genome reminiscent of the 21-mer superclusters. Interestingly, examination of tissue specificity showed that virtually all of the small RNAs from both the phased 21-mer and the phased 24-mer clusters are differentially expressed, accumulating almost exclusively in the developing inflorescence (Table 2). Mature leaf tissue contains the next highest number of 21-mer clusters, but none of these small RNA clusters are phased.
A 22-nt sequence motif is offset by exactly 12 nt from the main in-phase subset of the phased 21-mer clusters
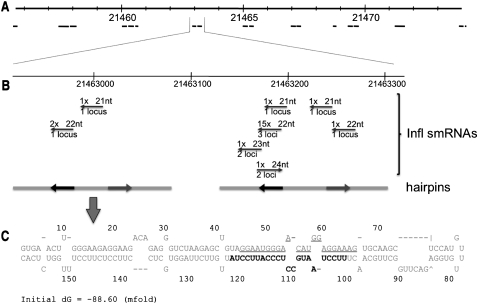
The phase of tasiRNAs is set by small RNA directed cleavage of the tasiRNA-producing RNA. To examine the possibility that a similar strategy is used to set the phase for the phased inflorescence small RNAs detected here, or that a common motif is associated with these clusters, the 21-mer cluster sequences plus an extra 100 nt on either side were scanned for potential motifs using the program MEME (Bailey and Elkan 1994), demanding at least a total of 500 motifs on either strand among the 1029 expanded 21-mer cluster regions. This analysis resulted in the detection of a 22-nt motif (Fig. 4) with a combined E-value across 809 of the 1029 21-mer clusters of 7.3 × 10−2746 (see Methods). The vast majority of these clusters, 89%, were associated with just one copy of the motif. The striking result is that 705 (85%) of the 831 phased clusters are associated with a copy of the motif that is offset by exactly 12 nt from the main phase of small RNAs in the cluster (Fig. 4A,B). In these cases, the motif is directed inwards toward the phased cluster and is more frequent near the edges of the clusters. The second possible position away from the edge of the small RNA clusters has the highest frequency of matches to the motif (Fig. 4C). This flanking motif could serve the same function as the miRNA or tasiRNA target sites found in tasiRNA transcripts, namely to serve as a target for small RNA directed cleavage that establishes phasing of the cluster.
Figure 4.
The 22-nt motif associated with the phased 21-mer clusters. (A) The phased and unphased small RNAs comprising a representative 21-mer cluster are shown. The location of the motif is shown by the arrow at the top, below “motif.” The polarity of the small RNAs is indicated by the arrows, and the number of times the small RNA was sequenced is indicated above the arrow. This 21-mer cluster is located at chr1:34554118..34554411. The probability that this cluster is phased by chance alone is 2.6 × 10−17, and the probability that the indicated motif matches the consensus by chance alone is 2.4 × 10−8. (B) The approximate probability of a nucleotide occurring at specific positions of the motif is shown in the top matrix, and the consensus sequence of the motif is shown below the matrix. (C) The positions of the 705 motifs that are offset 12 nt from the phase of the 21-mer clusters are shown.
We searched our small RNA libraries for RNAs that could potentially target the sense or antisense sequence of the 22-nt motif and direct phasing of the 21-mer phased inflorescence small RNAs. Twelve 22-nt small RNAs were identified that could target the motif using rules established for miRNA targeting (Table 3; also see Methods; Llave et al. 2002; Jones-Rhoades and Bartel 2004; Allen et al. 2005). Both strands of the motif were included during the search for motif-targeting small RNAs to reveal any bias in the strand that could be targeted. The 12 potential motif-targeting 22-nt small RNAs are predicted to induce cleavage after the 12th nucleotide of the motif, producing a discrete end that could set the phase of these 21-mer small RNA clusters. Furthermore, these 12 small RNAs all target the sense strand of the motif and each can target a number of the clusters (Table 3). Collectively the 12 motif-targeting small RNAs are capable of targeting 210 of the 705 phased 21-mer clusters associated with the motif using fairly stringent conditions for miRNA induced cleavage, which may not exactly apply in this situation. Lowering the stringency would likely identify additional small RNA/target pairs. In addition, our small RNA libraries are not saturated and further in-depth small RNA sequencing might uncover more motif-targeting small RNAs.
Table 3.
Small RNAs that may target the 22-nt motif associated 21-mer phased clusters
aThe tissues or sequencing runs from which the small RNA sequences originate. Run 1 and 2 are sequencing runs from mixed tissues and include material from inflorescence.
bThe nucleotide of the small RNA (mostly position 10) after which cleavage in the motif (after position 12) is predicted to occur consistent with the setting of the phase of the 21-mer clusters.
cThe strand of the motif that the indicated small RNA is predicted to target.
dThe number of cluster motifs that are targeted by the indicated small RNA.
