Abstract
Interactions between viruses and phytoplankton, the main primary producers in the oceans, affect global biogeochemical cycles and climate. Recent studies are increasingly revealing possible cases of gene transfers between cyanobacteria and phages, which might have played significant roles in the evolution of cyanobacteria/phage systems. However, little has been documented about the occurrence of horizontal gene transfer in eukaryotic phytoplankton/virus systems. Here we report phylogenetic evidence for the transfer of seven genes involved in the sphingolipid biosynthesis pathway between the cosmopolitan eukaryotic microalga Emiliania huxleyi and its large DNA virus EhV. PCR assays indicate that these genes are prevalent in E. huxleyi and EhV strains isolated from different geographic locations. Patterns of protein and gene sequence conservation support that these genes are functional in both E. huxleyi and EhV. This is the first clear case of horizontal gene transfer of multiple functionally linked enzymes in a eukaryotic phytoplankton–virus system. We examine arguments for the possible direction of the gene transfer. The virus-to-host direction suggests the existence of ancient viruses that controlled the complex metabolic pathway in order to infect primitive eukaryotic cells. In contrast, the host-to-virus direction suggests that the serial acquisition of genes involved in the same metabolic pathway might have been a strategy for the ancestor of EhVs to stay ahead of their closest relatives in the great evolutionary race for survival.
Oceanic phytoplankton (cyanobacteria and eukaryotic microalgae) is responsible for over half of the Earth's primary production. The high level of biological production by microalgae is principally due to their fast turnover rate of 2–6 d on average, relative to 19 yr for land plants (Field et al. 1998). During the last two decades, oceanographers discovered that viral infection is one of the major causes of death of marine microorganisms, including microalgae (Suttle 2007; Nagasaki 2008). It is estimated that 20%–40% of microorganisms in surface sea waters are killed by viruses each day. Being basal components of marine microbial food webs, viruses thus significantly contribute to the cycling of energy and nutrient on a global scale.
Emiliania huxleyi is the most prominent modern coccolithophore, a group of photosynthetic marine unicellular eukaryotes that play a critical role in ocean biogeochemistry (de Vargas et al. 2007). E. huxleyi is a member of the Haptophyta, one of the deepest branching lineages of the eukaryotic tree of life. This microalgal species is known for its beautiful exoskeleton made of calcium carbonate scales (“coccoliths”), and its recurrent blooms turning extensive areas (>10,000 km2) of oceanic surface waters milky-white. E. huxleyi actively participates to CO2 exchange between the atmosphere, seawater, and the lithosphere, through the synthesis of coccoliths and by driving massive sinking of organic and inorganic carbon into the deep sea; thus, it plays a critical role on global carbon cycling and climate change (Charlson et al. 1987; Westbroek et al. 1994). E. huxleyi blooms suddenly terminate with a sharp increase in the abundance of giant viruses (“coccolithoviruses”) that infect and lytically kill the microalgae (Bratbak et al. 1993; Delille et al. 2005). Coccolithoviruses are large double-stranded DNA viruses, and form a monophyletic group within the virus family Phycodnaviridae (Schroeder et al. 2002; Allen et al. 2006c). E. huxleyi virus 86 (EhV-86) is the type species of the genus Coccolithovirus and was originally isolated from a seawater sample collected from a dying E. huxleyi bloom in the English Channel. The 407-kbp genome of EhV-86 is predicted to encode 472 proteins (Wilson et al. 2005) and is the second largest among sequenced eukaryotic viral genomes (Raoult et al. 2004).
Along with the sequencing of the EhV-86 genome came the discovery, for the first time in a virus, of a series of enzyme-coding genes predicted to be involved in the biosynthesis of sphingolipids (Wilson et al. 2005). Sphingolipids are membrane lipids present in all eukaryotes, and some prokaryotes, that are involved in the regulation of various cellular processes (Futerman and Hannun 2004). Sphingolipid metabolism has been mostly studied in mammalian and yeast cells, where it was found to play key roles in signal transductions (Hannun et al. 2001). The de novo sphingolipid biosynthesis leads to the production of ceramide (Merrill 2002), which serves as the backbone for all complex sphingolipids and has a fundamental role in coordinating eukaryotic cell stress responses, including activation of apoptosis (Hannun 1996; Yang et al. 2004; Guenther et al. 2008).
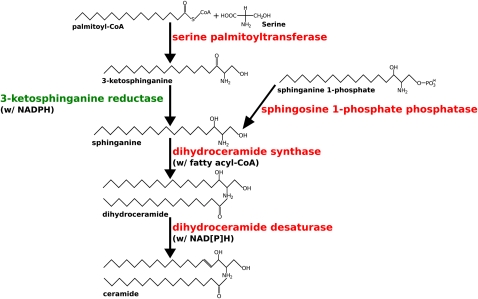
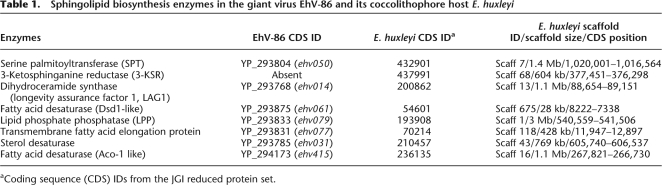
The genome of EhV-86 encodes at least seven enzymes predicted to be involved in the biosynthesis of sphingolipids (Wilson et al. 2005). These include four enzymes central to this metabolic pathway (Fig. 1; Merrill 2002): serine palmitoyltransferase (SPT), dihydroceramide synthase (longevity assurance factor 1 [LAG1]), dihydroceramide desaturase (Dsd1-like fatty acid desaturase [FAD]), and sphingosine 1-phosphate phosphatase (lipid phosphate phosphatase [LPP]). The remaining viral enzymes related to sphingolipid biosynthesis are a sterol desaturase, a transmembrane fatty acid elongation protein, and an Aco1-like FAD (Table 1). Together, these enzymes constitute an almost entire de novo sphingolipid biosynthesis pathway. The seven viral genes are dispersed in the EhV-86 genome, which does not encode any obvious homolog for 3-ketosphinganine reductase (3-KSR), the enzyme catalyzing the second step of the sphingolipid biosynthesis pathway.
Figure 1.
A model of de novo sphingolipid/ceramide biosynthesis pathway. The enzymes found in both EhV-86 and E. huxleyi are indicated in red. The enzyme present only in E. huxleyi is indicated in green.
Table 1.
Sphingolipid biosynthesis enzymes in the giant virus EhV-86 and its coccolithophore host E. huxleyi
aCoding sequence (CDS) IDs from the JGI reduced protein set.
