Abstract
Genomic imprinting arises from allele-specific epigenetic modifications that are established during gametogenesis and that are maintained throughout somatic development. These parental-specific modifications include DNA methylation and post-translational modifications to histones, which create allele-specific active and repressive domains at imprinted regions. Through the use of a high-density genomic tiling array, we generated DNA and histone methylation profiles at 11 imprinted gene clusters in the mouse from DNA and from chromatin immunoprecipitated from sperm, heart, and cerebellum. Our analysis revealed that despite high levels of differential DNA methylation at non-CpG islands within these regions, imprinting control regions (ICRs) and secondary differentially methylated regions (DMRs) were identified by an overlapping pattern of H3K4 trimethylation (active chromatin) and H3K9 trimethylation (repressive chromatin) modifications in somatic tissue, and a sperm differentially methylated region (sDMR; sperm ≠ somatic tissue). Using these features as a common signature of DMRs, we identified 11 unique regions that mapped to known imprinted genes, to uncharacterized genes, and to intergenic regions flanking known imprinted genes. A common feature among these regions was the presence of a CpG island and an array of tandem repeats. Collectively, this study provides a comprehensive analysis of DNA methylation and histone H3K4me3 and H3K9me3 modifications at imprinted gene clusters, and identifies common epigenetic and genetic features of regions regulating genomic imprinting.
Genomic imprinting is a phenomenon found in plants and in therian mammals. Thus, in animals, it is governed by recently evolved genetic characteristics (Hore et al. 2007). These genetic features dictate the establishment of germline-specific epigenetic modifications, many of which are stably maintained throughout somatic development, with the ability to direct allele-specific expression patterns (Ferguson-Smith and Surani 2001; Reik and Walter 2001). Currently there are about 100 known imprinted genes in the mouse, including protein coding genes, noncoding RNA transcripts, and micro-RNAs (www.har.mrc.ac.uk/research/genomic_imprinting). Most of these genes are found in clusters, and in most cases the domains harbor a CpG rich imprinting control region (ICR) that imparts the imprint throughout the genes in the cluster. ICRs are essential cis-acting elements that direct differential allelic expression patterns on multiple genes within a cluster depending on their epigenetic state (Li et al. 1993). In somatic tissue, the ICRs typically exhibit different methylation patterns between the parental alleles (Delaval et al. 2007). Furthermore, imprinted gene clusters contain secondary differentially methylated regions (DMRs) that regulate or modify genomic imprinting at individual genes (Edwards and Ferguson-Smith 2007). The majority of DMRs are modified during gametogenesis when most are methylated in the maternal germline and not in the paternal germline, although the reciprocal scenario has been observed at least three ICRs (Delaval and Feil 2004). A unique property of imprinting DMRs is their ability to maintain their respective gametic marks during the early stages of embryonic development when there are dramatic genome-wide reprogramming events of both parental genomes (Monk et al. 1987; Howlett and Reik 1991; Oswald et al. 2000; Santos et al. 2002). During development, these differential methylation patterns are thought to create unique epigenetic states on the parental chromosomes that mediate differential expression of genes on the maternal and paternal alleles.
Recent evidence supports a role for opposite patterns of heterochromatic and euchromatic associated histone modifications on the paternal and maternal chromosomes at DMRs that facilitate the differential expression of imprinted genes (Delaval et al. 2007; Mikkelsen et al. 2007). Trimethylation of histone H3 at lysine 4 (H3K4me3) and trimethylation of histone H3 at lysine 9 (H3K9me3) are mutually exclusive histone modifications that associate with the active and repressed chromosomal alleles, respectively, at most ICRs (Yang et al. 2003; Lewis et al. 2006; Regha et al. 2007; Wang et al. 2008). At the Igf2r locus in the mouse, where silencing of the paternal allele is mediated by the Airn antisense transcript, the DMR is marked by mutually exclusive H3K4me3 on the paternal and H3K9me3 on the maternal alleles (Regha et al. 2007). The active Airn promoter on the paternal chromosome lies within the DMR, which in turn is within a CpG island that is hypomethylated. Conversely, the repressed Airn promoter on the maternal chromosome is modified with H3K9me3 and the CpG island is methylated. Therefore, these opposite modifications on the two alleles represent active and repressive chromatin states of homologous chromosomes at imprinted regions.
In order to investigate the epigenetic landscape at imprinted gene clusters, we determined the DNA and histone methylation profiles at 11 clusters in the mouse using a high-density genomic tiling array. By comparing the DNA methylation patterns between sperm, heart, and cerebellum, we identified 422 sperm-specific DMRs (sDMR; e.g., sperm ≠ heart/cerebellum) and 147 tissue-specific DMRs (tDMR; e.g., heart ≠ cerebellum). We also determined the H3K4me3 and H3K9me3 modification profiles and found that they are both present (presumably on opposite alleles) at ICRs, secondary imprinting DMRs, a subset of imprinted genes lacking known DMRs, and at many other intragenic and intergenic regions. By identifying regions that had both a sDMR and overlapping H3K4me3 and H3K9me3 modifications in somatic tissue, we identified 11 regions that may represent new DMRs involved in genomic imprinting within these imprinted gene clusters. Furthermore, we found that regions bearing overlapping patterns of these epigenetic modifications included CpG rich and tandem repeat sequences, indicating distinctive genetic features of regions with these modifications.