The 22-nt small RNAs that potentially target the motif-associated 21-nt phased clusters constitute a new family of miRNAs, miR2118
The 12 small RNAs most likely to target the 22-nt motif could be mapped to only two regions of the genome, a large 20-kb region on chromosome 4 (chr4:21455568..21474171) (Fig. 5A) and a short 3-kb region on chromosome 11 (chr11:7802932..7806229). Interestingly, all of the motif-targeting small RNAs that are derived from chromosome 4 have the same polarity. This ∼20-kb region on chromosome 4 displays a highly unusual genomic organization and is comprised of nine repeats consisting of two to three copies of a ∼170-nt inverted repeat separated by ∼2 kb of sequence (Fig. 5A). The other potential motif-targeting small RNA locus on chromosome 11 also has at least three copies of these inverted repeats. RNA copies of the inverted repeats are predicted to fold into a hairpin structure having the properties of miRNA precursors with the motif-targeting small RNAs located in highly base-paired regions (Fig. 5A; Supplemental Fig. 5A). Furthermore, small RNAs having the properties of a miRNA* were detected for four of these inverted repeats (Supplemental Fig. 5A; Meyers et al. 2008). Therefore, the 22-nt small RNAs that likely target the 22-nt motif constitute a new family of miRNAs, miR2118. Because these miRNAs are 22 nt in length, which is the size of Arabidopsis DCL2 products, these data raise the possibility that excision of the motif-targeting miRNA is mediated by the rice homolog of Arabidopsis DCL2 rather than that of DCL1.
Figure 5.
The genomic origins of the motif-targeting small RNAs, miR2118, and potential folding of transcripts containing these miRNAs. (A) The ∼20-kb region that codes for most of the motif-targeting small RNAs. This region contains nine degenerate repeats each comprised of two to three inverted repeats. Inverted repeats are indicated by black lines below. (B) An enlargement of the indicated region. The small RNAs potentially derived from this region are displayed. The polarity of the small RNAs is indicated by the arrows, and the number of times the small RNA was sequenced and the size of the small RNA are indicated above the arrow. The number of loci potentially encoding each small RNA is shown below the arrow. The positions of the motif-targeting small RNA (black) and the motif-targeting small RNA* (gray) are shown on the gray line representing the inverted repeat. The left inverted repeat contains the motif-targeting 22-nt small RNA (smRNA165692) while the right inverted repeat contains a 22-nt motif-targeting small RNA (smRNA165598) that maps to an additional two locations in the overall region. (C) The predicted hairpin structure of the indicated inverted repeat. The motif-targeting small RNA, miR2118, is shown in black and the motif-targeting RNA*, miR2118*, is underlined.
Small RNA libraries from rice and other plants were searched for small RNAs in the new miRNA family. No members of the miR2118 family were detected in other rice small RNA libraries or in libraries derived from Arabidopsis (Gustafson et al. 2005; Backman et al. 2008), Medicago (Szittya et al. 2008), tomato (Moxon et al. 2008), or Selaginella (Axtell et al. 2007). Seven members of the miR2118 family were detected in libraries derived from early developmental stages of maize anthers. All seven are 22 nt in length and occur in regions of sequence that can fold into hairpin structures resembling miRNA precursors; in addition, four have associated miRNA*s (Supplemental Fig. 5B). Six of the seven miRNA, but none of the miRNA*, sequences were also detected in maize libraries derived from immature ears (Nobuta et al. 2008). However, members of the miR2118 family were not in libraries derived from seedlings, root or shoot apices, and mature leaves (Johnson et al. 2007; Wang et al. 2009; C Johnson, V Vance, V Sundaresan, and L Bowman, unpubl.). Based on the conservation of the new miRNA family in rice and maize, separated by 50 million years, along with the nearly exclusive expression in inflorescences, we propose that this new miRNA family plays an important role in the development of reproductive structures in both cereals.
The phased 24-mer clusters are also flanked by a motif that likely sets the phase of the clusters
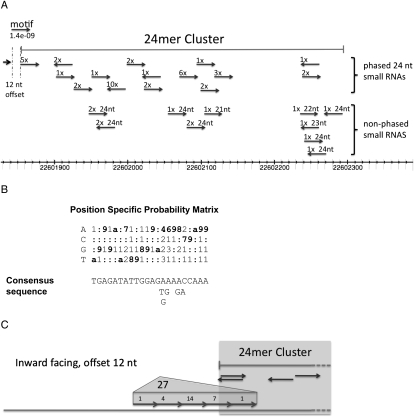
A similar search for a conserved motif was carried out for the 38 loci encoding phased 24-mer small RNAs, again using the program MEME. A 22-nt motif was identified upstream of 28 of the 38 loci encoding phased 24-mer small RNAs (Fig. 6A–C), and the vast majority are associated with only one copy of the motif. Strikingly, the motif is offset by 12 nt from the main phase of the 24-nt RNAs in 27 of these loci. Similar to the motif associated with the 21-mer clusters, the motif sequences for all 27 loci are pointed toward the cluster, and the second possible position away from the edge of the main phase has the highest frequency of matches to the motif (Fig. 6C). Although 22-nt motifs were identified upstream of loci encoding both 21-mer and 24-mer phased inflorescence small RNAs, the primary sequences of the motifs are unrelated to one another.
Figure 6.
The 22-nt motif associated with the phased 24-mer clusters. (A) The phased and unphased small RNAs that comprise a representative phased 24-mer cluster are shown, and the 22-nt motif is shown by the arrow at the top, below “motif.” The polarity of the small RNAs is indicated by the arrows, and the number of times the small RNA was sequenced is indicated above the arrow. This 24-mer cluster is located at chr11:22601855..22602293. The probability that this cluster is phased by chance alone is 6.8 × 10−15, and the probability that the indicated motif matches the consensus by chance alone is 1.4 × 10−9. (B) The probability of a nucleotide occurring at specific positions of the motif is shown in the top matrix, and the consensus sequence of the motif is shown below the matrix. The letter “a” represents a probability of 100%. (C) The positions of the 27 motifs that are offset 12 nt from the phase of the phased 24-mer clusters are shown.