Transcription of the viral genes involved in the sphingolipid pathway is coordinately regulated. It starts at 2 h post-infection, corresponding to an early stage of the viral replication cycle, which lasts from >4 h up to 2 d (Allen et al. 2006a). This timing coincides with the first expression of the viral RNA polymerase (Wilson et al. 2005; Allen et al. 2006a), suggesting that these viral genes may be transcribed by the virally encoded transcription machinery and thus expressed in the host cytoplasm (Allen et al. 2006a). The viral sphingolipid enzymes are not packaged in EhV-86 virions according to a proteomic survey (Allen et al. 2008). The sole EhV sphingolipid enzyme biochemically characterized to date is the SPT, which exhibits atypical domain fusion architecture. In most eukaryotes, SPTs are heterodimers composed of two aminotransferase subunits, the long chain base 1 (LCB1) and LCB2. Han et al. (2006) found that the EhV-86 gene ehv050 encodes a single polypeptide with N-terminal LCB2-like and C-terminal LCB1-like domains. They also found similar fusion proteins in EST libraries from E. huxleyi and Entamoeba histolytica. The viral SPT was further expressed in yeast, demonstrating its activity and unusual preference for myristoyl-CoA (C14) rather than palmitoyl-CoA (C16) (Han et al. 2006). Despite these recent efforts to characterize the EhV sphingolipid biosynthesis genes, their function in the virus replication cycle remains unknown. Based on the role of ceramide as an inducer of cell death in mammalian and yeast cells (Susin et al. 1997; Siskind 2005; Guenther et al. 2008), several investigators proposed that the viral sphingolipid/ceramide pathway may activate host cell death, thus helping disseminate newly generated virions in the host population (Wilson et al. 2005; Bidle et al. 2007).
The unique presence of sphingolipid enzyme genes in only EhV among all known viruses and their ubiquitous distribution in eukaryotes suggest the possibility of horizontal gene transfers (HGTs) of these functionally linked enzyme genes between ancestral virus and eukaryotic host lineages. Viruses are known to carry a variety of host genes. Recent genomics studies are increasingly revealing interesting cases of HGT between prokaryotic phytoplankton (cyanobacteria) and their viruses (cyanophages) (Lindell et al. 2004, 2005; Sullivan et al. 2006; Yoshida et al. 2008). From the observation of “host-like” genes in bacteriophage genomes, Hendrix and other authors proposed a modular theory of phage evolution, in which phages evolve through the stepwise acquisition of genes from diverse sources (Hendrix et al. 2000; Brussow and Hendrix 2002). Eukaryotic large DNA viruses also exhibit genes with homologs in cellular organisms, such as those related to the immune system in poxviruses (Hughes and Friedman 2005), and homologs of cellular genes found in the amoeba-infecting giant mimivirus; albeit with controversy on the timing, mechanism and frequency for possible gene transfers (Raoult et al. 2004; Claverie 2006; Filee et al. 2007; Ogata and Claverie 2007; Moreira and Brochier-Armanet 2008). Nevertheless, little has been documented about the occurrence of gene transfer in eukaryotic alga–virus systems due to the limited availability of genomic sequence data for such host–virus pairs. A recent comparative genomics study of the green alga Ostreococcus tauri and its virus OtV5 could reveal only one putative case of HGT for this eukaryotic alga–virus pair (Derelle et al. 2008). Here we test the hypothesis that HGT is at the origin of the EhV sphingolipid biosynthesis genes using the recently released draft genome sequences of E. huxleyi diploid strain CCMP1516 (7809 scaffolds, 168 Mb, 10× coverage) determined by the International E. huxleyi Genome Sequencing Consortium.
Results
Emiliania huxleyi possesses a full set of sphingolipid biosynthesis genes
Homologs of the seven viral proteins predicted to be involved in the sphingolipid/ceramide biosynthesis were readily identified (BLASTP, E-value < 10−10) in the E. huxleyi genome (Table 1). Remarkably, these E. huxleyi proteins (except for SPT) were the most similar to their viral counterparts, with which they shared from 26%–49% identical residues. In addition, a 3-KSR homolog, apparently missing in EhV-86 genome, was identified in the E. huxleyi genome using green plant and yeast 3-KSR sequences as queries. These host enzymes were found encoded in the middle of different scaffolds with various sizes from 28 kb up to 3 Mb and thus do not cluster in a small region of a host chromosome. The host and viral protein sequences were aligned with a wide phylogenetic array of homologs and were examined regarding the conservation of previously reported sequence features (Lindqvist et al. 1996; Mitchell and Martin 1997; Oh et al. 1997; Stukey and Carman 1997; Jiang et al. 1998; Winter and Ponting 2002). We confirmed the presence of most of the sequence motifs and conserved catalytic residues (Supplemental Fig. S1), suggesting that both viral and host enzymes are functional.
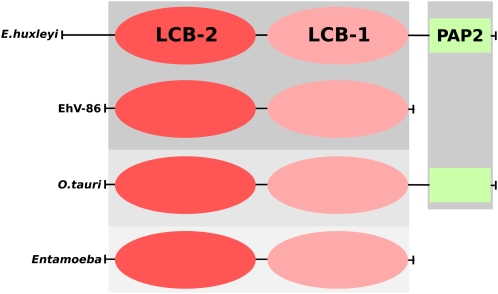
The viral LPP (ehv079) and its closest host homolog (JGI_193908) belong to the phosphatidic acid phosphatase type 2 (PAP2) superfamily (Pfam PF01569; E-value < 7 × 10−9). At least six PAP2 superfamily proteins were found encoded in the E. huxleyi genome. Notably, one of the PAP2 sequences was located in the C terminus of JGI_432901 corresponding to the enzyme SPT (E-value = 0.0096), indicating a fusion of three domains for the protein (LCB2, LCB1, and PAP2) (Fig. 2). We found the same tri-domain architecture in the homologous protein from the green alga O. tauri.
Figure 2.
Domain architectures of serine palmitoyltransferases from E. huxleyi, EhV-86, O. tauri, and Entamoeba spp. LCB2-like domains correspond to red ovals; LCB1-like domain, pink ovals; and PAP2 domains, green rectangles. Background gray scales correspond to the level of sequence similarity from the E. huxleyi SPT sequence (darker gray for higher BLAST scores).
E. huxleyi thus possesses a complete set of enzymes for the sphingolipid biosynthesis. We attempted to detect a distant 3-KSR homolog in EhV-86, using the newly identified host 3-KSR sequence and a Pfam profile for 3-KSRs (PF00106) as queries, but no such homolog could be found in the viral genome.