Results
DNA methylation profiles at imprinted gene clusters
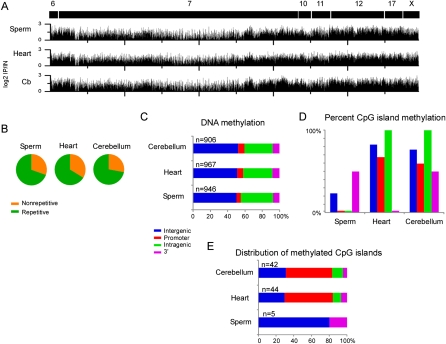
We determined the methylation profiles at each imprinted gene cluster in DNA isolated from sperm, heart, and cerebellum using the methylated DNA immunoprecipitation (MeDIP) method. The overall methylation profiles between samples were similar, showing enrichment for methylated DNA over ∼30% of the oligos on the array (Fig. 1A; Supplemental Fig. 1). The measurements for each sample were highly reproducible between biological replicates (two sperm samples, Pearson's r = 0.87; five cerebellum samples, r = 0.83; three heart samples, r = 0.88) (Supplemental Fig. 2). We also examined the technical variation in MeDIP by performing two replicates on one cerebellum sample, which highly correlated between experiments (r = 0.91). The methylation patterns between tissues were moderately similar (sperm vs. heart, r = 0.71, sperm vs. cerebellum, r = 0.73, and heart vs. cerebellum, r = 0.81), indicating that most of the methylated regions were conserved between samples. Next, we determined the sequence characteristics of the methylated regions and found that most contained repetitive sequences, with SINEs and simple repeats comprising most of the repetitive sites (Fig. 1B; Table 1). The hybridizing oligonucleotides are not within repetitive sequences, but the repetitive sequences are imbedded within blocks of methylated DNA. In addition, we found that intergenic and intragenic regions were heavily methylated compared to the promoter and 3′ ends of genes (Fig. 1C). Examination of methylated CpG islands indicated that they were frequently methylated in heart (71.7%) and cerebellum (65%), but not in sperm (8.5%) (Fig. 1D; Supplemental Table 2). Of the methylated CpG islands, most were located at or near the promoter (Fig. 1E), and as expected, corresponded to ICRs or DMRs. Collectively, these data indicate that intergenic and intragenic regions consisting primarily of repetitive sequences show moderately conserved patterns of methylation between sperm, heart, and cerebellum; however in contrast, CpG island methylation differs between sperm and somatic tissue.
Figure 1.
DNA methylation patterns at imprinted gene clusters. (A) MeDIP was performed on DNA isolated from sperm, heart, and cerebellum and the enriched (positive log2 IP/IN) values are plotted for each imprinted gene cluster. Chromosomal regions are marked as black boxes with corresponding chromosome numbers. (B) Sequence characterization of methylated regions in sperm, heart, and cerebellum showed that the imprinted gene clusters were primarily methylated at repetitive DNA based on repeat masker. (C) Plots showing the genomic location of methylated regions relative to the RefSeq gene annotation. (D) CpG island methylation was less frequent in sperm samples compared to the heart and cerebellum, particularly for the promoter and intragenic regions. (E) The majority of methylated CpG islands in heart and cerebellum were present in the promoters of known imprinted genes.
Table 1.
Percentage of methylated regions associated with repetitive elements
Differential DNA methylation between samples corresponds to CpG rich imprinted DMRs and to nonrepetitive regions
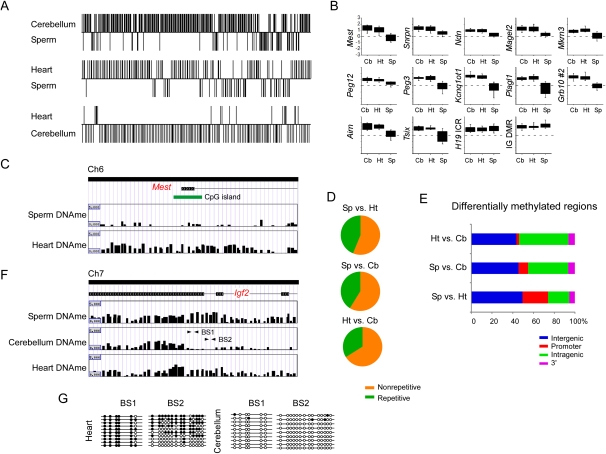
Next we identified sperm-specific DMRs (sDMRs) and tissue-specific DMRs (tDMRs) that exhibited significant (P < 0.00001) differences in the methylation profiles between samples. In all we detected 127 sDMRs when comparing sperm (Sp) to heart (Ht) (Ht > Sp, n = 91; Sp > Ht, n = 36), 295 sDMRs between sperm and cerebellum (Cb) (Cb > Sp, n = 232; Sp > Cb, n = 63), and 147 tDMRs between heart and cerebellum (Ht > Cb, n = 15; Cb > Ht, n = 132) (Fig. 2A; Supplemental Fig. 3; see Methods). As expected, we detected sDMRs at each known imprinted DMR (Arima et al. 2006; Geuns et al. 2007), which provided a positive control for the specificity of the MeDIP method in our analysis. At these regions, there was an absence of methylation at each ICR in sperm, except for the H19 ICR and Dlk1–Meg3 intergenic (IG) DMR, which displayed a higher level of enrichment for methylated DNA relative to the somatic tissue samples (Fig. 2B). Furthermore, the majority (>75%) of DMRs were located within regions that were highly conserved in other species (Supplemental Table 3). For both sDMRs and tDMRs, the overall majority mapped to nonrepetitive sequences (Fig. 2D). Of the DMRs mapping to repetitive elements, the sequence characteristics consisted primarily of SINEs and simple repeats (Table 2).
Figure 2.
Characterization of sperm-specific and tissue-specific differentially methylated regions at imprinted gene clusters. (A) A sliding window t-test was performed between sperm and heart (Sp vs. Ht; sDMR), sperm and cerebellum (Sp vs. Cb; sDMR), and heart and cerebellum (Ht vs. Cb; tDMR) using the log2 IP/IN ratios obtained from MeDIP to determine sperm and tissue-specific differentially methylated regions (sDMR and tDMR). The y-axis indicates regions that have significantly different (P < 0.00001) methylation profiles between samples and the direction of methylation. (B) The mean log2 IP/IN of probes was calculated over each known DMR in cerebellum, heart, and sperm. Box plots show less methylation in sperm DNA at each DMR, except at the H19 ICR and IG-DMR, which are methylated in sperm and differentially methylated in heart and cerebellum. Box plots represent the 25th and 75th percentile for each averaged sample. Whiskers show 10th and 90th percentiles. (C) An example showing the different patterns observed between sperm DNA methylation (Sperm DNAme) and heart DNA methylation (Heart DNAme) at the Mest DMR. (D) Sequence characterization of sDMR and tDMR showed that nonrepetitive DNA is primarily differentially methylated at imprinted gene clusters. (E) Genomic location of sDMRs and tDMRs relative to the RefSeq gene annotation. (F) An example of a tDMR at the 3′ end of the Igf2 gene. (G) Bisulfite sequencing confirmed loss of methylation in the cerebellum DNA at the 3′ end of the Igf2 gene. Arrows in F indicate regions amplified for bisulfite sequencing.
