Abstract
DNA methylation is an important component of epigenetic modifications, which influences the transcriptional machinery aberrant in many human diseases. In this study we present the first genome-wide integrative analysis of promoter methylation and gene expression for the identification of methylation markers in melanoma. Genome-wide promoter methylation and gene expression of eight early-passage human melanoma cell strains were compared with newborn and adult melanocytes. We used linear mixed effect models (LME) in combination with a series of filters based on the localization of promoter methylation relative to the transcription start site, overall promoter CpG content, and differential gene expression to discover DNA methylation markers. This approach identified 76 markers, of which 68 were hyper- and eight hypomethylated (LME, P < 0.05). Promoter methylation and differential gene expression of five markers (COL1A2, NPM2, HSPB6, DDIT4L, MT1G) were validated by sequencing of bisulfite-modified DNA and real-time reverse transcriptase PCR, respectively. Importantly, the incidence of promoter methylation of the validated markers increased moderately in early and significantly in advanced-stage melanomas, using early-passage cell strains and snap-frozen tissues (n = 18 and n = 24, respectively) compared with normal melanocytes and nevi (n = 11 and n = 9, respectively). Our approach allows robust identification of methylation markers that can be applied to other studies involving genome-wide promoter methylation. In conclusion, this study represents the first unbiased systematic effort to determine methylation markers in melanoma and revealed several novel genes regulated by promoter methylation that were not described in cancer cells before.
Aberrant epigenetic modification is frequent in several human diseases, including cancer. In particular, altered patterns of histone modifications and DNA methylation have been documented (Esteller 2007; Feinberg 2007; Goldberg et al. 2007; Jones and Baylin 2007). These epigenetic changes are the focus of intensive studies due to their role in chromatin structure and gene expression, and their potential use as marks for disease onset and progression. Classification of histone marks, however, remains a challenge, because histones undergo a variety of post-translational modifications under different conditions. On the other hand, DNA methylation is an attractive biomarker candidate due to its stability and its potential diagnostic value (Esteller 2003).
Methylation of cytosine residues at CpG dinucleotides is a well-described epigenetic DNA modification known to have profound effects on the regulation of gene expression (Bird and Wolffe 1999). Alterations in tumor DNA methylation include generalized genome-wide hypomethylation and locus-specific hypermethylation (Esteller 2007; Jones and Baylin 2007). Genomic hypomethylation occurs early in cellular transformation, a process that affects genome stability and the expression of imprinted genes (Feinberg 2004). Locus-specific hypermethylation occurs primarily in CpG islands that are often associated with gene regulatory regions. This, in turn, can lead to transcriptional inactivation of downstream gene(s), followed by changes in cellular functions. In cancer cells, locus-specific hypermethylation often involves the promoter of tumor suppressor genes, and therefore, is one of the key events in tumorigenesis.
Melanoma is a fatal skin cancer that is increasing in incidence. The 5-yr overall survival rate of 72% (ranging from 10.5% for stage IV to 92.1% for stage I patients) has not improved over many decades, despite the recent advances in surgical and adjuvant therapies (Gimotty et al. 2005; Ross 2006). Genetic alterations, such as chromosomal deletions/amplifications and mutations, as well as epigenetic events that promote the malignant phenotype, have been described (Houghton and Polsky 2002; Dahl and Guldberg 2007; Rothhammer and Bosserhoff 2007). Among the aberrantly hypermethylated and silenced genes in melanoma are known tumor suppressor genes. However, so far, only limited genomic regions were evaluated in melanoma, and high incidence of methylation was reported for only a few genes, e.g., RARB (72%), RASSF1A (55%), and PYCARD (50%) (Spugnardi et al. 2003; Hoon et al. 2004; Furuta et al. 2006). Moreover, the timing of promoter methylation changes and silencing of tumor suppressor genes in melanoma development remain poorly understood.
Several recent advances in technology, such as DNA tiling microarray and high-throughput sequencing, allow for an unbiased genome-wide analysis of epigenomic events (Shames et al. 2006; Weber et al. 2007; Cokus et al. 2008; Meissner et al. 2008). In our studies we used the methylated DNA immunoprecipitation (MeDIP) approach, which generates an enrichment of methylated genomic fragments by means of an antibody specific to 5′-methylcytosine (Weber et al. 2005) combined with hybridization of the fragments to a whole-genome promoter array. We then subjected the hybridization data to the MEDME (modeling experimental data with MeDIP enrichment) post-processing routine (Pelizzola et al. 2008) in order to determine the absolute and relative methylation levels in normal human melanocytes and melanoma cells. The data revealed potential methylation markers, and using additional gene expression experiments, revealed promoter features that appeared to be relevant for transcriptional regulation. These features were used to further filter the selected promoters and identify a set of 76 methylation markers in melanoma.
Results
Determination of genome-wide promoter methylation
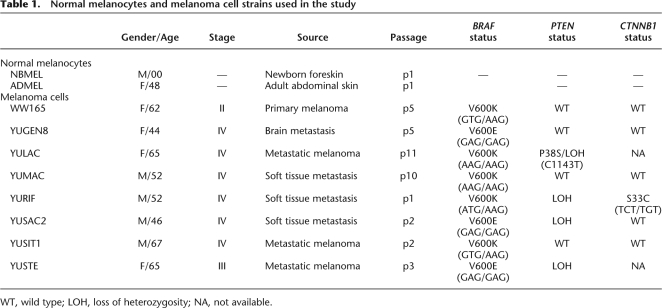
We performed comprehensive DNA methylation profiling of gene promoters in normal newborn and adult melanocytes and eight melanoma cell strains (Table 1). DNA methylation was determined using MeDIP (Weber et al. 2005) followed by hybridization to NimbleGen tiling microarray, which probes 24,103 RefSeq human promoters. MEDME (Pelizzola et al. 2008) was applied to the MeDIP hybridization data to determine the absolute and relative methylation scores (AMS and RMS, respectively). Briefly, AMS is an estimate of the probe-level absolute number of mCpG, and RMS is an estimate of the relative probe-level methylation (mCpG/CpG). Pairwise comparison of genome-wide promoter AMS across all cell strains showed that normal newborn and adult melanocytes correlated reasonably well with each other (Spearman's rank correlation coefficient P = 0.77), but not with the melanoma cells (Supplemental Fig. S1). The difference between NBMEL and ADMEL was expected and it is likely to be due mainly to alteration of epigenetic marks during aging (Fraga and Esteller 2007). We consider important the inclusion of both normal cell strains in this study, since the considered melanoma samples are derived from adult patients. This experimental design will avoid selection of melanoma markers whose differential methylation can also be found in adult normal cells and can more easily be explained as an aging effect. Finally, the eight melanoma cell strains show a good level of correlation (P > 0.80).