A search of our small RNA libraries identified a 22-nt small RNA, UUUGGUUUCCUCCAAUAUCUCA, that is predicted, based on rules established for miRNAs, to direct cleavage of the 22-nt motif associated with the phased 24-mer clusters, thereby setting their phase. As is the case for the small RNAs that could direct phasing of the 21-mer clusters, this small RNA also targets the sense strand of the motif and is preferentially expressed in developing inflorescences. Furthermore, an RNA sequence containing the 22-nt small RNA would be expected to fold into a stem–loop structure with the 22-nt small RNA located in the stem (Supplemental Fig. 6A). Interestingly, closely related small RNAs were detected in maize anthers. These also have the properties of miRNAs, and miRNA* sequences accumulate for some of these miRNAs (see Supplemental Fig. 6B and legend). Therefore, these RNAs constitute a new miRNA family, miR2775. The identified rice small RNA is predicted to target only three of the motif sequences associated with the phased 24-mer clusters, and the number of reads for this small RNA is low (Supplemental Fig. 6A). Therefore, the evidence for a potential role of this small RNA in setting the phasing of the 24-mer clusters is currently weak.
Discussion
The present work has identified two major types of small RNA clusters in rice, one characterized by 24-nt small RNAs and the other by 21-nt small RNAs. These two types of clusters are arranged differently in the genome and have different expression patterns. The vast majority of the clusters (29,765 of 36,780 or 81%) are of the 24-mer variety, a size class that has previously been implicated in transcriptional silencing of repeated regions of the genome often associated with transposons. These rice 24-mer clusters are distributed more or less uniformly over the genome and nearly half of them overlap with genome repeats, consistent with a role in directing silencing of elements associated with the repeats. The great majority of the 24-mer clusters (94%) are not significantly differentially expressed in the four tested tissues, suggesting that these small RNAs perform housekeeping functions in the silencing of many repeats in all tissues. Interestingly, the observation that some of the 24-mer clusters are expressed in a tissue-specific manner opens the possibility that transcriptional silencing of some genomic regions is developmentally regulated. Of the four tissues we examined, the greatest number and percentage of 24-mer clusters that were differentially expressed was found in mature leaf, which was the only terminally differentiated tissue examined.
Although the 21-mer clusters constituted only a small fraction of the total number of clusters (1029 or 2.8%), these small RNAs were striking in several respects. In contrast to the 24-mer clusters, the distribution of 21-mer clusters in the genome is nonrandom: The majority could be grouped into 31 superclusters, by definition containing at least 10 clusters each. The supercluster regions have properties distinct from other parts of the genome. Although they contain repeats at the same frequency as the rest of the genome, they have a significantly higher frequency of certain types of repeats such as “CACTA” and “MITE-adh-10-like” and a lower concentration of likely protein coding gene loci. This result raises the possibility that these classes of transposons play some role in the biogenesis of this class of small RNA.
Perhaps the most surprising feature of the 21-mer clusters is that the majority of them (∼80%) contain phased small RNAs, virtually all of which are highly preferential for developing inflorescence tissues. Furthermore, we identified a degenerate 22-nt motif, which flanked 85% of the phased 21-mer small RNA clusters, in each case located upstream of the cluster in a position that is offset by 12 nt from the phase of the 21-mer small RNAs. Examination of our database identified a new family of 22-nt miRNAs, miR2118, members of which could potentially direct cleavage of the cluster motifs, thereby setting the phase of the cluster. Consistent with the nearly exclusive accumulation of the 21-mer phased small RNAs in the immature inflorescence, we observed that the novel miRNAs that putatively set the phasing of the clusters also accumulate preferentially in the inflorescence.
The motif-targeting small RNAs were identified as a new rice miRNA family based on their origin from predicted hairpin precursors and the accumulation of both the miRNA and miRNA* species (Meyers et al. 2008). In addition to their presence in rice inflorescence small RNA libraries, both miRNA and miRNA* species were also detected in small RNA libraries derived from maize anthers, and the miRNA but not the miRNA* was found in immature ears. The conservation of the miRNA in rice and maize, as well as the conserved expression in reproductive tissues, suggest that it plays an important role in monocot reproductive development. The fact that the new miRNAs are 22 nt instead of 21 nt as is the case for most miRNAs raises the possibility that the motif-targeting small RNAs are produced by the activity of a DCL other than the rice homolog of Arabidopsis DCL1. The most likely candidate would be a rice DCL homolog of Arabidopsis DCL2, which is known to produce a 22-nt product.
Our results point to similarities between the rice phased inflorescence small RNAs and the previously reported class of phased small RNAs, the tasiRNAs. Although the genetic requirements for the biogenesis of the phased small RNAs identified here are not yet known, the current data suggest that their biogenesis resembles that of tasiRNAs, in that the phase is likely set by a miRNA-directed cleavage. There are several differences between the phased inflorescence small RNAs in rice and currently identified tasiRNAs. tasiRNAs arise from only a few loci and are expressed in most tissues, whereas the phased inflorescence small RNAs identified here derive from hundreds of loci, and these are arranged in superclusters and expressed almost exclusively in developing inflorescence tissue. Finally, whereas tasiRNA loci, at least in Arabidopsis, are often flanked on both sides by small RNA target sites (“two-hit” event), we detected a motif flanking only one side of the cluster in our analysis of both 21-mer and 24-mer small RNA generating loci, suggesting that phasing in these clusters is a one-hit event similar to that for TAS1 and TAS2 in Arabidopsis in which the miRNA target site is 5′ of the tasiRNAs produced (Allen et al. 2005). Furthermore, for all loci producing 21- and 24-mer phased inflorescence small RNAs for which a cleavage-motif was identified, the polarity of processing of the presumptive transcript matches that observed for TAS1/2.