Evidence of HGTs between giant DNA virus and its eukaryotic host
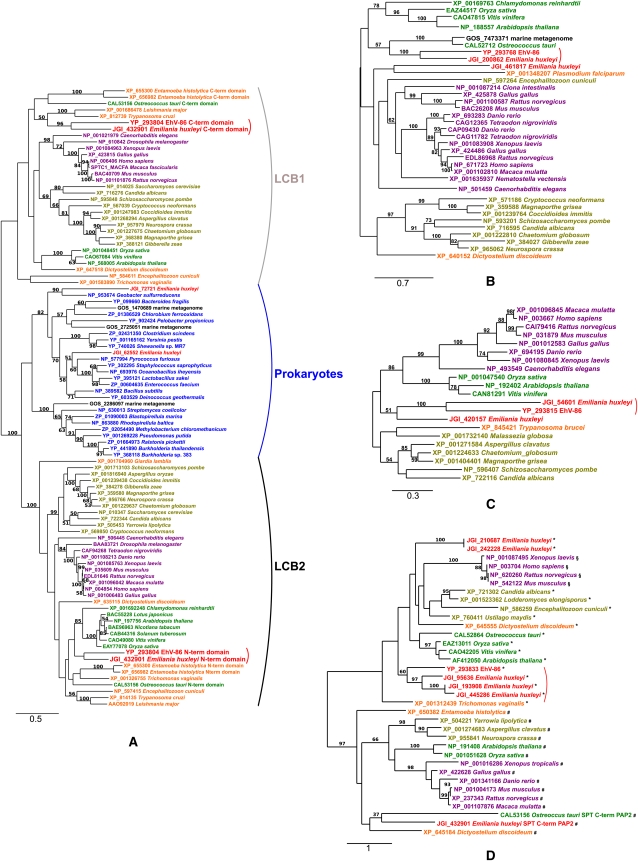
Maximum likelihood phylogenetic trees were reconstructed for the seven sphingolipid biosynthesis enzymes shared by EhV-86 and E. huxleyi, including the widest taxonomic range of homologs available in GenBank (Fig. 3; Supplemental Fig. S2). In all cases, the trees displayed a monophyletic grouping of EhV-86 and E. huxleyi sequences, including their respective SPT's LCB1 and LCB2 domains (Fig. 3A). In five cases, >90% bootstrap values supported the clustering of EhV-86 and E. huxleyi protein sequences (96% for the C-terminal LCB1 domain [Fig. 3A]; 100% for the LAG1 [Fig. 3B]; 100% for the Dsd1-like FAD [Fig. 3D]; 100% for the sterol desaturase, [Supplemental Fig. S2A]; and 91% for the transmembrane fatty acid elongation protein, [Supplemental Fig. S2C]). The branching positions of the EhV/E. huxleyi sequence groups for these genes are generally compatible with a deep phylogenetic origin of E. huxleyi within eukaryotes, except for the Aco1-like FADs (Supplemental Fig. S2B). These results strongly suggest that the seven sphingolipid biosynthesis genes were horizontally transferred between the eukaryotic and viral lineages leading to E. huxleyi and EhV-86, respectively. The EhV/E. huxleyi Aco1-like homologs were more similar to bacterial homologs than to eukaryotic homologs in terms of both domain organization and sequence; fungal homologs have two delta-9-desaturase domains and an additional chytochrome b5-domain (Sperling et al. 2003), while a single delta-9-desaturase domain was identified for EhV, E. huxleyi, and bacterial homologs.
Figure 3.
Maximum likelihood phylogenetic trees based on the amino acid sequences of the four central enzymes in the sphingolipid biosynthesis pathway. (A) Serine palmitoyltransferase LCB1 and LCB2 domain sequences and their homologs. (B) Dihydroceramide synthases (LAG1). (C) Dsd1-like fatty acid desaturases. (D) Lipid phosphate phosphatase (LPP), the PAP2-domain sequence from the E. huxleyi SPT, and their homologs. These trees are unrooted per se, although we have arbitrarily chosen a root (mostly by mid-point rooting) for each tree only for visualization purpose. The number of substitutions per site is indicated under the scale bar. In D, (#) sequences best hitting to sphingosine 1-phosphate phosphatases (cd03388) after NCBI/CDD searches; (*) sequences best hitting to phosphatidic acid phosphatases (cd03390); (§) sequences best hitting to the wunen subfamily sequences (a family of membrane associated phosphatidic acid phosphatases; cd03384).
Viral sphingolipid biosynthesis genes are widespread and functional
To further assess the presence and function of the sphingolipid/ceramide biosynthesis genes in the E. huxleyi/EhV system, we looked for six of them in various E. huxleyi and EhV strains covering a wide geographic range (Supplemental Table S1). We could successfully PCR amplify and sequence most of the genes in the host strains and all of them, except for 3-KSR, in the 11 virus strains. The sphingolipid biosynthesis genes thus appear to be prevalent in different E. huxleyi and EhV strains. The amplified gene sequences from the host strains were highly similar (Supplemental Fig. S3). In contrast, the viral sequences exhibited substantial interstrain variation and were further used to investigate the functional status of the encoded enzymes by assessing evolutionary rates and selection pressure at the sequence level.
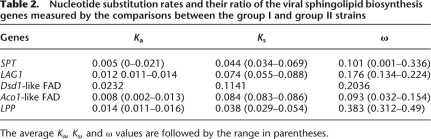
Maximum parsimony trees for the five genes from the 11 viral strains showed topologies compatible with each other and revealed a clustering of the strains into two groups, I and II (Supplemental Fig. S4). This clustering is consistent with previous reports (Allen et al. 2007) and correlates with the isolation time points and/or geographical origins of the strains (Supplemental Table S1). Group I is composed of three strains isolated from the English Channel in 1999 and three strains isolated during mesocosm experiments in Norway in 2000 and 2003, while group II comprises exclusively strains isolated from the English Channel in 2001. We computed synonymous (Ks) and nonsynonymous (Ka) substitution rates, and their ratio (Ka/Ks = ω) between group I and group II sequences (Table 2). The average ω values were substantially below 1 ranging from 0.09 for Aco1-like FAD to 0.43 for LPP.
Table 2.
Nucleotide substitution rates and their ratio of the viral sphingolipid biosynthesis genes measured by the comparisons between the group I and group II strains
The average Ka, Ks, and ω values are followed by the range in parentheses.