Table 1.
Normal melanocytes and melanoma cell strains used in the study
WT, wild type; LOH, loss of heterozygosity; NA, not available.
DNA methylation “code” and transcriptional regulation
The correlation between promoter methylation and transcriptional repression of downstream gene(s) has been established by many studies based on a limited set of loci (Bird and Wolffe 1999; Eckhardt et al. 2006; Suzuki and Bird 2008). Recently, Weber et al. (2007) reported that promoters with weak CpG islands unbound to RNA polymerase II (Pol II) were frequently methylated. However, the relevance of specific promoter features of DNA methylation, such as the distance of DNA methylation relative to the transcription start site (TSS), or the role of absolute and relative levels of mCpG to the transcriptional repression of downstream genes(s) remain poorly understood.
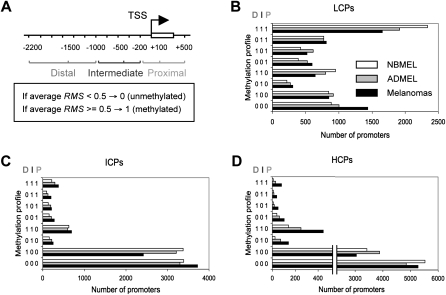
Profiling genome-wide promoters is expected to produce different patterns of DNA methylation. In order to classify these patterns, we divided each promoter into three regions relative to TSS: proximal (−200 to +500 bp), intermediate (−200 to −1000 bp), and distal (−1000 to −2200 bp) as presented in Figure 1A. Each region was then defined as highly methylated or unmethylated based on the average probe-level RMS (higher or lower than 0.5, respectively). Promoters were classified into groups according to their methylation profiles (for example, 111: fully methylated, 000: extensively unmethylated, and 001: proximally methylated). We also took into account the overall promoter CpG content as described (Weber et al. 2007). Promoters were divided into three groups based on their CpG ratio: low CpG (LCPs), intermediate CpG (ICPs), and high CpG content promoters (HCPs). The number of promoters with a specific methylation profile within each group is shown in Figure 1B–D. In general, almost 90% of ICPs and HCPs were either unmethylated (000 profile) or exclusively distally methylated (100 profile), while LCPs showed heterogeneous methylation patterns. LCPs in melanoma cells were hypomethylated compared with those in normal melanocytes, as shown by a 29% decrease in the 111 profile (fully methylated promoter; χ2 P = 3 × 10−29) and a 62% increase in the 000 profile (fully demethylated promoter; P = 1 × 10−31). Conversely, ICPs and HCPs methylated in intermediate and/or proximal regions (X10, X01, and X11 profiles) were consistently more represented in melanoma cells than in normal melanocytes (+66% and +613%, on average, in ICPs and HCPs, respectively; 7 × 10−2 < P < 1 × 10−36). Taken together, the results suggested that differential methylation of promoters in melanoma cells is dependent on overall CpG content and is enriched in specific regions relative to the TSS.
Figure 1.
Distribution of promoter methylation in normal melanocytes and melanoma cells. (A) Diagram showing the three promoter regions relative to the TSS: Proximal (−200 to +500 bp), Intermediate (−200 to −1000 bp), and Distal (−1000 to −2200 bp). Each region is labeled as highly methylated (1) or unmethylated (0) if its average probe-level RMS is higher or lower than 0.5, respectively. (B) Number of LCPs for each cell type (NBMEL, ADMEL, and melanomas) and for each methylation profile. (e.g., 111 fully methylated, 001 proximally methylated, and combination thereof). (C,D) The same as B, for ICPs and HCPs.
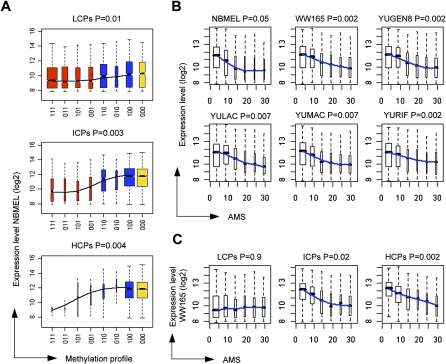
Gene expression was evaluated for each methylation profile in order to assess the site of promoter methylation relevant to transcriptional repression. Figure 2A and Supplemental Figure S2 show the expression of each gene according to its promoter methylation profile and CpG content for normal melanocytes and melanoma cells. The results indicated that the expression of genes under the control of LCPs is not dependent on the promoter methylation profile in both cell types (trend P-values 0.01 and 0.3 in normal melanocytes and melanoma cells, respectively). ICPs with proximal methylation seem to be less transcriptionally active than those without proximal methylation (NBMEL P = 0.003, and melanomas P = 0.005). Finally, the location of methylation relative to the TSS in HCPs significantly correlated with transcriptional repression of downstream genes (NBMEL P = 0.004, and melanomas P = 0.005). These findings suggest that transcriptional repression driven by promoter methylation is inversely related to the distance of mCpGs from the TSS and increases with promoter CpG content. Interestingly, this pattern is slightly more prominent in normal melanocytes than in melanoma cells, suggesting a possible degeneration of the epigenetic “code” and/or transcriptional machinery in malignant cells.
Figure 2.
Promoter methylation and transcriptional repression. (A) Gene expression in normal melanocytes (NBMEL) under the control of promoters with each methylation profile in each promoter category; red and blue groups indicate promoters with and without proximal methylation (XX1 and XX0 profiles, respectively); yellow indicates unmethylated promoters; the groups range from fully methylated to unmethylated from left to right, and the order is based on the progressive absence of methylation from distal to proximal regions; smoothing over the median for each group is shown. (B) Gene expression in normal melanocytes (NBMEL) and five melanoma cell strains as a function of the probe-level AMS of proximal promoters; smoothing over the median for each group is shown. (C) Gene expression of WW165 primary melanoma cells for each promoter category as a function of the probe-level AMS of proximal promoters; smoothing over the median for each group is shown. For each panel the trend P-value is indicated (see Methods).
We also compared the absolute levels of DNA methylation with transcriptional intensity to evaluate the possibility of any quantitative relationship between the two. All cell strains showed a clear correlation between AMS and transcriptional repression (Fig. 2B). The same analysis was repeated, segregating the promoters on the basis of their CpG content to further confirm the relevance of the level of promoter methylation in the setting of overall CpG content. Again, only ICPs and HCPs demonstrated association with methylation, whereas genes under the control of LCPs were expressed almost independently of the methylation status (Fig. 2C). Taken together, localization and density of mCpG, as well as overall promoter CpG content, were highly predictive of the degree of transcriptional repression. Notably, we did not find the lack of association between HCPs methylation and transcriptional repression reported by Weber et al. (2007) based on Poll II binding data.