Table 2.
Percentage of sDMRs and tDMRs associated with repetitive elements
Sp, sperm; Ht, heart; Cb, cerebellum.
We also found sDMRs at numerous CpG islands (heart, n = 22; cerebellum, n = 15) within imprinted genes and uncharacterized regions. However, we did not detect any tDMR that corresponded to a CpG island. Consistent with the methylation patterns of known DMRs, the sperm samples were predominantly hypomethylated relative to the somatic tissue samples. In comparisons between heart and cerebellum, there were more methylated regions in cerebellum than in the heart. Further analysis of both sDMRs and tDMRs indicated that most were present at sites away from the 5′ and 3′ ends of genes (Fig. 2E).
To further confirm our MeDIP results, we utilized an HpaII-PCR assay to assess the loss of methylation at three sDMRs. For each sDMR, the PCR reaction failed to generate an amplicon in the sperm DNA digested with HpaII, whereas cerebellum DNA digested with HpaII generated an amplicon of the expected size (Supplemental Fig. 3). In addition, we utilized bisulfite sequencing to assess the CpG methylation in the last intron of the Igf2 gene, where we observed a tDMR. Interestingly, hypomethylation of this region was specific to the cerebellum as both heart and sperm showed enrichment for methylated DNA (Fig. 2F). Bisulfite sequencing revealed that the heart sample was heavily methylated in the bisulfite sequence 1 (BS1) region, but was nearly completely unmethylated in the corresponding region in the cerebellum sample. In the bisulfite sequence 2 (BS2) region the heart sample was partially methylated, whereas the cerebellum was almost completely unmethylated (Fig. 2G). Collectively, these data indicate that there are numerous sDMRs and tDMRs within imprinted gene clusters that consist mostly of nonrepetitive DNA located within intergenic and intragenic regions; however, there were a limited number of sDMRs unique to CpG islands within imprinted DMRs and uncharacterized regions.
Overlapping patterns of H3K4me3 and H3K9me3 are common at imprinted genes, imprinted DMRs, and imprinting control regions
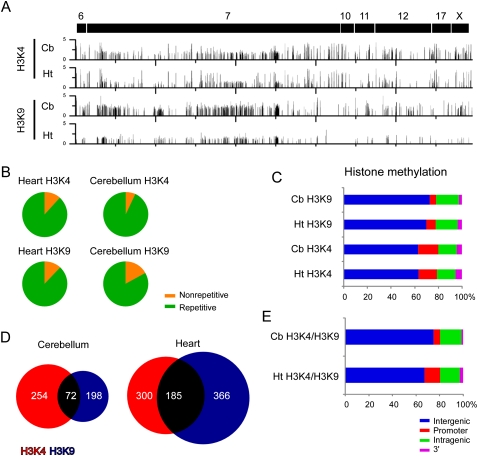
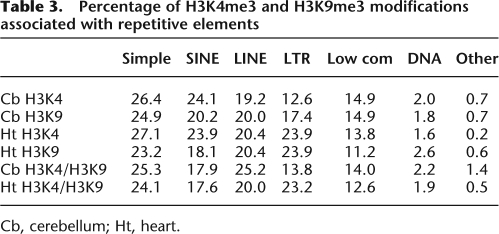
Next, we used chromatin immunoprecipitation (ChIP) to identify regions that were enriched for the H3K4me3 and H3K9me3 modifications using chromatin isolated from heart and cerebellum. H3K4me3 and H3K9me3 modifications were detected as discrete foci along each chromosomal region (Fig. 3A). The histone methylation profiles between male and female mice were identical at autosomal chromosomes, but displayed different patterns on the X chromosome (Supplemental Fig. 4). Sequence characterization of the H3K4me3 and H3K9me3 enriched regions in heart and cerebellum indicated that the majority was detected at repetitive elements (Fig. 3B; Table 3), and at intergenic regions (Fig. 3C). Next, we determined the genomic positions of the H3K4me3 modification and found 58 unique genes in heart, and 52 unique genes in cerebellum with modified regions in the gene body (e.g., promoter, intragenic, or 3′ untranslated region [UTR]). The modification appeared to be equally present at nonimprinted (heart, n = 24; cerebellum, n = 23) and imprinted (heart, n = 34; cerebellum, n = 35) genes. Within the gene body, H3K4me3 was primarily detected at the promoter (heart, 60.0%; cerebellum, 68.3%), although we did not correct for alternative promoters or for promoters of antisense transcripts that initiate within intragenic regions (e.g., Airn and Kcnq1ot1 antisense transcripts). We detected the H3K9me3 modification in the gene body of 42 unique genes in heart and 23 unique genes in cerebellum. Again, the modification appeared to be equally distributed to both imprinted (heart, n = 28; cerebellum, n = 12) and nonimprinted genes (heart, n = 14; cerebellum, n = 11). Relative to the H3K4me3 modification, we detected less H3K9me3 at the promoter (heart, 26.4%; cerebellum, 20.4%) although we detected similar levels in the intragenic and 3′ UTR (Supplemental Table 4).
Figure 3.
H3K4me3 and H3K9me3 enriched regions at imprinted gene clusters. (A) ChIP-chip profiles of histone modifications for H3K4me3 and H3K9me3 in heart and cerebellum. Y-axis values represent log2 IP/IN. (B) Sequence characterization of regions enriched for H3K4me3 and H3K9me3 showed that each modification was present primarily at repetitive sequences. (C) Genomic location of H3K4me3 and H3K4me3 modifications relative to the RefSeq gene annotation. (D) Venn diagram showing overlapping H3K4me3 and H3K9me3 regions in both heart and cerebellum. (E) Genomic locations of overlapping H3K4me3 and H3K9me3 modifications relative to the RefSeq gene annotations.
Table 3.
Percentage of H3K4me3 and H3K9me3 modifications associated with repetitive elements
Cb, cerebellum; Ht, heart.