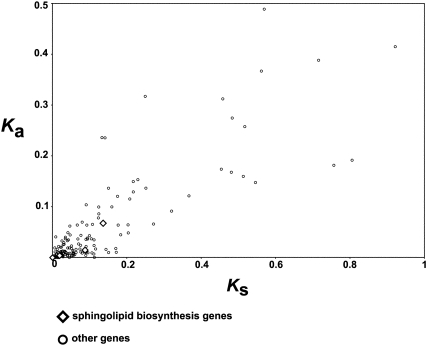
Finally, we assessed the evolutionary rates of the viral sphingolipid biosynthesis genes relative to other EhV genes by computing Ka and Ks values for orthologous gene pairs between two viral strains, EhV-86 and EhV-163 (Allen et al. 2006b). The substitution rates of the sphingolipid biosynthesis-related genes were found comparable to those of other EhV genes (Fig. 4). Overall, our results suggest that a negative selection acted on these viral genes, again supporting that they are fully functional.
Figure 4.
Synonymous (Ks) and nonsynonymous (Ka) substitution rates for the orthologous sequences between EhV-86 and EhV-163. Of the seven EhV-86 sphingolipid enzyme genes, EhV-163 orthologs were found for four genes (SPT, LAG1, Dsd1-like FAD, LPP), which are indicated as diamonds.
Discussion
We have provided here clear evidence supporting HGT(s) between E. huxleyi and EhV for seven genes probably involved in the biosynthesis of sphingolipids. Given the previously reported cotranscription of the viral genes (Allen et al. 2006a) and the validated enzymatic activity of the viral SPT (Han et al. 2006), all of these viral enzymes are likely to be functional. The prevalence of those genes in a wide range of host and virus strains and the pattern of their amino acid and DNA sequence conservation revealed by this study further support that these enzymes are functional in both E. huxleyi and EhVs.
The presence of conserved sequence motifs and catalytic residues in the viral and host enzyme sequences, and their phylogenetic positions within individual enzyme families are generally compatible with the current annotations of their enzymatic functions. Regarding the classification of these putative enzymes, the PAP2 superfamily sequences are worth mentioning. The enzyme LPP belongs to the PAP2 superfamily, which includes a variety of phosphatase subfamilies (Pfam PF01569). Our phylogenetic analysis (Fig. 3C) indicates that the viral LPP (ehv079) and its closest host homolog (JGI_193908) are more similar to phosphatidic acid phosphatases than to sphingosine 1-phosphate phosphatases. Thus the bona fide substrate of these LPPs may not be sphingosine 1-phosphate. By contrast, the PAP2 domain embedded in the C-terminal region of the E. huxleyi SPT sequence (JGI_432901) appears more similar to many sphingosine 1-phosphate phosphatases than other phosphatases. This algal protein may thus be involved in two distinct steps shown in the sphingolipid/ceramide biosynthesis pathway model in Figure 1.
Wilson et al. (2005) originally proposed that the viral enzymes would be part of a viral lysis strategy to kill the host cell. This hypothesis was based on the role of ceramide as an inducer of cell death in mammalian and yeast cells (Guenther et al. 2008). The presence of eight protease genes in the EhV-86 genome (Wilson et al. 2005) parallels the known connection between protease activation and ceramide-induced programmed cell death in mammalian cells (Susin et al. 1997; Siskind 2005). Bidle et al. (2007) recently demonstrated the activation of programmed cell death of E. huxleyi cells upon EhV infection and concomitant induction of host metacaspases. Notably, the EhV-86 genome encodes eight proteins with caspase cleavage recognition sequence motifs. Wilson et al. (2009) also suggested that the viral sphingolipid biosynthesis pathway might act to temporarily inhibit cell death process to prolong the length of infection. Given the wide variety of eukaryotic signaling pathways known to be induced by sphingolipids, however, possible biological roles of the viral sphingolipid pathway may not be necessarily linked to the control of lysis. For example, the viral sphingolipid biosynthesis may have a direct role in the highly specific cell/virus recognition processes involving membrane interactions (Allen et al. 2008). Sphingolipids have fundamental functions in host–pathogen membrane interactions (Riethmuller et al. 2006), through the organization of membrane domains (called membrane rafts) where different sphingolipids, cholesterol, receptors, and signaling molecules are recruited to coordinate the dynamics of membrane structures. EhV virions are thought to contain a lipid membrane layer within their capsid, which in turn is enveloped by another lipid membrane upon their release from cells (Allen et al. 2008). The EhV sphingolipid biosynthesis enzymes may thus modulate the lipid profile of the host and/or viral membranes (Han et al. 2006) to facilitate the release of newly formed virions by membrane budding (Allen et al. 2008) or to enhance the host–virus recognition process upon infection. It is also worth noting a recently revealed connection between EhV infectivity and the host's life cycle. Frada et al. (2008) showed that EhVs are capable of infecting the diploid phase of E. huxleyi cells but not the haploid phase of this alga. Intriguingly, sphingolipids are known to be involved in the regulation of meiotic division in higher animals (Strum et al. 1995; Yang et al. 2004). Given this new connection, it is tempting to speculate that EhVs may take advantage of their own sphingolipid pathway to control a host's sexual life cycle.
Mixing of genetic pools between viruses and their hosts by horizontal transfer or through symbiotic association might have played significant roles in the evolution of viruses as well as of their hosts. The “eukaryogenesis” theory (Bell 2001; Takemura 2001) proposes that viruses might have been at the origin of the nucleus and that viruses have provided several “viral” features to primitive cells, which are now seen as properties of modern eukaryotic cells. Early and possibly bidirectional (Claverie 2006) genetic exchanges between viruses and their hosts through such a process could lead to a situation where homologs of certain viral genes were found in all or most of modern eukaryotic species (Roossinck 2005; Villarreal 2005; Claverie 2006), as previously proposed, for instance, for the eukaryotic DNA polymerase (Villarreal and DeFilippis 2000; Forterre 2006). However, such early evolutionary processes prior to the divergence of major eukaryotic lineages do not readily explain the HGT(s) of the sphingolipid biosynthesis genes analyzed in this study, since the viral sphingolipid genes show a much higher sequence similarity to the E. huxleyi homologs than to the homologs in other eukaryotes; the HGT(s) are likely to have occurred after the separation of the lineage leading to E. huxleyi from other major lineages of eukaryotes.