Identification of melanoma methylation markers
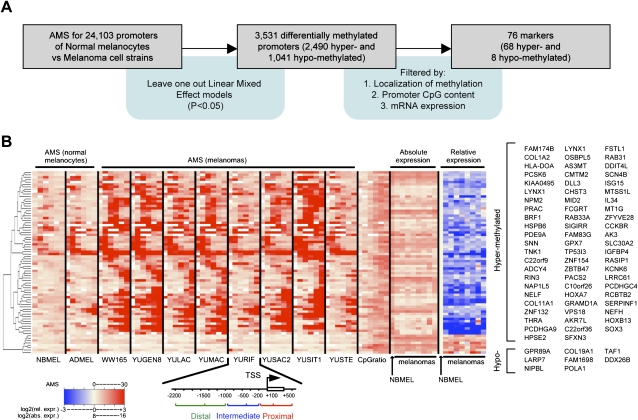
We compared AMS profiles of normal melanocytes with melanoma cells in order to identify methylation markers associated with malignant transformation. The linear mixed effect models (LME) was applied to identify differentially methylated promoters to accommodate the heterogeneity of DNA methylation across the cell types. This method identified 3531 differentially methylated promoters with P < 0.05 (2490 hypermethylated and 1041 hypomethylated in melanoma cells compared with normal melanocytes). As expected from the results shown in Figure 1B, there was overrepresentation of LCPs in hypomethylated promoters. We then used proximal methylation level, promoter CpG content, and gene expression to select methylation markers (Fig. 3A). These filters increased the likelihood of identifying promoters with dysregulated methylation pattern causally related to the differential expression of downstream genes. As a result, 76 promoters (68 hypermethylated and eight hypomethylated in melanomas) were selected as methylation markers (Fig. 3B; Supplemental Tables S1, S2). Analysis of Gene Ontology functional categories for the 68 hypermethylated candidates genes revealed overrepresentation of functional categories common in cancer progression, such as skeletal development (P = 1.4 × 10−4) and cell–cell adhesion (P = 3.6 × 10−3), although no prominent enrichment was observed (Supplemental Fig. S3). Also, several genes that were previously reported as melanoma markers were identified (Supplemental Tables S1, S2). For example, we previously showed that the expression of RAB33A, a member of the small GTPase superfamily, is suppressed in melanoma cells by proximal promoter methylation in a process that recapitulated silencing of X-linked genes (Cheng et al. 2006). To check for robustness of the selected markers with respect to different data processing, we repeated the pipeline displayed in Figure 3A based on the Batman methylation scores (Down et al. 2008). Despite the fact that Batman provides estimates of the relative rather then the absolute methylation level, 32 of the 67 hypermethylated and one of the eight hypomethylated markers identified based on the MEDME AMS were confirmed.
Figure 3.
Selection of promoter methylation markers. (A) Outline of the pipeline used to identify markers. (B) Heat-map of the selected markers; promoter AMS for newborn melanocytes (NBMEL), adult melanocytes (ADMEL), and eight melanoma cell strains, and CpG ratio in each of six regions is represented; absolute gene expression as well the expression relative to newborn melanocytes (NBMEL) are displayed; gene symbols of differentially expressed genes are shown on the right side in order of significance, top to bottom from the left column. Two alternative promoters are displayed for LYNX1 (see Supplemental Table S1).
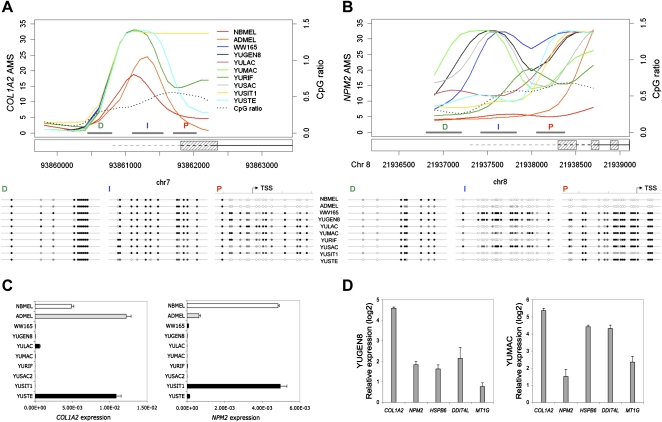
Five genes were selected for further validation of promoter methylation and gene expression (COL1A2, NPM2, HSPB6, DDIT4L, and MT1G). These genes, except for COL1A2 and DDIT4L, were not previously reported to be aberrantly methylated in melanomas. Figure 4, A and B show the AMS profiles of COL1A2 and NPM2 promoters. The methylation estimates were validated by sequencing of bisulfite (BS) converted DNA (Fig. 4A,B). In particular, we confirmed the methylation status of promoter regions expected to be hypermethylated in melanoma cells and unmethylated in normal melanocytes. Quantitative real-time RT-PCR validated the inverse correlation between gene expression and promoter methylation (Fig. 4C). Moreover, these genes were reactivated after treatment with 0.2 μM 5-Aza-2′-deoxycytidine (Aza) (Fig. 4D), confirming the causal relationship between methylation and gene expression. Analysis of HSPB6, DDIT4L, and MT1G showed similar results (Supplemental Fig. S4). The reactivation was even stronger after treatment with 1.0 μM Aza (data not shown).
Figure 4.
Promoter methylation and gene expression for selected markers. (A) Probe-level AMS, CpG ratio, and sequence of BS-modified genomic DNA for the proximal (P), intermediate (I), and distal (D) promoter regions of COL1A2 in newborn (NBMEL) and adult (ADMEL) melanocytes and eight melanoma cell strains; gray bars indicate the amplicons of BS sequencing; D, distal, I, intermediate, and P, proximal for each promoter region; 1 kb upstream region (dashed line), exon (dashed box), and coding sequences (solid black line) for each RefSeq in the locus are displayed; CpGs are represented as circles, and white, gray, and black shades refer to the average of mCpG/CpG (0 to 1) for each sample. (B) Same as A, for NPM2. (C) COL1A2 and NPM2 expression levels measured by real-time RT-PCR. Expression levels were normalized to that of ACTB. (D) Restoration of gene expression (COL1A2, DDIT4L, NPM2, and MT1G) after treatment with Aza (0.2 μM). Gene transcripts were measured by real-time RT-PCR, and expression levels were normalized to that of ACTB. The histogram shows log2 increase after Aza treatment for YUGEN8 and YUMAC melanoma cells.