We next identified regions with both overlapping H3K4me3 and H3K9me3 modifications (Fig. 3D). We detected numerous regions in heart (n = 185) and cerebellum (n = 72) with the overlapping blocks of the H3K4me3 and H3K9me3 modifications, which mapped primarily to intergenic regions (heart, 67.6%; cerebellum, 72%), but also to intragenic regions (heart, 16.8%; cerebellum, 21.3%), to promoters (heart, 13.5%; cerebellum, 5.3%), and to the 3′ UTR of genes (heart, 2.2%; cerebellum, 1.3%). Two intergenic overlapping regions mapped to the H19 ICR on chromosome 7 and the IG-DMR on chromosome 12 (data not shown). Within the gene body, we found that 62.8% and 70% of the overlapping histone modifications in heart and cerebellum, respectively, mapped to known imprinted genes including the Airn ICR, although there were a number of genes on the X chromosome and genes not previously characterized as imprinted that possessed both modifications (Supplemental Table 5). Interestingly, we detected many olfactory receptor genes, which undergo allelic exclusion, bearing overlapping modifications.
These data indicate that there are unique patterns of histone modifications for H3K4me3 and H3K9me3 in heart and cerebellum that localize to discrete regions within intragenic and intergenic regions at imprinted gene clusters. Sites where the two modifications overlap correspond to imprinted DMRs, imprinted genes, genes on the X chromosome, and olfactory receptor genes. Modifications at these regions are consistent with differential active and repressive chromatin states on homologous chromosomes. Collectively, these data show that domains with differential active and repressive chromatin states are modified with H3K4me3 and H3K9me3, respectively.
Sperm-specific DMRs with overlapping H3K4me3 and H3K9me3 histone modifications are a common signature for imprinted DMRs and ICRs
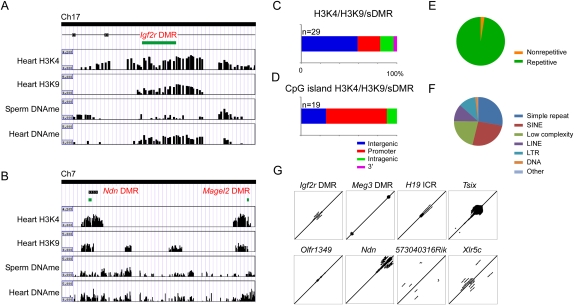
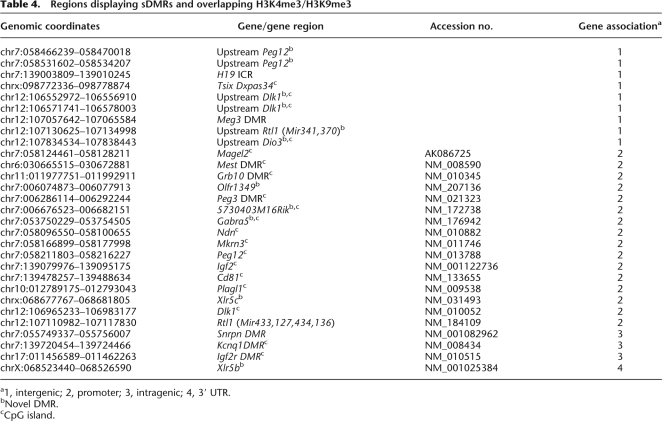
Our analysis of sDMRs identified each previously known ICR and DMR on the array (Fig. 2B), and also identified numerous other sDMRs (Fig. 2E; Supplemental Table 3). In addition, we identified many intergenic and intragenic regions with overlapping H3K4me3 and H3K9me3 modifications, a feature present at each ICR and at secondary DMRs, as well as many other imprinted genes and unique regions (Fig. 3E). Therefore, we decided to identify regions within the imprinted gene clusters that were sDMRs (i.e., sperm vs. heart; P < 0.00001) and that had overlapping H3K4me3 and H3K9me3 histone modifications (i.e., H3K4me3/HeK9me3 overlapping in heart; see Methods). We detected 29 regions that met the criteria (Supplemental Table 6). As expected, we identified each known ICR (Fig. 4A) except the IG-DMR which was excluded due to a sDMR P-value greater than 0.00001 (IG-DMR, P = 0.0001). In addition, we detected 11 regions that corresponded to unique sites (Fig. 4B; Table 4). Six of the new sites mapped to intergenic regions flanking known imprinted genes and five to intragenic regions (promoter, n = 4; 3′ UTR, n = 1). Collectively, there was a bias away from the gene body (Fig. 4C). Nineteen of the 29 regions corresponded to CpG islands, and the majority of these regions mapped to promoters (Fig. 4D). For all of the regions identified, except the Xlr5b locus, the modified region corresponded to a repetitive element that was comprised mostly of simple repeats, SINEs, and low complexity repeats (Fig. 4E,F). Further analysis of the genomic sequence directly underlying the differential modifications indicated that the majority (83%) contained tandem repeats (Fig. 4G). We examined CpG islands within both imprinted genes that lacked an ICR, as well as nonimprinted genes in the region, and we did not detect similar levels of tandem repeats within these regions, indicating that tandem repeats are a common genetic feature of sDMRs with overlapping H3K4me3 and H3K9me3 modifications. Collectively, these data indicate that overlapping H3K4me3 and H3K9me3 modifications, specifically at sDMRs, are a common mark for imprinting control regions, and by selecting for regions that possess these epigenetic features, we detected regions that may represent novel DMRs regulating genomic imprinting.
Figure 4.
Overlapping patterns of H3K4me3 and H3K9me3 at sDMRs identified imprinting control regions. (A) An example of the Igf2r DMR in intron 2 of the Igf2r locus. The CpG island (green box) indicates the DMR, which contains both H3K4me3 and H3K9me3 modifications and differentially methylated DNA between sperm and heart. (B) Examples of the Ndn and Magel2 DMR showing overlapping H3K4me3 and H3K9me3, as well as sDMR between sperm and heart. (C) Genomic locations of regions with a sDMR and overlapping H3K4me3 and H3K9me3. (D) Genomic locations of CpG islands with a sDMR and overlapping H3K4me3 and H3K9me3. (E) Sequence characterization of regions with a sDMR and overlapping H3K4me3 and H3K9me3 indicated regions are highly repetitive and (F) are comprised mostly of simple repeats, SINES, and low complexity repeats. (G) Examples of sequence alignments for known ICRs (e.g., Igf2r DMR, Meg3 DMR, H19 ICR, and Tsix) and putative ICRs (e.g., Olfr1349, Upstream Ndn, 5730403M16Rik, and Xlr5C) showing repeated elements.
Table 4.
Regions displaying sDMRs and overlapping H3K4me3/H3K9me3
a1, intergenic; 2, promoter; 3, intragenic; 4, 3′ UTR.
bNovel DMR.
cCpG island.