We also identified a small number of phased 24-mer clusters, a size class of phased small RNAs that has not been previously reported. Whereas the majority of 21-mer clusters were phased, the phased 24-mer clusters constituted only a very small fraction of the 24-mer clusters (38 of 36,780 total or 0.128%). These phased 24-mers were similar to the phased 21-mer RNAs in some respects: They were expressed almost exclusively in inflorescence tissues and flanked by a degenerate 22-nt motif located in a position offset from the main phase of the cluster. Furthermore, a motif-targeting small RNA was identified in our small RNA library and expressed preferentially in inflorescence. Finally, the motif-targeting small RNA for the 24-mer clusters derives from an RNA sequence that is predicted to fold into a stem–loop structure reminiscent of miRNA precursors and is a member of another new miRNA family, miR2775. The identified 22-nt small RNA is predicted to target only three of the 28 motifs, whereas the 22-nt miRNAs that set the phase for the 21-mer clusters are predicted to target 30% of these clusters. Although we identified small RNAs related to the rice small RNA that putatively sets the phase for the 24-mer cluster in maize anthers, deeper sequencing is required to determine the significance of this motif-targeting small RNA.
Many groups have undertaken an analysis of small RNA populations in rice (Johnson et al. 2007; Nobuta et al. 2007; Heisel et al. 2008; Lu et al. 2008; Sunkar et al. 2008; Zhu et al. 2008; Zhou et al. 2009). Of these studies, three have identified phased small RNAs. An abundant set of phased small RNAs was detected in rice grain, but these are derived from a limited set of long hairpin precursors and, therefore, have a different biogenesis pathway than the phased small RNA detected here (Heisel et al. 2008; Zhu et al. 2008). In addition, six novel tasiRNA-generating loci that produce predominately 21-nt phased small RNAs were proposed by Lu et al. (2008), but we were unable to confirm these loci using our small RNA sequence data (see Supplemental Table 5).
Although the preferential accumulation of phased 21-nt small RNAs and the phase-setting miRNAs in reproductive tissues suggest that these small RNAs function in a conserved process (or processes) that is unique to reproductive development, their precise function is unclear. One possibility is that the phased 21-nt small RNAs mediate a massive reprogramming of gene expression associated with the onset of flowering. Because the phased small RNAs are primarily 21-mers rather than the 24-mer size class associated with transcriptional silencing, the change in gene expression would likely be at the post-transcriptional level. Computer searches for targets of these phased inflorescence small RNAs, using rules established for miRNA targeting, identified hundreds of potential targets. The down-regulation of some of these targets might be essential for development of the inflorescence preceding the initiation of gametogenesis. Using Gene Ontology (GO) annotation (Ashburner et al. 2000) we were unable to identify any particular class of genes that is preferentially targeted by the phased 21-mer small RNAs. Only one group of genes is preferentially targeted by the phased 24-mer small RNAs. Three of the six genes targeted by the phased 24-mer small RNAs encode a eukaryotic initiation factor 5A (EIF5A). A phased 24-mer cluster overlaps the 5′ end of one gene in the EIF5A gene family, and these small RNAs are predicted to target other members of the family. Further experimentation is required to determine the significance of this result.
The rice phased inflorescence RNAs are similar in some respects to the piRNAs, a group of small RNAs that are associated with the piwi members of the argonaute family in animals (for reviews, see Lin 2007; Klattenhoff and Theurkauf 2008). Both classes of small RNA are preferentially expressed in cell lineages connected with developing germ cells or cells associated with reproduction, and are derived from genome clusters rather than being evenly distributed across the genome. The biogenesis of the two kinds of small RNAs is different: The rice phased inflorescence RNAs are likely the product of dicer digestion, whereas piRNAs are not phased and are generated in a dicer-independent manner. The relationship, if any, between these two classes of small RNAs with respect to their genomic functions remains to be elucidated, although it is intriguing that in both plants and animals there may be classes of small RNAs associated primarily with reproduction.
While this manuscript was under review the following manuscript reported the detection of members of miR2118 miRNA family in Phaseolus vulgaris (Arenas-Huertero et al. 2009).
Methods
Production of small RNA sequence data
Rice tissues were harvested from plants grown in a Conviron Environmental Chamber at high light intensity using both high pressure sodium and metal halide lamps for 10.5 h at 28°C and for 13.5 h at 26°C in the dark. RNA samples were extracted from four different tissues: root apex, shoot apex, developing inflorescence, and leaf.