Regarding the possible direction of the sphingolipid pathway gene transfer, the direction from viruses to the ancestors of E. huxleyi (“V2H direction”) has an advantage in that it would easily minimize the number of required evolutionary events by invoking genome or en bloc gene transfer. The sphingolipid genes are dispersed in both the EhV-86 and E. huxleyi genomes. Transfer of all or part of a relatively small viral genome to a larger host genome (by unknown mechanism), probably followed by the elimination of the original eukaryotic homologs, would require fewer evolutionary steps than the transfer of a set of genes in the reverse direction. The V2H scenario predicts that the E. huxleyi homologs may retain viral homolog-like properties that are missing in other protist lineages. In this regard, the atypical fusion of the LCB2/LCB1 domains in SPT (Fig. 2) would deserve future investigation. The domain fusion was observed not only in EhVs and E. huxleyi but also in other eukaryotes such as O. tauri and Entamoeba clearly placed outside the group of EhV/E. huxleyi sequences in the LCB2/LCB1 tree (Fig. 3A). Notably, an additional PAP2-domain was found in the SPTs of E. huxleyi and O. tauri. Whether these SPT domain organizations support the V2H scenario is unclear due to the lack of estimate for the relative frequency between this sort of domain fusion/deletion and gene transfer among viruses and eukaryotes. Sequencing of other protists, including other members of the Haptophyta, may provide important clues to better understand the origin of the SPT domain organizations and to assess the likeliness of the V2H hypothesis. The V2H hypothesis implies that sphingolipid biosynthesis pathway genes were present in the genomes of very ancestral viruses infecting primitive eukaryotic cells.
The direction from the ancestors of E. huxleyi or their relatives to the viral lineages leading to EhV (“H2V direction”) is also possible and appears more parsimonious than the V2H direction in several aspects. First, these sphingolipid genes are ubiquitous in eukaryotes. The V2H scenario would require an additional and earlier HGT for a set of these genes between ancestral eukaryotes and ancestral viruses (e.g., an earlier HGT prior to the divergence of eukaryotic lineages). Furthermore, the branching positions of the EhV/E. huxleyi sequence groups for these genes are globally compatible with a deep phylogenetic origin of E. huxleyi within eukaryotes, with one exception for the viral and host Aco1-like FADs (Fig. S2B). The V2H scenario would require an additional evolutionary mechanism (or constraint) forcing the sequences of the EhV homologs to be placed near the basis of eukaryotic trees without long branches. It should be noted that branches longer for viruses than for their eukaryotic hosts are often obtained by phylogenetic tree reconstruction (Moreira and Brochier-Armanet 2008; Claverie et al. 2009; Forterre and Gadelle 2009). In the H2V scenario, the viral acquisition of the sphingolipid enzyme genes was probably gradual, through multiple HGT events, rather than through a single en bloc transfer of multiple genes. However, it is difficult to reliably assess the relative timing of these HGT events from the current sequence data due to the large sequence divergence between the viral and host homologs. The initial acquisition of one of the genes of this metabolic pathway, for instance, the enzyme SPT (the rate limiting step of this pathway), might have been sufficient for the virus to start modulating its host's life span or lipid profile, thus giving this altered virus a selective advantage on other viral strains. The later acquisitions of additional genes could have further enhanced the viral capacity to modulate the cellular metabolism. This type of serial gene acquisition by a virus could be a possible way to increase its fitness, and might be a driving force in the Red Queen evolution of viral strains infecting the same host species.
Methods
Sphingolipid biosynthesis gene sequences from E. huxleyi CCMP1516
The genome sequence data of E. huxleyi CCMP1516 strain were produced by the International E. huxleyi Genome Sequencing Consortium in collaboration with the U.S. Department of Energy Joint Genome Institute (http://www.jgi.doe.gov/). The genome sequence data are being analyzed by the consortium members and will be published elsewhere. The amino acid sequences corresponding to the seven EhV-86 sphingolipid biosynthesis genes (Table 1) were used to identify their homologs in the E. huxleyi genome sequences, using BLASTP searches (Altschul et al. 1997) against the host's ORFeome (the JGI reduced protein set as of April 4, 2008; E-value < 10−20). For the detection of E. huxleyi 3-KSR homolog, 3-KSR homologs from green plants (Arabidopsis thaliana and Ostreococcus tauri) were used as TBLASTN queries.
PCR-amplification and sequencing of sphingolipid biosynthesis genes from host and virus strains
Six E. huxleyi and 11 EhV strains were chosen by taking into account their distant geographical origins (Supplemental Table S1) and distinct behavior regarding susceptibility to EhV infection (data not shown). To extract E. huxleyi DNA, 250 mL of late exponential growing cultures were harvested by centrifugation (14,000 rpm for 2 min). A 0.5-mL pellet was recovered and initially treated with proteinase K (5 mg/mL) in a lysis buffer containing 20 mM EDTA (pH 8) and 0.5% SDS (w/v) for 1 h at 65°C. Major cell debris was removed by adding 600 μL of phenol to each sample and centrifuging at maximum speed for 10 min. The top layer was recovered, and the DNA was extracted using an equal volume of chloroform:isoamyl alcohol (24:1). The DNA was precipitated with the addition of 0.5× volume 7.5 M ammonium acetate (pH 7.5), and 2.5× volume absolute ethanol. The pellet was washed three times in 300 μL of ice-cold 70% ethanol, after which it was dried and resuspended in 30 μL of DNase free water. The virus isolates were directly used as DNA template for PCR without prior DNA purification. The on-line application Primer3 (Rozen and Skaletsky 2000) was used to design primers that target homologous regions in both host and viral genes (Supplemental Table S2). The PCR reaction was set up as follows: 1 μL of DNA template (extracted DNA in case of the hosts, viral isolate in the case of the virus) was added to a 25 μL reaction mixture that contained: 1 U Taq DNA polymerase (Promega), 1× PCR reaction buffer (Promega), BSA, 0.25 mM dNTPs, 2.5 mM MgCl2, and 10 pmol of each primer. The PCR was conducted in a PTC-100 cycler (MJ Research) with an initial denaturing step of 95°C (5 min), followed by 35 cycles of denaturing at 95°C (60 sec), annealing at 56°C (60 sec), and extension at 74°C (60 sec). A SequiTherm EXCEL II DNA Sequencing Kit-LC (EpicentreTechnologies) with a LI-COR Automated DNA Sequencer was used to sequence the PCR products.
EhV-86/EhV-163 orthologs
We extracted open reading frames (≥ 60 amino acids) from the partial genome sequence data of EhV-163 (Allen et al. 2006b) using EMBOSS/GETORF software (Rice et al. 2000). EhV-86/EhV-163 orthologous sequence pairs were determined using the reciprocal BLASTP best hit criterion.