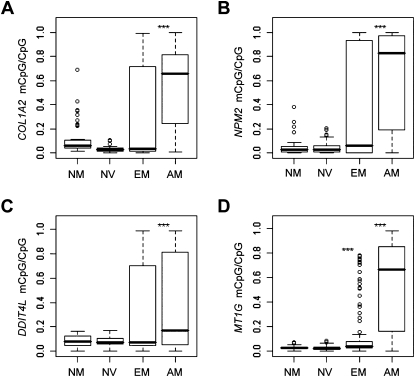
The methylation status of the proximal promoter of four of these genes (COL1A2, NPM2, DDIT4L, and MT1G) was evaluated by sequencing of BS-modified DNA on an additional set of early-passage (passage 0–5) cells. Figure 5 displays the validation of this additional set of samples in combination with those used for microarray hybridization. In total, 18 melanoma cell strains (seven early and 11 advanced-stage melanomas at Stage I/II and III/IV, respectively) and 11 normal cell strains (five normal melanocytes and six nevi) were evaluated. The incidence of promoter methylation was slightly higher in early-stage melanomas and significantly increased in advanced-stage melanomas (P < 0.001 for the four markers) in comparison to normal melanocytes, whereas no methylation was observed in nevi.
Figure 5.
Promoter methylation of selected markers in early-passage cells results from sequencing of BS. Modified DNA was used to determine the methylation level of each CpG dinucleotide within each proximal promoter. The distribution of CpG-relative methylation is displayed in normal melanocytes (NM, n = 5), nevi (NV, n = 6), early-stage melanomas (EM, n = 8), and advanced-stage melanomas (AM, n = 10). P-values for each group were determined relative to NM using a two-sample nonparametric Wilcoxon test. 0.05 < *P ≤ 0.01, 0.01 < **P ≤ 0.001, ***P < 0.001. (A) COL1A2; (B) NPM2; (C) DDIT4L; (D) MT1G.
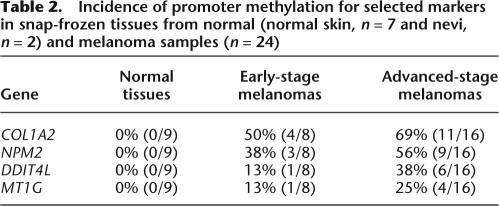
Finally, to address whether promoter methylation was not an artifact of culture conditions and can be detected in clinical specimens, we examined nine skin samples (seven normal skin cells and two nevi), and 24 snap-frozen melanoma tumors (eight early and 16 advanced-stage tumors) from different individuals. The data show that promoter methylation was predominantly detected in advanced-stage tumors, whereas there was no methylation in benign tissue samples, consistent with our observations using early-passage melanoma cell strains (Table 2). These results suggest that methylation of COL1A2, NPM2, DDIT4L, and MT1G can be useful for assessing tumor progression.
Table 2.
Incidence of promoter methylation for selected markers in snap-frozen tissues from normal (normal skin, n = 7 and nevi, n = 2) and melanoma samples (n = 24)
Discussion
The goal of our study was to discover, in an unbiased fashion, DNA methylation markers that contribute to transcriptional dysregulation in melanoma cells. Although several hyper- and hypomethylated candidate genes have been identified in melanomas, few are suitable for clinical use as methylation markers (Muthusamy et al. 2006; Rothhammer and Bosserhoff 2007). The reasons for this shortcoming could be attributed to: (1) artifacts of cell culture (Catalina et al. 2008; Meissner et al. 2008), and (2) heterogeneity across individual tumor cells (Eckhardt et al. 2006; Feinberg et al. 2006). Although tissue samples are preferable, the presence of normal cells and DNA damage typical of formalin-fixed paraffin-embedded material reduces the quality of the DNA for analysis. To avoid changes introduced in long-term cell cultures, we used cells derived from freshly cultured normal or benign skin and tumors, and went on to validate the results in snap-frozen lesions. Furthermore, we applied a selection pipeline for robust identification of methylation markers to overcome heterogeneity across individual tumors. First, we used MEDME methylation estimates rather than raw MeDIP enrichment values. Second, we integrated epigenomics data with gene expression profiling to identify promoters that showed an association between methylation status and expression of downstream genes. To this end, the localization of promoter methylation and overall promoter CpG content, two features that we have shown to be highly predictive of the intensity of transcriptional repression, were considered in the analysis. In this regard, it is important to compare our findings with those reported by two recent studies. Irizarry et al. (2009) found that most of the differentially methylated regions in colon cancer are located within 2 kb of CpG islands, in regions named CpG island shores. This supports our decision to focus on a wide region across the TSS to determine differential methylation expanding the analysis beyond CpG islands. At the same time, this does not contradict our finding showing that the methylation levels in regions proximal to the TSS are strongly associated with transcriptional repression. A second study from Ball et al. (2009) showed a stronger correlation between DNA methylation levels and transcriptional repression in ICPs compared with HCPs and LCPs. Rather, we found a clear association between both ICPs and HCPs methylation and transcriptional repression of the downstream genes. We explain this apparent discrepancy, noting that we focused here on the absolute rather than the relative methylation level used in that study. Indeed, similar amounts of variation in the absolute number of mCpG in ICPs and HCPs (Fig. 2C) can be obscured in the latter when considering the relative methylation level because of the their high CpG content.