Discussion
Here we report the use of a high-density genomic tiling array to determine the DNA and histone methylation patterns at 11 imprinted gene clusters in mouse sperm, heart, and cerebellum. Our analysis revealed that despite substantial levels of differential DNA and histone modifications within imprinted clusters, ICRs and DMRs are characterized by specific overlapping patterns of epigenetic modifications that encompass unique genetic elements. Using these features as a signature for DMRs associated with genomic imprinting, we identified 11 regions that may represent new imprinting control elements within these clusters. Collectively, these data provide a high-density analysis of DNA and histone methylation at imprinted genes in the mouse.
In our analysis we found that the methylated regions in all samples were primarily at repetitive elements located in intergenic and intragenic regions, which is consistent with previous large-scale methylation studies (Weber et al. 2005; Eckhardt et al. 2006). In addition, we detected numerous CpG islands at imprinted DMRs that were methylated in heart and cerebellum, but not in sperm. This was expected given the design of the array, and allowed us to confirm the specificity of MeDIP to detect both qualitative and quantitative changes in DNA methylation. Our findings that most DMRs, excluding those associated with genomic imprinting, were present at nonrepetitive intergenic and intragenic regions located away from CpG islands is important as most studies to date have focused on the methylation patterns of promoters and CpG islands (Shiota et al. 2002; Schilling and Rehli 2007; Suzuki et al. 2007; Weber et al. 2007). This finding is supported by Irizarry et al. (2009) who have shown that at least 76% of tissue-specific DMRs in human tissues occur at “CpG island shores,” which are regions that are adjacent to, but are not located within, CpG islands. Similar findings in the mouse have been reported using restriction landmark genomic scanning (Oakes et al. 2007). Collectively these data suggest that the tissue-specific DMRs may play an important role in gene regulation and potentially as modifiers of genomic imprinting, but that they consist of genetic elements that are different from imprinted DMRs.
The patterns of histone modifications we detected are also consistent with large scale ChIP with massively parallel sequencing (ChIP-seq) studies in ES cells that have shown that H3K4me3 and H3K9me3 are mutually exclusive modifications that mark the active and repressed alleles, respectively, at ICRs (Mikkelsen et al. 2007). This has also been shown at individual imprinted genes using tiling arrays (Regha et al. 2007) and at other imprinted loci through conventional ChIP-PCR (Sakamoto et al. 2004; Vu et al. 2004; Lewis et al. 2006). At least in the heart samples, our data indicate that overlapping H3K4me3 and H3K9me3 are common marks at ICRs and DMRs. Perhaps surprisingly, this was the case for Cd81, which is imprinted in placenta (Lewis et al. 2004b). In the cerebellum samples however, we observed a major reduction in the number of overlapping regions at both imprinted and nonimprinted genes, indicating dramatic changes in histone modifications between these tissues. This is not surprising given the number of genes that are exclusively imprinted in brain (e.g., Ube3a), or that undergo relaxation of imprinting in brain (e.g., Igf2r). Although we found overlapping H3K4me3 and H3K9me3 modifications at all of the known imprinted DMRs, this feature was not exclusive to them. Given this, we aligned sDMR regions for each tissue comparison to regions with overlapping H3K4me3 and H3K9me3 modifications in that same tissue. By doing so we detected each known imprinting ICR and DMR tiled on the array, with the exception of the IG-DMR on chromosome 12 (Dlk1–Dio3 imprinted cluster), which had a P-value slightly less than our very stringent threshold for detection. In addition, we identified 11 novel regions that mapped to intergenic sites flanking imprinted gene clusters or to uncharacterized genes. Thus, the presence of overlapping H3K4me3 and H3K9me3 modifications in somatic tissues and differential methylation between sperm and somatic tissue marks imprinted DMRs. A similar approach has been reported where overlapping histone H3 dimethyl lysine 4 (HeK4me2) and DNA methylation identifies imprinted gene regions in human T cells and immortalized lymphoblast cell lines. By including known CTCF binding sites (i.e., “triple hits”), the identification of imprinted genes is dramatically increased (Wen et al. 2008). Our “triple hit” analysis identified all but one imprinted DMR, which would have been detected in a less stringent statistical analysis.
The identification of the putative imprinting control elements prompted us to more closely examine the underlying genetic sequence between known imprinting DMRs and the new regions identified in our study. For each imprinting control element (i.e., sDMR/H3K4me3/H3K9me3) we found that most were present at CpG islands and consisted primarily of repetitive elements that included simple repeats, SINES, and low complexity repeats. However, the most common sequence feature among each region was the presence of tandem repeats, which were not as frequently detected at the promoters or CpG islands of other imprinted genes lacking DMRs. Tandem repeats at DMRs have been the focus of numerous studies as common genetic elements involved in genomic imprinting (Neumann et al. 1995; Moore et al. 1997; Reinhart et al. 2006). The presence of tandem repeats in the majority of DMRs in our analysis, including the unique regions we identified, indicates that they are usually associated with imprinted regions, although their role in genomic imprinting is unclear. The fact that tandem repeats are not present in every imprinted DMR suggests that there may be other as yet unidentified genetic components with which tandem repeats often, but not always, co-occur. At least one genetic study demonstrates that the tandem repeats located ∼5 kb upstream of the mouse H19 ICR are not necessary for genomic imprinting at the H19 and Igf2 genes (Lewis et al. 2004a), but it is difficult to attribute any specific role to these repeats as they are located outside the boundaries of the DMR. Regardless, our approach is more direct in that we only investigated experimentally identified regions with epigenetic features unique to imprinted DMRs. Thus, there does appear to be a tendency for tandem repeats to be associated with differentially methylated regions, which includes most of the novel regions identified in this study; however. further genetic analysis will be required to determine the specific role of these elements in genomic imprinting.