The shoot and root apices were harvested from plants grown for 13–14 d. The root apices were harvested from the root tip just covered by the root cap, ∼250 μm. For the shoot apices P4 primordium and above were dissected away from the meristems and where possible; P3 was also dissected away for a minority of the apices. The inflorescences were 1–2.5 cm in length, corresponding to early-to-mid Stage 7, during which the floral meristems have initiated organ primordia or have begun to form the floral organs (Itoh et al. 2005). For the mature leaf sample the entire leaves were harvested from plants after emergence of inflorescence and were ∼60 cm in length. Maize anthers from the W23 inbred lines were collected at the 1-mm (mitotic proliferation), 1.5-mm (post-mitotic), and 2.0-mm (prophase1 of meiosis) stages. RNA was isolated using Tri Reagent (Sigma) according to the supplied instructions, dissolved in 20 mM Tris-HCl (pH 7.4), 0.25 M NaCl, 1 mM EDTA, and 0.5% sodium dodecyl sulfate, extracted once with an equal volume of phenol and chloroform, and precipitated with ethanol. Small RNA sequences were obtained using the 454 pyrosequencing method (Margulies et al. 2005) using three different linkers (not shown) that were used to represent replicates. The total mapped reads from the individual replicates were 99,465, 62,381, and 112,802 from root apices, 76,679, 71,248, and 114,113 from shoot apices, 145,870, 81,361, and 136,406 from inflorescence, and 85,015, 65,521, and 110,655 from leaves.
Small RNA data handling
The sequence data from three “replicates” of four tissues, root apex, shoot apex, inflorescence, and leaf, make up a total of 12 data sets. These data sets are stored in a MySQL relational database from which most analysis has been performed using various Perl scripts. Small RNAs were mapped to the rice TIGR version 5 genome and the coordinates stored in the same relational database. All small RNAs belonging to the four tissues that map to a single site on the genome (i.e., genome-unique) were processed together and placed into margin-defined groups of small RNAs by demanding a maximum of 100 nt as the separation distance between small RNAs in the same group. Therefore, any small RNA that is ≤100 nt of another will be placed within the same margin-defined group. A cluster is defined as any margin-defined group in which there are at least two different sets of coordinates. This definition does not necessarily imply more than one distinct small RNA sequence (e.g., in the case of a tandem duplication) in all cases; however, since the small RNAs in this analysis were all genome-unique, this case never arose.
Characterization of clusters
Small RNA clusters were classified into three groups, depending on whether the 21-nt or 24-nt small RNAs were in the majority or whether neither the 21-nt nor the 24-nt small RNAs were in the majority. Only those in which 21 and 24-nt small RNAs were in the majority were used for further analysis. The tissue dominance of each cluster was determined by assessing the relative proportion of normalized reads arising from each tissue. The normalized reads were determined for each “replicate” by dividing the observed reads by the normalization factor (i.e., replicate library size divided by 10,000). The tissue normalized read count is the sum of that determined for the three replicates.
Differential expression of 21/24-mer clusters was determined assuming a normal distribution for random variation in normalized small RNA read counts between tissues. The probability that any cluster was differentially expressed across the four tissues was estimated using a four-celled χ2 test with the expected counts dependent on the proportions of total reads between each of the four tissue small RNA libraries (i.e., pooled across “replicates”), and, when <10,000 calculations were required for a multinomial (in this case a quadnomial), the precise P-value was also determined. Both tests were performed using Perl scripts that also updated the relational database. Clusters were considered differentially expressed if there was an available multinomial P-value that was less than or equal to the alpha cutoff of 0.001 (i.e., 99.9% confidence limit), and, when the multinomial was not available, the χ2 value was used.
Clusters in the 21/24-mer classes were classified as being phased or not phased using two different P-value cutoffs. The P-values were determined using the hypergeometric distribution (i.e., sampling without replacement—that is, each position can only be counted once as distinct positions rather than reads at positions, which implies replacement). The statistical test was performed using only the size class that had the main in-phase subset, and this main in-phase subset of small RNAs was used as the positive class and the remainder as the negative class. The test took into account the 2-nt 3′ overhang and the range adjustments thus required, and the available positions on both strands were used. It should be noted that the statistical test was performed following the removal of one small RNA from the main-phase group (since the first small RNA sets the frame and should not be counted twice).
Determining the optimal margin of separation for the regions of 21-mer clusters
If the 21-mer clusters were randomly positioned over the chromosomes, one might expect clustering of these into regions of 21-mer clusters by chance alone. In order to measure how clustered the observed arrangement of 21-mer clusters really is, a simulation was done 1000 times using the same number of 21-mer clusters (1029) and randomly placing them on the genome. For each of the 1000 simulations, the simulated cluster positions were then processed into regions of 21-mer clusters using margins ranging from 10 to 1000 kb and counting how many regions of 21-mer clusters resulted. This was also done on the observed data. For each of the margins, a ratio of the mean number of regions of 21-mers for the 1000 simulations to the observed number was produced and plotted (see Supplemental Fig. 3). The higher this ratio, the greater the difference between the observed and the simulated number of 21-mer cluster regions. The largest ratio indicates the optimal margin and gives the best compromise between the number of 21-mer clusters in any region and the number of regions.
Identifying motifs and potential targeting-small RNAs
The sequence overlapping the 21/24-mer clusters as well as an additional 100 nt of sequence on either side were searched for motifs using the MEME program (Bailey et al. 2006). For the 21-mer clusters, all 1029 clusters were searched asking for at least 500 motifs to be found, while for the 24-mer clusters only those having in-phase P-values ≤ 0.00012766 (FDR = 10%), which was 38, were used. A 22-nt motif was imposed as a refinement after first allowing MEME to determine the motif consensus. The MAST program (Bailey and Gribskov 1998) was used for finding matches of the 22-nt motif in the rice genome, and these results were processed using various Perl scripts in association with MySQL to identify the motif-cluster relationships.