We identified 76 markers in melanoma cells, of which 68 were hypermethylated and eight were hypomethylated. Sixteen of these markers were previously found to be affected by promoter methylation in human cells, and 12 were reported to be involved in cancer pathogenesis. Of these, only four were previously described in melanoma (COL1A2, RAB33A, DDIT4L, and HOXB13) (Cheng et al. 2006; Furuta et al. 2006; Muthusamy et al. 2006). Epigenetics control and involvement in cancer have not yet been established for the remaining 60 markers (Supplemental Tables S1, S2), even if several functional families related to cancer pathogenesis were over-represented. Five hypermethylated markers belonging to different functional groups that may have relevance to cancer pathogenesis were selected for further validation. COL1A2 (type I collagen alpha 2; P = 4.4 × 10−4, ranked second) is a member of a large family of extracellular matrix proteins that maintain cellular and tissue integrity and contributes to homeostasis of the human body (Myllyharju and Kivirikko 2004). DNA methylation in the first exon of COL1A2, the binding site for regulatory factor X 1 (RFX1), increases RFX1 binding and decreases COL1A2 transcription (Sengupta et al. 1999). There is also evidence for COL1A2 aberrant promoter methylation in genome-wide methylation studies of cancer cells, such as medulloblastoma (Anderton et al. 2008) and hepatoma (Chiba et al. 2005). NPM2 (nucleoplasmin 2; P = 0.0020, ranked seventh), a gene frequently methylated in leukemia cell lines and in patients with acute myeloid leukemia (Kroeger et al. 2008), and HSPB6 (heat-shock protein alpha-crystallin-related B6; P = 0.003, ranked 10th), which belongs to a class of proteins functioning as molecular chaperones, have been linked to cell malfunction when perturbed (Sun and MacRae 2005). MT1G (metallothionein 1G; P = 0.033, ranked 54th) belongs to a class of metal-binding proteins involved in several cellular processes. This gene was implicated as a putative tumor suppressor in thyroid tumorigenesis, even though its role in this process is not completely understood (Huang et al. 2003; Morris et al. 2003). Finally, DDIT4L (DNA-damage-inducible transcript 4-like; P = 0.029, ranked 49th) was reported to inhibit cell growth and to be up-regulated under conditions of ischemia and oxidative stress (Corradetti et al. 2005). Aberrant methylation of DDIT4L promoter was previously identified by a genome-wide search of melanoma cell lines using methylation-sensitive representational difference analysis (Furuta et al. 2006). In this study we validated its promoter methylation in early-passage melanoma cell strains and snap-frozen tumors.
We assessed the methylation status of COL1A2, NPM2, DDIT4L, and MT1G in normal melanocytes, nevi, early-stage and advanced-stage melanomas in both early-passage cell strains and snap-frozen tissues. Despite the small number of samples, the incidence of promoter hypermethylation significantly increased during melanoma development, implying that these specific markers could be associated with melanoma progression and be used to predict melanoma prognosis.
LCP methylation status was independent of downstream gene expression, although they were extensively hypomethylated in melanoma cells (Fig. 2A,C). For this reason, we filtered out LCPs in the marker identification process, selecting only eight hypomethylated markers in ICP and HCP promoters. Hypomethylation of cancer/testis antigens such as MAGE, BAGE, and GAGE has been shown to be associated with melanoma tumorigenesis (Chomez et al. 2001; Scanlan et al. 2002). Interestingly, many of these genes were hypomethylated in only a subset of the melanoma cell strains used in our study (MAGEA3 P = 0.028, MAGEA6 P = 0.23, PRAME P-value from 0.007 to 0.017 for alternative RefSeq promoters; Supplemental Fig. S5).
In summary, we have shown that integration of high-throughput data from different omics by means of a pipeline designed to accommodate the expected heterogeneity of methylation within promoter regions and between samples can identify new tumorigenic melanoma markers. The proposed methodology may be useful for detection of robust markers over highly heterogeneous samples in other disease states. Finally, we provided the first genome-wide integrative analysis of promoter methylation and gene expression in melanoma. The aberrant methylated promoters reported here have the potential to be relevant to the clinical arena as markers for melanoma progression.
Methods
Cells and tumors
Normal human melanocytes were isolated from newborn foreskins and adult skins. The cells were grown in OptiMEM with antibiotics, 5% fetal calf serum (basal medium), and growth supplements as described (Cheng et al. 2006). Melanoma tumors were excised to improve patient quality of life. They were collected with participants' informed written consent according to Health Insurance Portability and Accountability Act (HIPAA) regulations with Human Investigative Committee protocol. A part of the tumor was snap frozen in liquid nitrogen and another part was dissociated and cultured in basal medium (metastatic cells) or in medium supplemented with 0.1 mM 3-isobutyl-1-methyl xanthin (Sigma Chemical) (primary melanoma cells), a supplement required for primary melanoma cells proliferation. The melanoma cell strains used in the study were from short-term cultures, i.e., passage 1–15 (Table 1). All cell strains carried the BRAF activating mutation, none harbored the NRAS codon 61 mutation, four carried PTEN loss of heterozygosity (LOH), one a known PTEN variant (Pro38Ser), while one carried inactivating CTNNB1 mutation (Ser33Cys) (Halaban et al. 2009).
Methylation profiling by MeDIP
The MeDIP assay was performed as described (Weber et al. 2005). Briefly, genomic DNA was extracted by DNeasy Blood & Tissue Kit (QIAGEN), sheared into 300–1200-bp fragments by sonication, and aliquots (10 μg) were incubated with 20 μL of anti-5-methylcytidine mAb (Eurogentec) for 12 h at 4°C. Antibody-bound DNA was precipitated with 50 μL of Dynabeads (M-280 sheep antibodies to mouse IgG, Dynal Biotech) at 4°C for 2 h on a rotating wheel. Bound DNA was eluted two times with 200 μL of TE containing 1.0% and 0.67% SDS, respectively, and DNA was purified by standard proteinase K/phenol-chloroform procedure. The eluted DNA fractions and sonicated input DNA were differentially labeled with fluorescent dyes (Cy3 and Cy5, respectively) and hybridized to genomic promoter tiling arrays.
The methylation level of each cell strain was evaluated by one array hybridization. We decided to invest more in the number of cell strains rather than replicates because of the expected heterogeneity in the DNA methylation levels. Also, in order to obtain replicated hybridizations we would have needed to collect higher amounts of genomic DNA, likely increasing the number of passages in culture. This, in turn, would have been problematic because of the altered methylation patterns determined by cell culture (Meissner et al. 2008). Application of the same methodology for the selection of markers to other cancer types could be less problematic and take advantage of an increased replication level.
Array design, data processing, and probe annotation of the arrays for detection of genome-wide promoter methylation
We used NimbleGen C4226-00-01 promoter-tiling arrays that were designed based on the HG18 genome release. The array contains 390,000 probes with an average length of 60 bp tiled in 110-bp steps along the upstream promoter regions. We applied standard normalization methods for two-channel microarrays: within (Loess-based) and between (Quantile-based) normalization, available in the Limma Bioconductor library (Ihaka and Gentleman 1996; Smyth and Speed 2003; Gentleman et al. 2004; Smyth 2005). The probe-level log ratio was determined as the log2 of the cy3/cy5 channels and used as a measure of MeDIP enrichment. The position of the center of each probe on the array was compared with the transcription start site (TSS) of known RefSeqs retrieved from UCSC human genome annotations (hg18). Multiple annotations of a probe in association with different RefSeq IDs were allowed.