Methods
Development of a mouse genomic imprinting array
We designed a custom 105,000 oligonucleotide mouse genomic imprinting array (105K MIA) using the Agilent E-array platform (August 2005 mm7, build 35 genomic sequence). The array included sequence from nine autosomal imprinted gene clusters (chromosomes 6, 7, 10, 11, 12, and 17) and two X chromosome imprinted gene clusters (Supplemental Fig. 1; www.har.mrc.ac.uk/research/genomic_imprinting). We designed the array by tiling across each imprinted gene cluster and also included adjacent sequence up to, but not including, the nearest upstream and downstream non-imprinted gene. For each region, we tiled at approximately a 100 base pair (bp) resolution (approximately nine 60-mers per kilobase of DNA); the E-array design excludes repetitive elements and low copy repetitive regions on the same or other chromosomes. Collectively, the array spans ∼11 Mb of genomic sequence and includes 60 imprinted genes and 37 nonimprinted genes (Supplemental Table 1).
MeDIP and ChIP methods
Methylated DNA immunoprecipitation (MeDIP)
DNA was isolated from eight-week-old male and female cerebellum and heart by proteinase K digestion followed by phenol chloroform extraction and ethanol precipitation. DNA was isolated from adult male sperm by 0.1 mM DTT/proteinase K digestion followed by phenol chloroform extraction and ethanol precipitation. Twenty micrograms of DNA was diluted to a final volume of 100 μL with dH2O in a 0.6-mL Eppendorf tube. Sonication was performed on ice using an Ultrasonic Dismembrator Model 500 (Fisher Scientific). The samples were pulsed three times for 15 sec at 14% power between 30-sec intervals. An average DNA fragment size of ∼400 bp was verified on a 1.5% agarose gel. Four micrograms of sonicated DNA was then diluted to a final volume of 500 μL with IP buffer (10 mM sodium phosphate at pH 7.0, 140 mM NaCl, 0.05% Triton X) in a 1.5-mL Eppendorf tube. To remove potentially cross-reactive contaminants, a preclearance procedure was performed by adding 30 μL of prewashed Dynal M-280 sheep anti-mouse IgG beads (Invitrogen) to the sample and chilling at 4°C for 4 h with rotation. The precleared supernatant was transferred to a new tube and a 100 μL aliquot was removed and stored at −20°C for sample input. The samples were denatured at 95°C for 10 min and immediately transferred to ice. Ten microliters of anti-5-methyl cytosine antibody (#BI-MECY, Eurogentec) was added to the sample and chilled at 4°C for 2 h with rotation. Thirty microliters of prewashed Dynal M-280 sheep anti-mouse IgG beads were subsequently added to the sample and chilled at 4°C overnight with rotation. The supernatant was removed and beads were washed four times with 1mL of IP buffer. The DNA was eluted by adding 200 μL of elution buffer (100 mM sodium bicarbonate, 1% SDS) to the beads and rotating at room temperature for 15 min. Both IP and input samples were purified using the Qiagen Miniprep DNA Purification (Qiagen) protocol and eluted in 30 μL of dH2O. Both IP and Input samples were subsequently amplified using the WGA II Amplification (Sigma) protocol prior to labeling.
MNase chromatin immunoprecipitation (NChIP)
Freshly isolated cerebellum and heart samples (∼75–100 mg) were placed into a 1.5-mL Eppendorf tube containing 375 μL of 1× Douncing buffer (10 mM Tris-Cl at pH 7.5, 4 mM MgCl2, 2 mM CaCl2). The tissue was homogenized using a battery operated pestle for 30 sec. Micrococcal nuclease in the amount of 11.25 μL (Sigma) (0.2 units/μL in 0.1% BSA) was added to each tube, vortexed, and incubated at 37°C for 7 min. To stop the reaction, 4.61 μL of 1 M EDTA was added. The content was transferred into a 15-mL conical tube on ice containing 4.65 mL of hypotonic lysis solution (0.1 M DTT, 0.1 M EDTA, 0.01 M PMSF, 0.1 M benzamidine). The tubes were chilled on ice and mixed two to three times every 10 min for 1 h. Cellular debris was removed by centrifugation at 3000g for 10 min and the supernatant was aliquoted into four Eppendorf tubes containing 1100 μL each. Unused chromatin aliquots were stored at −70°C. For immunoprecipitation, 122 μL of 10× incubation buffer (1 M EDTA at pH 8.0, 1 M Tris-Cl, 1 M NaCl) was added to each tube and mixed thoroughly. Preclearing was performed by adding 100 μL of protein A anti-rabbit IgG bound beads (prewashed and prepared 1 d in advance) to each tube and chilling at 4°C with rotation for 1 h. The precleared supernatant was transferred to a new 1.5-mL Eppendorf tube and 100 μL of protein A conjugated to each antibody (anti-trimethyl H3 lysine 9, #07-442, Upstate Biotechnologies; anti-trimethyl H3 lysine 4, #05-745, Upstate Biotechnologies) bound beads (prewashed and prepared 1 d in advance) was added to each tube and chilled overnight at 4°C with rotation. The supernatant was removed and the beads were washed in the following order: 2× 1 mL of low salt solution (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 150 mM NaCl, 20 mM Tris-Cl at pH 8.0, 1× complete protease inhibitor), 2× 1 mL of high salt solution (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 500 mM NaCl, 20 mM Tris-Cl at pH 8.0, 1× complete protease inhibitor), 1× 1 mL of lithium chloride solution (0.25 M LiCl, 1% NP-40, 1% deoxycholate, 1 mM EDTA, 10 mM Tris-Cl at pH 8.0), and 2× 1 mL of TE solution (10 mM Tris-Cl at pH 8.0, 1 mM EDTA). DNA–protein complexes were eluted by adding 200 μL of elution buffer (100 mM sodium bicarbonate, 1% SDS) to the beads and rotating at room temperature for 15 min. Both IP and input samples were purified using the Qiagen Miniprep cleanup protocol and eluted in 30 μL of dH2O. Both IP and input samples were amplified using the WGA Amplification Protocol prior to labeling.
Qiagen Miniprep DNA purification
Qiagen Miniprep columns are used for all DNA purifications. The Qiagen Miniprep column allows smaller fragments of DNA to be retained when compared to Qiagen PCR purification columns. Additionally, we have experienced more consistent WGA amplification with greater yields when using this approach as opposed to phenol-chloroform extraction. DNA was mixed with PB buffer in a 1:5 ratio using centrifugation of >10,000g for 1 min. The column was washed twice with 800 μL using centrifugations of >10,000g for 1 min each. Residual wash buffer was removed through an additional centrifugation of >10,000g for 2 min. 30 μL high quality dH2O was applied to the center of the column and subsequently eluted via centrifugation of >10,000g for 1 min. Assessment of DNA quantity and quality was performed using a NanoDrop Spectrophotomer (Thermo Scientific NanoDrop Technologies).