Candidate targeting-small RNAs were identified by first coming up with a candidate short-list using the MAST program at low stringency (-ev 1000) to identify sequences with similarities to both the sense and antisense sequence of the motif. These small RNAs were then used in a reverse search against the sense and antisense motifs with an additional 100 nt of flanking sequence on either side using the FASTH program (Zuker 2003b) to identify potential RNA∷RNA duplexes. Perl scripts were used to reformat the FASTH results and then analyze the relationships of these candidates to the position of the cluster-associated motifs they are predicted to target. Those RNA∷RNA duplexes having normalized alignment scores (Johnson et al. 2007) of ≥1.4 were retained. These were further filtered based on miRNA-targeting criteria as previously reported (Sunkar et al. 2005; Archak and Nagaraju 2007; see supplementary material in Allen et al. 2005), with the exception of the minimum free energy ratio filter included in Allen et al. (2005). Secondary RNA structures were predicted using the online mfold program (Zuker 2003a).
Statistics
Statistics on large data sets was performed using Perl in association with a relational MySQL database, while individual and small scale statistics was performed using the R statistical package, in addition to creating charts. The estimation of the binomial P-value with the assumption of a normal distribution was calculated using the dbinom library. The Mann-Whitney tests were performed using the Wilcox.test R function.
Acknowledgments
This research was supported by a grant from the National Science Foundation (0501760) to L.H.B, V.V., and V.S. We thank Gail Pruss for helpful comments on the manuscript. Maize data generation was supported by a grant from the National Science Foundation (0701880) to V.W.
Footnotes
[Supplemental material is available online at www.genome.org. The rice small RNA sequence data have been submitted to NCBI Gene Expression Omnibus (GEO) (http://www.ncbi.nlm.nih.gov/geo/) repository under accession nos. GSE16248 and GSE16350. The miRNAs have been submitted to Rfam (http://rfam.sanger.ac.uk/) under submission no. 4a19f686 and to EMBL Nucleotide Sequence Database (http://www.ebi.ac.uk/embl/) under submission nos. FN397534–FN397564.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.089854.108.
References
- Alleman M, Sidorenko L, McGinnis K, Seshadri V, Dorweiler JE, White J, Sikkink K, Chandler VL. An RNA-dependent RNA polymerase is required for paramutation in maize. Nature. 2006;442:295–298. doi: 10.1038/nature04884. [DOI] [PubMed] [Google Scholar]
- Allen E, Xie Z, Gustafson AM, Carrington JC. microRNA-directed phasing during trans-acting siRNA biogenesis in plants. Cell. 2005;121:207–221. doi: 10.1016/j.cell.2005.04.004. [DOI] [PubMed] [Google Scholar]
- Ambros V, Chen X. The regulation of genes and genomes by small RNAs. Development. 2007;134:1635–1641. doi: 10.1242/dev.002006. [DOI] [PubMed] [Google Scholar]
- Archak S, Nagaraju J. Computational prediction of rice (Oryza sativa) miRNA targets. Genomics Proteomics Bioinformatics. 2007;5:196–206. doi: 10.1016/S1672-0229(08)60007-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arenas-Huertero C, Pérez B, Rabanal F, Blanco-Melo D, De la Rosa C, Estrada-Navarrete G, Sanchez F, Covarrubias AA, Reyes JL. Conserved and novel miRNAs in the legume Phaseolus vulgaris in response to stress. Plant Mol Biol. 2009;70:385–401. doi: 10.1007/s11103-009-9480-3. [DOI] [PubMed] [Google Scholar]
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. Gene Ontology: Tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axtell MJ, Jan C, Rajagopalan R, Bartel DP. A two-hit trigger for siRNA biogenesis in plants. Cell. 2006;127:565–577. doi: 10.1016/j.cell.2006.09.032. [DOI] [PubMed] [Google Scholar]
- Axtell MJ, Snyder JA, Bartel DP. Common functions for diverse small RNAs of land plants. Plant Cell. 2007;19:1750–1769. doi: 10.1105/tpc.107.051706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Backman TW, Sullivan CM, Cumbie JS, Miller ZA, Chapman EJ, Fahlgren N, Givan SA, Carrington JC, Kasschau KD. Update of ASRP: The Arabidopsis Small RNA Project Database. Nucleic Acids Res. 2008;36:D982–D985. doi: 10.1093/nar/gkm997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey TL, Elkan C. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc Int Conf Intell Syst Mol Biol. 1994;2:28–36. [PubMed] [Google Scholar]
- Bailey TL, Gribskov M. Methods and statistics for combining motif match scores. J Comput Biol. 1998;5:211–221. doi: 10.1089/cmb.1998.5.211. [DOI] [PubMed] [Google Scholar]
- Bailey TL, Williams N, Misleh C, Li WW. MEME: Discovering and analyzing DNA and protein sequence motifs. Nucleic Acids Res. 2006;34:369–373. doi: 10.1093/nar/gkl198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deleris A, Gallego-Bartolome J, Bao J, Kasschau KD, Carrington JC, Voinnet O. Hierarchical action and inhibition of plant Dicer-like proteins in antiviral defense. Science. 2006;313:68–71. doi: 10.1126/science.1128214. [DOI] [PubMed] [Google Scholar]