Array design, data processing, and probe annotation of the arrays for detection of genome-wide gene expression
We used NimbleGen 2005-04-20_Human_60mer_1in2 genome-wide human expression arrays that were designed based on the HG17 genome release. A total of 380,000 probes for almost 30,000 transcripts and 20,000 known genes are represented on this array. NimbleGen provided design and probe annotation. The QSPLINE nonlinear normalization method was applied on a data set of single-channel absolute values (Workman et al. 2002).
Determination of absolute and relative methylation estimates
Absolute and relative methylation scores (AMS and RMS, respectively) were determined using the MEDME Bioconductor library (Pelizzola et al. 2008; http://espresso.med.yale.edu/medme/ and Bioconductor repository http://www.bioconductor.org). MEDME was calibrated on the data as described before (Pelizzola et al. 2008) using a window size of 1 kb and linear weighting for smoothing of log ratios and determination of the probe CpG content.
Determination of trend P-value
Trend P-values in Figure 2 were determined based on the box plot medians, using the SAGx R library (http://www.r-project.org/).
Identification of biomarkers
Intermediate and proximal promoter regions of each RefSeq were divided into six regions based on the distance from the TSS (−500 to −350, −200 to −50, −50 to +100, +100 to +250, and +250 to +500 bp, with respect to the TSS). The AMS for a given region was determined as the average of the AMS for the probes in that genomic area. A linear mixed effects model was used in order to select promoters that are differentially methylated between melanoma cells and normal melanocytes. The cell type (melanoma vs. normal melanocytes) was included as a fixed effect; the individual melanoma strains were considered as random samples from the type populations and, thus, included as random effects. Quadratic and cubic terms for distance were included in the model due to the nonlinear relationship between the distance of the promoter regions from the TSS and AMS. The P-value was determined for the normal newborn and adult melanocytes in comparison to each possible group of seven out of eight melanoma cell strains, and the best P-value was retained for subsequent steps. Only promoters whose P-values were less than α = 0.05 were considered for further evaluation.
Finally, selected promoters were screened based on three filters designed to increase the likelihood of differential methylation and differential expression being causally related: (1) promoters with proximal RMS higher than 0.5 were retained; (2) promoters with low CpG content (as defined in Weber et al. 2007) were discarded; (3) only promoters whose downstream gene was differentially expressed at least 1.5-fold in at least six out of eight melanoma cell strains were considered. For the last filter, methylation markers in melanoma cells were required to display inverse relationship to gene expression, i.e., hyper- and hypomethylated being down- and up-regulated, respectively.
Sequencing of bisulfite modified DNA
Genomic DNA was extracted from normal melanocytes and melanoma cells and subjected to bisulfite (BS) modification (Epitect Bisulfite Kit, QIAGEN), followed by Sanger sequencing as described (Jacobsen et al. 2000). The target regions of the relevant genes and the primers used for amplification are listed in Supplemental Table S3. The cell strain methylation levels displayed in Figures 4 and 5 were calculated from the amplitude of cytosine and thymine within each CpG dinucleotide, C/(C+T), using the phred software (http://www.phrap.com/). Considering the likelihood of contamination from normal cells in clinical samples, the methylation level of snap-frozen tissues indicated in Table 2 was determined based on three CpGs that were highly discriminative between normal melanocytes and melanoma cells. Each promoter was then defined as methylated or unmethylated based on the presence of significant amplitude of cytosine or thymine, respectively, consistently on the three CpGs.
Treatment with Decitabine
Decitabine (5-Aza-2′-deoxycytidine, Sigma Chemical [Aza]) was dissolved in methanol as 10 mM stock solution, aliquoted and kept at −20°C. Melanoma cells were seeded in Petri dishes (∼5000 cells/cm2) in regular medium untreated or treated with Aza (0.2 μM) for 2 d, with fresh drug-supplemented medium on the second day. The cells were harvested after 1-d recovery in drug-free medium.
Gene expression by quantitative RT-PCR
Total RNA was isolated using RNeasy Mini Kit (QIAGEN), and 2-μg aliquots were reverse transcribed with Transcriptor First Strand cDNA Synthesis Kit (Roche Applied Science) according to the manufacturer's instruction. Quantitative real-time RT-PCR was then carried out in triplicate using ABI 7500 Fast Real-Time PCR Systems (Applied Biosystems). The primers used for each gene are listed in Supplemental Table S4. The expression of ACTB was used as a reference to normalize for input cDNA. The relative expression values were computed by the comparative Ct method (Pfaffl 2001).
Acknowledgments
We thank the Cell Culture Core facility of the Yale Skin Disease Research Core Center (YSDRCC) supported by NIAMS grant 5P30 AR 041942-12 (Robert Tigelaar) for providing normal human melanocytes and melanoma cells, to Antonella Bacchiocchi for collecting the tissues and growing the cells, and to Harriet Kluger for staging the melanoma tumors. This work was supported by the National Cancer Institute grant Yale SPORE in Skin Cancer number 1 P50 CA121974 (R.H.). M.K. was supported by the National Library of Medicine K22LM009255, and A.M.M. was supported by a National Cancer Institute grant K22CA123146-2.
Authors' contributions: Y.K. and M.P. designed the study. Y.K. and E.C. performed the experiments. M.P. and Y.K. analyzed the data and interpreted the results. M.K. and A.M.M. contributed bioinformatics and statistical analysis. R.H. and S.W. provided scientific leadership. S.A. and D.N. excised the tissues. M.S. is the oncologist who took care of the patients. Y.K., M.P., R.H., and S.W. wrote the manuscript.
Footnotes
[Supplemental material is available online at www.genome.org. The microarray data from this study have been submitted to Gene Expression Omnibus (GEO) (http://www.ncbi.nlm.nih.gov/geo) under accession no. GSE13706.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.091447.109.