Whole genome amplification (WGA) of MeDIP and NChIP DNA
Amplification of MeDIP and NChIP products was based on a protocol described elsewhere (O'Geen et al. 2006). All components for amplification were provided by the WGA2 Kit (Sigma). Between 10 and 100 ng of DNA was diluted to a final volume of 11 μL using high quality dH2O. To this, 2 μL of 1× library preparation buffer and 1 μL of library stabilization solution was added. The samples were thoroughly mixed and denatured at 95°C for 2 min and immediately transferred to ice. While on ice, 1 μL of library preparation enzyme was added, mixed and placed into a thermocycler for the following temperatures and times: 16°C for 20 min, 24°C for 20 min, 37°C for 20 min, and 75°C for 5 min followed by a hold at 4°C. The prepared library was then amplified by adding 60 μL of a master mix consisting of 7.5 μL of 10× amplification mix, 47.5 μL of nuclease-free H2O, and 5 μL of WGA polymerase. The samples were denatured for 3 min at 95°C and cycled 14 times for 15 sec at 94°C and 5 min at 65°C, followed by a hold at 4°C. The samples were purified using the Qiagen Miniprep DNA Purification protocol. DNA quantity and quality was assessed using a NanoDrop Spectrophotomer (Nanodrop Technologies).
MeDIP and NChIP labeling and purification
Labeling of both MeDIP and NChIP DNA products is based on a protocol provided by the Wellcome Trust Sanger Institute. Between 300 and 500 ng of DNA was diluted to a final volume of 48 μL using high quality dH2O. To this, 40 μL of random primers (Invitrogen Bioprime DNA Labeling System) was added. The samples were denatured at 100°C for 10 min and immediately transferred to ice. Ten microliters of 10× dCTP nucleotide mix (Bioprime), 4 μL of Cy3 or Cy5 labeled dCTP (GE Healthcare), and 2 μL of exo Klenow fragment (Bioprime) were added to each sample. The samples were incubated at 37°C overnight. The following morning, 10 μL of stop buffer (Bioprime) was added to each sample and mixed thoroughly. The labeled samples were purified using the Bioprime Array CGH Purification Module (Invitrogen). To each sample, 400 μL of buffer A was added and mixed thoroughly. The samples were placed in the provided purification column and centrifuged for 1 min at 8,000g. The columns were washed twice in 600 and 200 μL of buffer B and centrifuged for 1 and 2 min at 8,000g, respectively. Forty microliters of high quality dH2O was placed in each column and centrifuged for 1 min at full speed. Assessment of DNA quantity and dye incorporation was performed using the microarray function of a NanoDrop Spectrophotomer (NanoDrop Technologies).
MeDIP and NChIP array hybridization
Labeled MeDIP and NChIP DNA products were processed in 1.5-mL Eppendorf tubes for hybridization by adding 25 μL of Cot1 DNA (Invitrogen), 25 μL of 10× blocking agent (Agilent), and 125 μL of 2× hybridization buffer for (Agilent) to 75 μL of solution containing 2.5 μg of Cy5 labeled immunoprecipitated DNA and 2.5 μg of Cy3 labeled input DNA. Samples were denatured at 95°C for 3 min and immediately transferred to a 37°C water bath for 30 min. Tubes were spun at 13,000 rpm for 1 min and 240 μL of sample applied to the 2X105K custom mouse imprinting array as per Agilent instructions. Hybridizations were carried out at 65°C in an Agilent rotisserie oven with max rotation for 40 h.
MeDIP and NChIP filtering and annotation
Methylated regions were determined using a stringent simple enrichment filtering method (log2 IP/IN ≥ 0.5) that were within blocks of at least 10 cells, allowing no more than one cell with a log2 IP/IN ≤ 0.5 within the block. Differentially methylated regions (DMRs) were determined by averaging the log2 IP/IN values for each tissue (sperm, n = 2; cerebellum, n = 5; heart, n = 3) and then by performing a sliding window t-test (20 rows) between two averaged sample sets (sperm vs. heart; sperm vs. cerebellum; heart vs. cerebellum). The P-value for each window was centered within the 20-window range in order to accurately reflect the genomic coordinates of the significantly different log2 IP/IN values between tissues. DMRs were then identified as regions with P-values < 0.00001. To ensure that at least one sample was methylated, the log2 IP/IN values for each sample within the DMR were averaged over the region included in the DMR, and regions where both samples had negative log2 IP/IN values were removed. DMRs were then manually checked to ensure that at least one of the two samples had a log2 IP/IN ≥ 0.5 (methylated region). ChIP profiles were analyzed using the Agilent ChIP analytical software. The settings used were blank subtraction normalization, intra-array normalization, variance stabilization, and Predefined Peak shape detection (v 2.0) algorithms. ChIP regions with fewer than three positive oligos were also removed. Genomic positions of MeDIP and ChIP regions were determined using the mouse RefSeq gene annotation. Promoters were identified as regions within at least ±2 kb of the transcriptional start site and at the 3′ end as regions within at least ±2 kb of the 3′ UTR. CpG island annotation, simple repeat annotations, and 7× regulatory potential (evolutionary conserved regions) annotations were downloaded from the UCSC Genome Browser (http://genome.ucsc.edu/) and aligned against enriched regions. Tandem repeats were detected using two methods: First, regions were analyzed in the Tandem Repeats Finder (http://tandem.bu.edu/trf/trf.html) using the following advanced settings: alignment parameters = 2, 5, 7; minimum alignment = 80; and maximum period = 2000. Next, regions were self-aligned using PiPmaker (http://pipmaker.bx.psu.edu/pipmaker/). All MeDIP and ChIP data are freely available on the Genboree website (www.genboree.org). Copies of the scripts used for bioinformatic analysis of MeDIP and ChIP regions can be obtained by contacting the corresponding author.