Phylogenetic analysis
EhV-86 and E. huxleyi sphingolipid biosynthesis-related protein sequences were used as queries for BLASTP searches against the NCBI non-redundant database (Pruitt et al. 2007) (E-value < 10−3) to identify and retrieve their homologs. We generated multiple sequence alignments using MUSCLE (Edgar 2004). The SPT protein sequences from EhV-86, E. huxleyi, O. tauri, and E. histolytica were split into subsequences according to their particular domain architecture. All gap containing sites were removed from the alignments for the following phylogenetic analyses. Maximum-likelihood phylogenetic analyses were performed using PHYML (Guindon and Gascuel 2003) with the Jones-Taylor-Thornton substitution model (Jones et al. 1994) and with 100 bootstrap replicates. Maximum parsimony phylogenetic analyses were performed for PCR-amplified sequences using PHYLIP/DNAPARS (Felsenstein 2004).
Estimation of Ka and Ks
Each orthologous protein sequence was aligned by MUSCLE and then back-translated into codon alignments. The maximum likelihood computation of synonymous (Ks) and nonsynonymous (Ka) substitution rates and their ratio (ω) for each orthologous pair of sequences was performed using CODEML from the PAML 4 package (Yang 2007). For the comparison of EhV-86/EhV-163 orthologs, we discarded all the sequence pairs showing estimated evolutionary parameters (Ks, Ka) with >50% of standard errors and those having Ks > 1.0.
Acknowledgments
We thank the anonymous referees for their precious comments. The comments from one of the referees on an earlier version of the manuscript were helpful to improve the discussion about the direction of gene transfers. This study is part of the PhD thesis work of A.M. (supported by the EuroPathoGenomics European network of excellence) and is part of the PhD thesis work of A.P. (supported by a Marie-Curie Early Stage Training fellowship [EU-FP6] awarded to the Station Biologique Roscoff). A.M. has performed the reported bioinformatics analyses. AP has performed the reported experimental work. A.M. and A.P. wrote the initial draft of the manuscript. E. huxleyi genome sequence data were produced by the US Department of Energy Joint Genome Institute (http://www.jgi.doe.gov/) in collaboration with the International E. huxleyi Genome Sequencing Consortium community. M.J.A. and A.P. were supported by grants awarded to Dr. William H. Wilson from the Natural Environment Research Council (NERC) Environmental Genomics thematic program (nos. NE/A509332/1 and NE/D001455/1) and Oceans 2025. This work is part of the pluridisciplinary project BOOM (Biodiversity of Open Ocean Microcalcifiers, CdV) funded by the French Agence Nationale de la Recherche, grant ANR-05-BIODIV-004. This work was performed in part on the PACA-BioInfo IBISA Platform.
Footnotes
[Supplemental material is available online at www.genome.org. The sequence data from this study have been submitted to GenBank (http://www.ncbi.nlm.nih.gov/Genbank/) under accession nos. FJ531546–FJ531633.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.091686.109.
References
- Allen MJ, Forster T, Schroeder DC, Hall M, Roy D, Ghazal P, Wilson WH. Locus-specific gene expression pattern suggests a unique propagation strategy for a giant algal virus. J Virol. 2006a;80:7699–7705. doi: 10.1128/JVI.00491-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen MJ, Schroeder DC, Donkin A, Crawfurd KJ, Wilson WH. Genome comparison of two coccolithoviruses. Virol J. 2006b;3:15. doi: 10.1186/1743-422X-3-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen MJ, Schroeder DC, Holden MT, Wilson WH. Evolutionary history of the Coccolithoviridae. Mol Biol Evol. 2006c;23:86–92. doi: 10.1093/molbev/msj010. [DOI] [PubMed] [Google Scholar]
- Allen MJ, Martinez-Martinez J, Schroeder DC, Somerfield PJ, Wilson WH. Use of microarrays to assess viral diversity: From genotype to phenotype. Environ Microbiol. 2007;9:971–982. doi: 10.1111/j.1462-2920.2006.01219.x. [DOI] [PubMed] [Google Scholar]
- Allen MJ, Howard JA, Lilley KS, Wilson WH. Proteomic analysis of the EhV-86 virion. Proteome Sci. 2008;6:11. doi: 10.1186/1477-5956-6-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell PJ. Viral eukaryogenesis: Was the ancestor of the nucleus a complex DNA virus? J Mol Evol. 2001;53:251–256. doi: 10.1007/s002390010215. [DOI] [PubMed] [Google Scholar]
- Bidle KD, Haramaty L, Barcelos ERJ, Falkowski P. Viral activation and recruitment of metacaspases in the unicellular coccolithophore, Emiliania huxleyi. Proc Natl Acad Sci. 2007;104:6049–6054. doi: 10.1073/pnas.0701240104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bratbak G, Egge JK, Heldal M. Viral mortality of the marine alga Emiliania huxleyi (Haptophyceae) and termination of algal blooms. Mar Ecol Prog Ser. 1993;93:39–48. [Google Scholar]
- Brussow H, Hendrix RW. Phage genomics: Small is beautiful. Cell. 2002;108:13–16. doi: 10.1016/s0092-8674(01)00637-7. [DOI] [PubMed] [Google Scholar]
- Charlson RJ, Lovelock JE, Andreae MO, Warren SG. Oceanic phytoplankton, atmospheric sulphur, cloud albedo and climate. Nature. 1987;326:655–661. [Google Scholar]
- Claverie JM. Viruses take center stage in cellular evolution. Genome Biol. 2006;7:110. doi: 10.1186/gb-2006-7-6-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claverie JM, Abergel C, Ogata H. Mimivirus. Curr Top Microbiol Immunol. 2009;328:89–121. doi: 10.1007/978-3-540-68618-7_3. [DOI] [PubMed] [Google Scholar]
- Delille B, Harlay J, Zondervan I, Jacquet S, Chou L, Wollast R, Bellerby R, Frankignoulle M, Vieira Borges A, Riebesell U, et al. Response of primary production and calcification to changes of pCO2 during experimental blooms of the coccolithophorid Emiliania huxleyi. Global Biogeochem Cycles. 2005;19:GB2023. doi: 10.1029/2004GB002318. [DOI] [Google Scholar]