References
- Anderton JA, Lindsey JC, Lusher ME, Gilbertson RJ, Bailey S, Ellison DW, Clifford SC. Global analysis of the medulloblastoma epigenome identifies disease subgroup-specific inactivation of COL1A2. Neuro Oncol. 2008;10:981–994. doi: 10.1215/15228517-2008-048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ball MP, Li JB, Gao Y, Lee JH, LeProust EM, Park IH, Xie B, Daley GQ, Church GM. Targeted and genome-scale strategies reveal gene-body methylation signatures in human cells. Nat Biotechnol. 2009;27:361–368. doi: 10.1038/nbt.1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bird AP, Wolffe AP. Methylation-induced repression—belts, braces, and chromatin. Cell. 1999;99:451–454. doi: 10.1016/s0092-8674(00)81532-9. [DOI] [PubMed] [Google Scholar]
- Catalina P, Montes R, Ligero G, Sanchez L, de la Cueva T, Bueno C, Leone PE, Menendez P. Human ESCs predisposition to karyotypic instability: Is a matter of culture adaptation or differential vulnerability among hESC lines due to inherent properties? Mol Cancer. 2008;7:76. doi: 10.1186/1476-4598-7-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng E, Trombetta SE, Kovacs D, Beech RD, Ariyan S, Reyes-Mugica M, McNiff JM, Narayan D, Kluger HM, Picardo M, et al. Rab33A: Characterization, expression, and suppression by epigenetic modification. J Invest Dermatol. 2006;126:2257–2271. doi: 10.1038/sj.jid.5700386. [DOI] [PubMed] [Google Scholar]
- Chiba T, Yokosuka O, Fukai K, Hirasawa Y, Tada M, Mikata R, Imazeki F, Taniguchi H, Iwama A, Miyazaki M, et al. Identification and investigation of methylated genes in hepatoma. Eur J Cancer. 2005;41:1185–1194. doi: 10.1016/j.ejca.2005.02.014. [DOI] [PubMed] [Google Scholar]
- Chomez P, De Backer O, Bertrand M, De Plaen E, Boon T, Lucas S. An overview of the MAGE gene family with the identification of all human members of the family. Cancer Res. 2001;61:5544–5551. [PubMed] [Google Scholar]
- Cokus SJ, Feng S, Zhang X, Chen Z, Merriman B, Haudenschild CD, Pradhan S, Nelson SF, Pellegrini M, Jacobsen SE. Shotgun bisulphite sequencing of the Arabidopsis genome reveals DNA methylation patterning. Nature. 2008;452:215–219. doi: 10.1038/nature06745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corradetti MN, Inoki K, Guan KL. The stress-inducted proteins RTP801 and RTP801L are negative regulators of the mammalian target of rapamycin pathway. J Biol Chem. 2005;280:9769–9772. doi: 10.1074/jbc.C400557200. [DOI] [PubMed] [Google Scholar]
- Dahl C, Guldberg P. The genome and epigenome of malignant melanoma. APMIS. 2007;115:1161–1176. doi: 10.1111/j.1600-0463.2007.apm_855.xml.x. [DOI] [PubMed] [Google Scholar]
- Down TA, Rakyan VK, Turner DJ, Flicek P, Li H, Kulesha E, Graf S, Johnson N, Herrero J, Tomazou EM, et al. A Bayesian deconvolution strategy for immunoprecipitation-based DNA methylome analysis. Nat Biotechnol. 2008;26:779–785. doi: 10.1038/nbt1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckhardt F, Lewin J, Cortese R, Rakyan VK, Attwood J, Burger M, Burton J, Cox TV, Davies R, Down TA, et al. DNA methylation profiling of human chromosomes 6, 20 and 22. Nat Genet. 2006;38:1378–1385. doi: 10.1038/ng1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esteller M. Relevance of DNA methylation in the management of cancer. Lancet Oncol. 2003;4:351–358. doi: 10.1016/s1470-2045(03)01115-x. [DOI] [PubMed] [Google Scholar]
- Esteller M. Cancer epigenomics: DNA methylomes and histone-modification maps. Nat Rev Genet. 2007;8:286–298. doi: 10.1038/nrg2005. [DOI] [PubMed] [Google Scholar]
- Feinberg AP. The epigenetics of cancer etiology. Semin Cancer Biol. 2004;14:427–432. doi: 10.1016/j.semcancer.2004.06.005. [DOI] [PubMed] [Google Scholar]
- Feinberg AP. Phenotypic plasticity and the epigenetics of human disease. Nature. 2007;447:433–440. doi: 10.1038/nature05919. [DOI] [PubMed] [Google Scholar]
- Feinberg AP, Ohlsson R, Henikoff S. The epigenetic progenitor origin of human cancer. Nat Rev Genet. 2006;7:21–33. doi: 10.1038/nrg1748. [DOI] [PubMed] [Google Scholar]
- Fraga MF, Esteller M. Epigenetics and aging: The targets and the marks. Trends Genet. 2007;23:413–418. doi: 10.1016/j.tig.2007.05.008. [DOI] [PubMed] [Google Scholar]
- Furuta J, Nobeyama Y, Umebayashi Y, Otsuka F, Kikuchi K, Ushijima T. Silencing of Peroxiredoxin 2 and aberrant methylation of 33 CpG islands in putative promoter regions in human malignant melanomas. Cancer Res. 2006;66:6080–6086. doi: 10.1158/0008-5472.CAN-06-0157. [DOI] [PubMed] [Google Scholar]
- Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, et al. Bioconductor: Open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gimotty PA, Botbyl J, Soong SJ, Guerry D. A population-based validation of the American Joint Committee on Cancer melanoma staging system. J Clin Oncol. 2005;23:8065–8075. doi: 10.1200/JCO.2005.02.4976. [DOI] [PubMed] [Google Scholar]
- Goldberg AD, Allis CD, Bernstein E. Epigenetics: A landscape takes shape. Cell. 2007;128:635–638. doi: 10.1016/j.cell.2007.02.006. [DOI] [PubMed] [Google Scholar]
- Halaban R, Krauthammer M, Pelizzola M, Cheng E, Kovacs D, Sznol M, Ariyan S, Narayan D, Bacchiocchi A, Molinaro A, et al. Integrative analysis of epigenetic modulation in melanoma cell response to decitabine: Clinical implications. PloS One. 2009;4:e4563. doi: 10.1371/journal.pone.0004563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoon DS, Spugnardi M, Kuo C, Huang SK, Morton DL, Taback B. Profiling epigenetic inactivation of tumor suppressor genes in tumors and plasma from cutaneous melanoma patients. Oncogene. 2004;23:4014–4022. doi: 10.1038/sj.onc.1207505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houghton AN, Polsky D. Focus on melanoma. Cancer Cell. 2002;2:275–278. doi: 10.1016/s1535-6108(02)00161-7. [DOI] [PubMed] [Google Scholar]