- Dorweiler JE, Carey CC, Kubo KM, Hollick JB, Kermicle JL, Chandler VL. Mediator of paramutation1 is required for establishment and maintenance of paramutation at multiple maize loci. Plant Cell. 2000;12:2101–2118. doi: 10.1105/tpc.12.11.2101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eamens A, Wang MB, Smith NA, Waterhouse PM. RNA silencing in plants: Yesterday, today, and tomorrow. Plant Physiol. 2008;147:456–468. doi: 10.1104/pp.108.117275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farazi TA, Juranek SA, Tuschl T. The growing catalog of small RNAs and their association with distinct Argonaute/Piwi family members. Development. 2008;135:1201–1214. doi: 10.1242/dev.005629. [DOI] [PubMed] [Google Scholar]
- Fusaro AF, Matthew L, Smith NA, Curtin SJ, Dedic-Hagan J, Ellacott GA, Watson JM, Wang MB, Brosnan C, Carroll BJ, et al. RNA interference-inducing hairpin RNAs in plants act through the viral defence pathway. EMBO Rep. 2006;7:1168–1175. doi: 10.1038/sj.embor.7400837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girard A, Hannon GJ. Conserved themes in small-RNA-mediated transposon control. Trends Cell Biol. 2008;18:136–148. doi: 10.1016/j.tcb.2008.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goff SA, Ricke D, Lan TH, Presting G, Wang R, Dunn M, Glazebrook J, Sessions A, Oeller P, Varma H, et al. A draft sequence of the rice genome (Oryza sativa L. ssp. japonica) Science. 2002;296:92–100. doi: 10.1126/science.1068275. [DOI] [PubMed] [Google Scholar]
- Gustafson AM, Allen E, Givan S, Smith D, Carrington JC, Kasschau KD. ASRP: The Arabidopsis Small RNA Project Database. Nucleic Acids Res. 2005;33:D637–D640. doi: 10.1093/nar/gki127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heisel SE, Zhang Y, Allen E, Guo L, Reynolds TL, Yang X, Kovalic D, Roberts JK. Characterization of unique small RNA populations from rice grain. PLoS One. 2008;3:e2871. doi: 10.1371/journal.pone.0002871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson IR, Jacobsen SE. Epigenetic inheritance in plants. Nature. 2007;447:418–424. doi: 10.1038/nature05917. [DOI] [PubMed] [Google Scholar]
- Huettel B, Kanno T, Daxinger L, Bucher E, van der Winden J, Matzke AJ, Matzke M. RNA-directed DNA methylation mediated by DRD1 and Pol IVb: A versatile pathway for transcriptional gene silencing in plants. Biochim Biophys Acta. 2007;1769:358–374. doi: 10.1016/j.bbaexp.2007.03.001. [DOI] [PubMed] [Google Scholar]
- Itoh J, Nonomura K, Ikeda K, Yamaki S, Inukai Y, Yamagishi H, Kitano H, Nagato Y. Rice plant development: From zygote to spikelet. Plant Cell Physiol. 2005;46:23–47. doi: 10.1093/pcp/pci501. [DOI] [PubMed] [Google Scholar]
- Johnson C, Bowman L, Adai AT, Vance V, Sundaresan V. CSRDB: A small RNA integrated database and browser resource for cereals. Nucleic Acids Res. 2007;35:D829–D833. doi: 10.1093/nar/gkl991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones-Rhoades MW, Bartel DP. Computational identification of plant microRNAs and their targets, including a stress-induced miRNA. Mol Cell. 2004;14:787–799. doi: 10.1016/j.molcel.2004.05.027. [DOI] [PubMed] [Google Scholar]
- Klattenhoff C, Theurkauf W. Biogenesis and germline functions of piRNAs. Development. 2008;135:3–9. doi: 10.1242/dev.006486. [DOI] [PubMed] [Google Scholar]
- Lin H. piRNAs in the germ line. Science. 2007;316:397. doi: 10.1126/science.1137543. [DOI] [PubMed] [Google Scholar]
- Liu B, Chen Z, Song X, Liu C, Cui X, Zhao X, Fang J, Xu W, Zhang H, Wang X, et al. Oryza sativa Dicer-like4 reveals a key role for small interfering RNA silencing in plant development. Plant Cell. 2007;19:2705–2718. doi: 10.1105/tpc.107.052209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llave C, Xie Z, Kasschau KD, Carrington JC. Cleavage of Scarecrow-like mRNA targets directed by a class of Arabidopsis miRNA. Science. 2002;297:2053–2056. doi: 10.1126/science.1076311. [DOI] [PubMed] [Google Scholar]
- Lu C, Jeong DH, Kulkarni K, Pillay M, Nobuta K, German R, Thatcher SR, Maher C, Zhang L, Ware D, et al. Genome-wide analysis for discovery of rice microRNAs reveals natural antisense microRNAs (nat-miRNAs) Proc Natl Acad Sci. 2008;105:4951–4956. doi: 10.1073/pnas.0708743105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallory AC, Elmayan T, Vaucheret H. MicroRNA maturation and action—the expanding roles of ARGONAUTEs. Curr Opin Plant Biol. 2008;11:560–566. doi: 10.1016/j.pbi.2008.06.008. [DOI] [PubMed] [Google Scholar]
- Margulies M, Egholm M, Altman WE, Attiya S, Bader JS, Bemben LA, Berka J, Braverman MS, Chen YJ, Chen Z, et al. Genome sequencing in microfabricated high-density picolitre reactors. Nature. 2005;437:376–380. doi: 10.1038/nature03959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyers BC, Axtell MJ, Bartel B, Bartel DP, Baulcombe D, Bowman JL, Cao X, Carrington JC, Chen X, Green PJ, et al. Criteria for annotation of plant microRNAs. Plant Cell. 2008;20:3186–3190. doi: 10.1105/tpc.108.064311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mlotshwa S, Pruss GJ, Vance V. Small RNAs in viral infection and host defense. Trends Plant Sci. 2008;13:375–382. doi: 10.1016/j.tplants.2008.04.009. [DOI] [PubMed] [Google Scholar]