- Derelle E, Ferraz C, Escande ML, Eychenie S, Cooke R, Piganeau G, Desdevises Y, Bellec L, Moreau H, Grimsley N. Life-cycle and genome of OtV5, a large DNA virus of the pelagic marine unicellular green alga Ostreococcus tauri. PLoS One. 2008;3:e2250. doi: 10.1371/journal.pone.0002250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vargas C, Aubry MP, Probert I, Young J, Falkowski P, Knoll AH. Origin and evolution of coccolithophores: From coastal hunters to oceanic farmers. In: Falkowski P, Knoll AH, editors. Evolution of aquatic photoautotrophs. Elsevier Academic; New York: 2007. pp. 251–281. [Google Scholar]
- Edgar RC. MUSCLE: A multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics. 2004;5:113. doi: 10.1186/1471-2105-5-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein J. Department of Genome Sciences; University of Washington, Seattle: 2004. PHYLIP (Phylogeny Inference Package), version 3.6. Distributed by the author. http://evolution.genetics.washington.edu/phylip.html. [Google Scholar]
- Field CB, Behrenfeld MJ, Randerson JT, Falkowski P. Primary production of the biosphere: Integrating terrestrial and oceanic components. Science. 1998;281:237–240. doi: 10.1126/science.281.5374.237. [DOI] [PubMed] [Google Scholar]
- Filee J, Siguier P, Chandler M. I am what I eat and I eat what I am: Acquisition of bacterial genes by giant viruses. Trends Genet. 2007;23:10–15. doi: 10.1016/j.tig.2006.11.002. [DOI] [PubMed] [Google Scholar]
- Forterre P. Three RNA cells for ribosomal lineages and three DNA viruses to replicate their genomes: A hypothesis for the origin of cellular domain. Proc Natl Acad Sci. 2006;103:3669–3674. doi: 10.1073/pnas.0510333103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forterre P, Gadelle D. Phylogenomics of DNA topoisomerases: Their origin and putative roles in the emergence of modern organisms. Nucleic Acids Res. 2009;37:679–692. doi: 10.1093/nar/gkp032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frada M, Probert I, Allen MJ, Wilson WH, de Vargas C. The “Cheshire Cat” escape strategy of the coccolithophore Emiliania huxleyi in response to viral infection. Proc Natl Acad Sci. 2008;105:15944–15949. doi: 10.1073/pnas.0807707105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Futerman AH, Hannun YA. The complex life of simple sphingolipids. EMBO Rep. 2004;5:777–782. doi: 10.1038/sj.embor.7400208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guenther GG, Peralta ER, Rosales KR, Wong SY, Siskind LJ, Edinger AL. Ceramide starves cells to death by downregulating nutrient transporter proteins. Proc Natl Acad Sci. 2008;105:17402–17407. doi: 10.1073/pnas.0802781105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- Han G, Gable K, Yan L, Allen MJ, Wilson WH, Moitra P, Harmon JM, Dunn TM. Expression of a novel marine viral single-chain serine palmitoyltransferase and construction of yeast and mammalian single-chain chimera. J Biol Chem. 2006;281:39935–39942. doi: 10.1074/jbc.M609365200. [DOI] [PubMed] [Google Scholar]
- Hannun YA. Functions of ceramide in coordinating cellular responses to stress. Science. 1996;274:1855–1859. doi: 10.1126/science.274.5294.1855. [DOI] [PubMed] [Google Scholar]
- Hannun YA, Luberto C, Argraves KM. Enzymes of sphingolipid metabolism: From modular to integrative signaling. Biochemistry. 2001;40:4893–4903. doi: 10.1021/bi002836k. [DOI] [PubMed] [Google Scholar]
- Hendrix RW, Lawrence JG, Hatfull GF, Casjens S. The origins and ongoing evolution of viruses. Trends Microbiol. 2000;8:504–508. doi: 10.1016/s0966-842x(00)01863-1. [DOI] [PubMed] [Google Scholar]
- Hughes AL, Friedman R. Poxvirus genome evolution by gene gain and loss. Mol Phylogenet Evol. 2005;35:186–195. doi: 10.1016/j.ympev.2004.12.008. [DOI] [PubMed] [Google Scholar]
- Jiang JC, Kirchman PA, Zagulski M, Hunt J, Jazwinski SM. Homologs of the yeast longevity gene LAG1 in Caenorhabditis elegans and human. Genome Res. 1998;8:1259–1272. doi: 10.1101/gr.8.12.1259. [DOI] [PubMed] [Google Scholar]
- Jones DT, Taylor WR, Thornton JM. A mutation data matrix for transmembrane proteins. FEBS Lett. 1994;339:269–275. doi: 10.1016/0014-5793(94)80429-x. [DOI] [PubMed] [Google Scholar]
- Lindell D, Sullivan MB, Johnson ZI, Tolonen AC, Rohwer F, Chisholm SW. Transfer of photosynthesis genes to and from Prochlorococcus viruses. Proc Natl Acad Sci. 2004;101:11013–11018. doi: 10.1073/pnas.0401526101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindell D, Jaffe JD, Johnson ZI, Church GM, Chisholm SW. Photosynthesis genes in marine viruses yield proteins during host infection. Nature. 2005;438:86–89. doi: 10.1038/nature04111. [DOI] [PubMed] [Google Scholar]
- Lindqvist Y, Huang W, Schneider G, Shanklin J. Crystal structure of Δ9 stearoyl-acyl carrier protein desaturase from castor seed and its relationship to other di-iron proteins. EMBO J. 1996;15:4081–4092. [PMC free article] [PubMed] [Google Scholar]
- Merrill AH., Jr De novo sphingolipid biosynthesis: A necessary, but dangerous, pathway. J Biol Chem. 2002;277:25843–25846. doi: 10.1074/jbc.R200009200. [DOI] [PubMed] [Google Scholar]
- Mitchell AG, Martin CE. Fah1p, a Saccharomyces cerevisiae cytochrome b5 fusion protein, and its Arabidopsis thaliana homolog that lacks the cytochrome b5 domain both function in the alpha-hydroxylation of sphingolipid-associated very long chain fatty acids. J Biol Chem. 1997;272:28281–28288. doi: 10.1074/jbc.272.45.28281. [DOI] [PubMed] [Google Scholar]
- Moreira D, Brochier-Armanet C. Giant viruses, giant chimeras: The multiple evolutionary histories of Mimivirus genes. BMC Evol Biol. 2008;8:12. doi: 10.1186/1471-2148-8-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagasaki K. Dinoflagellates, diatoms, and their viruses. J Microbiol. 2008;46:235–243. doi: 10.1007/s12275-008-0098-y. [DOI] [PubMed] [Google Scholar]