- Huang Y, de la Chapelle A, Pellegata NS. Hypermethylation, but not LOH, is associated with the low expression of MT1G and CRABP1 in papillary thyroid carcinoma. Int J Cancer. 2003;104:735–744. doi: 10.1002/ijc.11006. [DOI] [PubMed] [Google Scholar]
- Ihaka R, Gentleman R. R: A language for data analysis and graphics. J Comput Graph Stat. 1996;4:299–314. [Google Scholar]
- Irizarry RA, Ladd-Acosta C, Wen B, Wu Z, Montano C, Onyango P, Cui H, Gabo K, Rongione M, Webster M, et al. The human colon cancer methylome shows similar hypo- and hypermethylation at conserved tissue-specific CpG island shores. Nat Genet. 2009;41:178–186. doi: 10.1038/ng.298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobsen SE, Sakai H, Finnegan EJ, Cao X, Meyerowitz EM. Ectopic hypermethylation of flower-specific genes in Arabidopsis. Curr Biol. 2000;10:179–186. doi: 10.1016/s0960-9822(00)00324-9. [DOI] [PubMed] [Google Scholar]
- Jones PA, Baylin SB. The epigenomics of cancer. Cell. 2007;128:683–692. doi: 10.1016/j.cell.2007.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroeger H, Jelinek J, Estecio MR, He R, Kondo K, Chung W, Zhang L, Shen L, Kantarjian HM, Bueso-Ramos CE, et al. Aberrant CpG island methylation in acute myeloid leukemia is accentuated at relapse. Blood. 2008;112:1366–1373. doi: 10.1182/blood-2007-11-126227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meissner A, Mikkelsen TS, Gu H, Wernig M, Hanna J, Sivachenko A, Zhang X, Bernstein BE, Nusbaum C, Jaffe DB, et al. Genome-scale DNA methylation maps of pluripotent and differentiated cells. Nature. 2008;454:766–770. doi: 10.1038/nature07107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris MR, Hesson LB, Wagner KJ, Morgan NV, Astuti D, Lees RD, Cooper WN, Lee J, Gentle D, Macdonald F, et al. Multigene methylation analysis of Wilms' tumour and adult renal cell carcinoma. Oncogene. 2003;22:6794–6801. doi: 10.1038/sj.onc.1206914. [DOI] [PubMed] [Google Scholar]
- Muthusamy V, Duraisamy S, Bradbury CM, Hobbs C, Curley DP, Nelson B, Bosenberg M. Epigenetic silencing of novel tumor suppressors in malignant melanoma. Cancer Res. 2006;66:11187–11193. doi: 10.1158/0008-5472.CAN-06-1274. [DOI] [PubMed] [Google Scholar]
- Myllyharju J, Kivirikko KI. Collagens, modifying enzymes and their mutations in humans, flies and worms. Trends Genet. 2004;20:33–43. doi: 10.1016/j.tig.2003.11.004. [DOI] [PubMed] [Google Scholar]
- Pelizzola M, Koga Y, Urban AE, Krauthammer M, Weissman S, Halaban R, Molinaro AM. MEDME: An experimental and analytical methodology for the estimation of DNA methylation levels based on microarray derived MeDIP-enrichment. Genome Res. 2008;18:1652–1659. doi: 10.1101/gr.080721.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:e45. doi: 10.1093/nar/29.9.e45. http://nar.oxfordjournals.org/cgi/content/full/29/9/e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross MI. Early-stage melanoma: Staging criteria and prognostic modeling. Clin Cancer Res. 2006;12:2312s–2319s. doi: 10.1158/1078-0432.CCR-05-2643. [DOI] [PubMed] [Google Scholar]
- Rothhammer T, Bosserhoff AK. Epigenetic events in malignant melanoma. Pigment Cell Res. 2007;20:92–111. doi: 10.1111/j.1600-0749.2007.00367.x. [DOI] [PubMed] [Google Scholar]
- Scanlan MJ, Gure AO, Jungbluth AA, Old LJ, Chen YT. Cancer/testis antigens: An expanding family of targets for cancer immunotherapy. Immunol Rev. 2002;188:22–32. doi: 10.1034/j.1600-065x.2002.18803.x. [DOI] [PubMed] [Google Scholar]
- Sengupta PK, Ehrlich M, Smith BD. A methylation-responsive MDBP/RFX site is in the first exon of the collagen α2(I) promoter. J Biol Chem. 1999;274:36649–36655. doi: 10.1074/jbc.274.51.36649. [DOI] [PubMed] [Google Scholar]
- Shames DS, Girard L, Gao B, Sato M, Lewis CM, Shivapurkar N, Jiang A, Perou CM, Kim YH, Pollack JR, et al. A genome-wide screen for promoter methylation in lung cancer identifies novel methylation markers for multiple malignancies. PLoS Med. 2006;3:e486. doi: 10.1371/journal.pmed.0030436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth GK. Bioinformatics and computational biology solutions using R and bioconductor, Limma: Linear models for microarray data. Springer; New York: 2005. [Google Scholar]
- Smyth GK, Speed T. Normalization of cDNA microarray data. Methods. 2003;31:265–273. doi: 10.1016/s1046-2023(03)00155-5. [DOI] [PubMed] [Google Scholar]
- Spugnardi M, Tommasi S, Dammann R, Pfeifer GP, Hoon DS. Epigenetic inactivation of RAS association domain family protein 1 (RASSF1A) in malignant cutaneous melanoma. Cancer Res. 2003;63:1639–1643. [PubMed] [Google Scholar]
- Sun Y, MacRae TH. The small heat shock proteins and their role in human disease. FEBS J. 2005;272:2613–2627. doi: 10.1111/j.1742-4658.2005.04708.x. [DOI] [PubMed] [Google Scholar]
- Suzuki MM, Bird A. DNA methylation landscapes: Provocative insights from epigenomics. Nat Rev Genet. 2008;9:465–476. doi: 10.1038/nrg2341. [DOI] [PubMed] [Google Scholar]
- Weber M, Davies JJ, Wittig D, Oakeley EJ, Haase M, Lam WL, Schubeler D. Chromosome-wide and promoter-specific analyses identify sites of differential DNA methylation in normal and transformed human cells. Nat Genet. 2005;37:853–862. doi: 10.1038/ng1598. [DOI] [PubMed] [Google Scholar]
- Weber M, Hellmann I, Stadler MB, Ramos L, Paabo S, Rebhan M, Schubeler D. Distribution, silencing potential and evolutionary impact of promoter DNA methylation in the human genome. Nat Genet. 2007;39:457–466. doi: 10.1038/ng1990. [DOI] [PubMed] [Google Scholar]
- Workman C, Jensen LJ, Jarmer H, Berka R, Gautier L, Nielser HB, Saxild HH, Nielsen C, Brunak S, Knudsen S. A new non-linear normalization method for reducing variability in DNA microarray experiments. Genome Biol. 2002;3:research0048. doi: 10.1186/gb-2002-3-9-research0048. [DOI] [PMC free article] [PubMed] [Google Scholar]