- Moxon S, Jing R, Szittya G, Schwach F, Rusholme Pilcher RL, Moulton V, Dalmay T. Deep sequencing of tomato short RNAs identifies microRNAs targeting genes involved in fruit ripening. Genome Res. 2008;18:1602–1609. doi: 10.1101/gr.080127.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nobuta K, Venu RC, Lu C, Belo A, Vemaraju K, Kulkarni K, Wang W, Pillay M, Green PJ, Wang GL, et al. An expression atlas of rice mRNAs and small RNAs. Nat Biotechnol. 2007;25:473–477. doi: 10.1038/nbt1291. [DOI] [PubMed] [Google Scholar]
- Nobuta K, Lu C, Shrivastava R, Pillay M, De Paoli E, Accerbi M, Arteaga-Vazquez M, Sidorenko L, Jeong DH, Yen Y, et al. Distinct size distribution of endogeneous siRNAs in maize: Evidence from deep sequencing in the mop1-1 mutant. Proc Natl Acad Sci. 2008;105:14958–14963. doi: 10.1073/pnas.0808066105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouyang S, Buell CR. The TIGR Plant Repeat Databases: A collective resource for the identification of repetitive sequences in plants. Nucleic Acids Res. 2004;32:D360–D363. doi: 10.1093/nar/gkh099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peragine A, Yoshikawa M, Wu G, Albrecht HL, Poethig RS. SGS3 and SGS2/SDE1/RDR6 are required for juvenile development and the production of trans-acting siRNAs in Arabidopsis. Genes & Dev. 2004;18:2368–2379. doi: 10.1101/gad.1231804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajagopalan R, Vaucheret H, Trejo J, Bartel DP. A diverse and evolutionarily fluid set of microRNAs in Arabidopsis thaliana. Genes & Dev. 2006;20:3407–3425. doi: 10.1101/gad.1476406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramachandran V, Chen X. Small RNA metabolism in Arabidopsis. Trends Plant Sci. 2008;13:368–374. doi: 10.1016/j.tplants.2008.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schauer SE, Jacobsen SE, Meinke DW, Ray A. DICER-LIKE1: Blind men and elephants in Arabidopsis development. Trends Plant Sci. 2002;7:487–491. doi: 10.1016/s1360-1385(02)02355-5. [DOI] [PubMed] [Google Scholar]
- Sunkar R, Girke T, Jain PK, Zhu JK. Cloning and characterization of microRNAs from rice. Plant Cell. 2005;17:1397–1411. doi: 10.1105/tpc.105.031682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunkar R, Zhou X, Zheng Y, Zhang W, Zhu JK. Identification of novel and candidate miRNAs in rice by high throughput sequencing. BMC Plant Biol. 2008;8:25. doi: 10.1186/1471-2229-8-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szittya G, Moxon S, Santos DM, Jing R, Fevereiro MP, Moulton V, Dalmay T. High-throughput sequencing of Medicago truncatula short RNAs identifies eight new miRNA families. BMC Genomics. 2008;9:593. doi: 10.1186/1471-2164-9-593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vazquez F, Vaucheret H, Rajagopalan R, Lepers C, Gasciolli V, Mallory AC, Hilbert JL, Bartel DP, Crete P. Endogenous trans-acting siRNAs regulate the accumulation of Arabidopsis mRNAs. Mol Cell. 2004;16:69–79. doi: 10.1016/j.molcel.2004.09.028. [DOI] [PubMed] [Google Scholar]
- Vazquez F, Blevins T, Ailhas J, Boller T, Meins F., Jr Evolution of Arabidopsis MIR genes generates novel microRNA classes. Nucleic Acids Res. 2008;36:6429–6438. doi: 10.1093/nar/gkn670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Elling AA, Li X, Li N, Peng Z, He G, Sun H, Qi Y, Liu XS, Deng XW. Genome-wide and organ-specific landscapes of epigenetic modifications and their relationships to mRNA and small RNA transcriptomes in maize. Plant Cell. 2009;21:1053–1069. doi: 10.1105/tpc.109.065714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodhouse MR, Freeling M, Lisch D. Initiation, establishment, and maintenance of heritable MuDR transposon silencing in maize are mediated by distinct factors. PLoS Biol. 2006;4:e339. doi: 10.1371/journal.pbio.0040339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshikawa M, Peragine A, Park MY, Poethig RS. A pathway for the biogenesis of trans-acting siRNAs in Arabidopsis. Genes & Dev. 2005;19:2164–2175. doi: 10.1101/gad.1352605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, Sunkar R, Jin H, Zhu JK, Zhang W. Genome-wide identification and analysis of small RNAs originated from natural antisense transcripts in Oryza sativa. Genome Res. 2009;19:70–78. doi: 10.1101/gr.084806.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu QH, Spriggs A, Matthew L, Fan L, Kennedy G, Gubler F, Helliwell C. A diverse set of microRNAs and microRNA-like small RNAs in developing rice grains. Genome Res. 2008;18:1456–1465. doi: 10.1101/gr.075572.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003a;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuker M. Predicting nucleic acid hybridization and melting profiles. Genome inform. 2003b;14:266–268. [Google Scholar]