- Ogata H, Claverie JM. Unique genes in giant viruses: Regular substitution pattern and anomalously short size. Genome Res. 2007;17:1353–1361. doi: 10.1101/gr.6358607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh CS, Toke DA, Mandala S, Martin CE. ELO2 and ELO3, homologues of the Saccharomyces cerevisiae ELO1 gene, function in fatty acid elongation and are required for sphingolipid formation. J Biol Chem. 1997;272:17376–17384. doi: 10.1074/jbc.272.28.17376. [DOI] [PubMed] [Google Scholar]
- Pruitt KD, Tatusova T, Maglott DR. NCBI reference sequences (RefSeq): A curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 2007;35:D61–D65. doi: 10.1093/nar/gkl842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raoult D, Audic S, Robert C, Abergel C, Renesto P, Ogata H, La Scola B, Suzan M, Claverie JM. The 1.2-megabase genome sequence of Mimivirus. Science. 2004;306:1344–1350. doi: 10.1126/science.1101485. [DOI] [PubMed] [Google Scholar]
- Rice P, Longden I, Bleasby A. EMBOSS: The European Molecular Biology Open Software Suite. Trends Genet. 2000;16:276–277. doi: 10.1016/s0168-9525(00)02024-2. [DOI] [PubMed] [Google Scholar]
- Riethmuller J, Riehle A, Grassme H, Gulbins E. Membrane rafts in host-pathogen interactions. Biochim Biophys Acta. 2006;1758:2139–2147. doi: 10.1016/j.bbamem.2006.07.017. [DOI] [PubMed] [Google Scholar]
- Roossinck MJ. Symbiosis versus competition in plant virus evolution. Nat Rev Microbiol. 2005;3:917–924. doi: 10.1038/nrmicro1285. [DOI] [PubMed] [Google Scholar]
- Rozen S, Skaletsky H. Primer3 on the WWW for general users and for biologist programmers. Methods Mol Biol. 2000;132:365–386. doi: 10.1385/1-59259-192-2:365. [DOI] [PubMed] [Google Scholar]
- Schroeder DC, Oke J, Malin G, Wilson WH. Coccolithovirus (Phycodnaviridae): Characterisation of a new large dsDNA algal virus that infects Emiliana huxleyi. Arch Virol. 2002;147:1685–1698. doi: 10.1007/s00705-002-0841-3. [DOI] [PubMed] [Google Scholar]
- Siskind LJ. Mitochondrial ceramide and the induction of apoptosis. J Bioenerg Biomembr. 2005;37:143–153. doi: 10.1007/s10863-005-6567-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sperling P, Ternes P, Zank TK, Heinz E. The evolution of desaturases. Prostaglandins Leukot Essent Fatty Acids. 2003;68:73–95. doi: 10.1016/s0952-3278(02)00258-2. [DOI] [PubMed] [Google Scholar]
- Strum JC, Swenson KI, Turner JE, Bell RM. Ceramide triggers meiotic cell cycle progression in Xenopus oocytes. A potential mediator of progesterone-induced maturation. J Biol Chem. 1995;270:13541–13547. doi: 10.1074/jbc.270.22.13541. [DOI] [PubMed] [Google Scholar]
- Stukey J, Carman GM. Identification of a novel phosphatase sequence motif. Protein Sci. 1997;6:469–472. doi: 10.1002/pro.5560060226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan MB, Lindell D, Lee JA, Thompson LR, Bielawski JP, Chisholm SW. Prevalence and evolution of core photosystem II genes in marine cyanobacterial viruses and their hosts. PLoS Biol. 2006;4:e234. doi: 10.1371/journal.pbio.0040234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Susin SA, Zamzami N, Larochette N, Dallaporta B, Marzo I, Brenner C, Hirsch T, Petit PX, Geuskens M, Kroemer G. A cytofluorometric assay of nuclear apoptosis induced in a cell-free system: Application to ceramide-induced apoptosis. Exp Cell Res. 1997;236:397–403. doi: 10.1006/excr.1997.3733. [DOI] [PubMed] [Google Scholar]
- Suttle CA. Marine viruses: Major players in the global ecosystem. Nat Rev Microbiol. 2007;5:801–812. doi: 10.1038/nrmicro1750. [DOI] [PubMed] [Google Scholar]
- Takemura M. Poxviruses and the origin of the eukaryotic nucleus. J Mol Evol. 2001;52:419–425. doi: 10.1007/s002390010171. [DOI] [PubMed] [Google Scholar]
- Villarreal LP. Viruses and the evolution of life. ASM Press; Washington, DC: 2005. [Google Scholar]
- Villarreal LP, DeFilippis VR. A hypothesis for DNA viruses as the origin of eukaryotic replication proteins. J Virol. 2000;74:7079–7084. doi: 10.1128/jvi.74.15.7079-7084.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westbroek P, van Hinte JE, Brummer GJ, Veldhuis M, Brownlee C, Green JC, Harris R, Heimdal BR. Emiliania huxleyi as a key to biosphere–geosphere interactions. In: Green J, Leadbeater B, editors. The haptophyte algae. Oxford University Press; Oxford, UK: 1994. pp. 321–334. [Google Scholar]
- Wilson WH, Schroeder DC, Allen MJ, Holden MT, Parkhill J, Barrell BG, Churcher C, Hamlin N, Mungall K, Norbertczak H, et al. Complete genome sequence and lytic phase transcription profile of a coccolithovirus. Science. 2005;309:1090–1092. doi: 10.1126/science.1113109. [DOI] [PubMed] [Google Scholar]
- Wilson WH, Van Etten JL, Allen MJ. The Phycodnaviridae: The story of how tiny giants rule the world. Curr Top Microbiol Immunol. 2009;328:1–42. doi: 10.1007/978-3-540-68618-7_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter E, Ponting CP. TRAM, LAG1 and CLN8: Members of a novel family of lipid-sensing domains? Trends Biochem Sci. 2002;27:381–383. doi: 10.1016/s0968-0004(02)02154-0. [DOI] [PubMed] [Google Scholar]
- Yang Z. PAML 4: Phylogenetic analysis by maximum likelihood. Mol Biol Evol. 2007;24:1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- Yang J, Yu Y, Sun S, Duerksen-Hughes PJ. Ceramide and other sphingolipids in cellular responses. Cell Biochem Biophys. 2004;40:323–350. doi: 10.1385/CBB:40:3:323. [DOI] [PubMed] [Google Scholar]
- Yoshida T, Nagasaki K, Takashima Y, Shirai Y, Tomaru Y, Takao Y, Sakamoto S, Hiroishi S, Ogata H. Ma-LMM01 infecting toxic Microcystis aeruginosa illuminates diverse cyanophage genome strategies. J Bacteriol. 2008;190:1762–1772. doi: 10.1128/JB.01534-07. [DOI] [PMC free article] [PubMed] [Google Scholar]