Bisulfite sequencing and HpaII analysis of methylated regions
For bisulfite sequencing, DNA isolated from heart and cerebellum and samples were sodium bisulfite treated, following the instructions provided by the manufacturer (Zymo Research). For amplification of the 3′ end of the Igf2 gene two primer sets were designed to amplify the region displaying differential methylation between heart and cerebellum by MeDIP (build 35, within coordinates chr 7: 139,081,171–139,081,721). The primers used to amplify the BS1 region were (F:CGGGTACGTAGGAGGGTAGG; R:CGACACCTAAATAAAAACTCAAACT) and for the BS2 were (F:GAGGTTGATAGTAAAATGTGTGAGG; R:AAACTTCCAAACAAACCTTCAAA). PCR for the BS1 and BS2 primer sets was conducted under the following conditions: 3 μL of sodium bisulfite treated DNA, 5 μL of 5× GoTaq buffer (Promega), 2 μL of 2 mM MgCl2, 1.0 μL of 2.5 mM dNTPs, 1.5 μL of 3 μM forward primer, 1.5 μL of 3 μM reverse primer, 0.125 μL of Taq (Promega), and 10.875 μL of dH2O. The cycling conditions were as follows: 95°C for 5 min; 35 cycles of 95°C for 30 sec, 54.5°C for 30 sec, and 72°C for 1 min; and a final extension at 72°C for 7 min. Amplicons were then cloned into the PCR2.1 Sequencing vector using a TOPOTA cloning kit (Invitrogen). Plasmids positive for the insert were sequenced using the M13R primer. Sequences were aligned and the number of unconverted cytosines adjacent to guanines was calculated. For the HpaII methylation analysis, DNA was isolated from sperm and cerebellum from the same male mouse. A total of 5 μg of DNA was cut with HpaII overnight at 37°C. Control samples of equal concentration, containing the HpaII reaction buffers without the HpaII enzyme were incubated overnight and run in parallel with the digested DNA. The HpaII enzyme was then heat inactivated at 65°C for 20 min and the reactions were then diluted to a concentration of 50 ng/μL. The digested DNA was then amplified using the following primers: Upstream Dcn (F:TTAAGGCCTTGGGAACACAG; R:GCAGAAACCCTGTGGAATGT), Mest DMR (F:AACCGCGAACGATAGGAAAT; R:CCGAAGCACCAGCATCTAAG), and upstream Gabra5 (F:TGTGACCTAGGTGCTTGTCC; R:CTCAGAAAAACCCCCAAAGG). PCR was conducted under the following conditions: 2 μL of cut and uncut DNA, 5 μL of 5× GoTaq buffer (Promega), 2 μL of 2 mM MgCl2, 1.0 μL of 2.5 mM dNTPs, 1.5 μL of 3 μM forward primer, 1.5 μL of 3 μM reverse primer, 0.125 μL of Taq (Promega), and 11.875 μL of dH2O. The cycling conditions were as follows: 95°C for 5 min; 30 cycles of 95°C for 30 sec, 55°C for 30 sec, and 72°C for 1 min; and a final extension at 72°C for 7 min. Amplicons were then resolved on a 1.5% agarose gel.
Acknowledgments
We thank Alan Harris, Andrew Jackson, and the Genboree team (www.genboree.org) for experimental data visualization and the publication of the supplemental data, as well as Jan Bressler and Paul Samollow for reading the manuscript.
Footnotes
[Supplemental material is available online at www.genome.org. The microarray data from this study have been submitted to Gene Expression Omnibus (GEO) (http://www.ncbi.nlm.nih.gov/geo/) under accession no. GSE16588.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.089185.108.
References
- Arima T, Yamasaki K, John RM, Kato K, Sakumi K, Nakabeppu Y, Wake N, Kono T. The human HYMAI/PLAGL1 differentially methylated region acts as an imprint control region in mice. Genomics. 2006;88:650–658. doi: 10.1016/j.ygeno.2006.07.005. [DOI] [PubMed] [Google Scholar]
- Delaval K, Feil R. Epigenetic regulation of mammalian genomic imprinting. Curr Opin Genet Dev. 2004;14:188–195. doi: 10.1016/j.gde.2004.01.005. [DOI] [PubMed] [Google Scholar]
- Delaval K, Govin J, Cerqueira F, Rousseaux S, Khochbin S, Feil R. Differential histone modifications mark mouse imprinting control regions during spermatogenesis. EMBO J. 2007;26:720–729. doi: 10.1038/sj.emboj.7601513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckhardt F, Lewin J, Cortese R, Rakyan VK, Attwood J, Burger M, Burton J, Cox TV, Davies R, Down TA, et al. DNA methylation profiling of human chromosomes 6, 20 and 22. Nat Genet. 2006;38:1378–1385. doi: 10.1038/ng1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards CA, Ferguson-Smith AC. Mechanisms regulating imprinted genes in clusters. Curr Opin Cell Biol. 2007;19:281–289. doi: 10.1016/j.ceb.2007.04.013. [DOI] [PubMed] [Google Scholar]
- Ferguson-Smith AC, Surani MA. Imprinting and the epigenetic asymmetry between parental genomes. Science. 2001;293:1086–1089. doi: 10.1126/science.1064020. [DOI] [PubMed] [Google Scholar]
- Geuns E, De Temmerman N, Hilven P, Van Steirteghem A, Liebaers I, De Rycke M. Methylation analysis of the intergenic differentially methylated region of DLK1-GTL2 in human. Eur J Hum Genet. 2007;15:352–361. doi: 10.1038/sj.ejhg.5201759. [DOI] [PubMed] [Google Scholar]
- Hore TA, Rapkins RW, Graves JA. Construction and evolution of imprinted loci in mammals. Trends Genet. 2007;23:440–448. doi: 10.1016/j.tig.2007.07.003. [DOI] [PubMed] [Google Scholar]
- Howlett SK, Reik W. Methylation levels of maternal and paternal genomes during preimplantation development. Development. 1991;113:119–127. doi: 10.1242/dev.113.1.119. [DOI] [PubMed] [Google Scholar]
- Irizarry RA, Ladd-Acosta C, Wen B, Wu Z, Montano C, Onyango P, Cui H, Gabo K, Rongione M, Webster M, et al. The human colon cancer methylome shows similar hypo- and hypermethylation at conserved tissue-specific CpG island shores. Nat Genet. 2009;41:178–186. doi: 10.1038/ng.298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis A, Mitsuya K, Constancia M, Reik W. Tandem repeat hypothesis in imprinting: Deletion of a conserved direct repeat element upstream of H19 has no effect on imprinting in the Igf2-H19 region. Mol Cell Biol. 2004a;24:5650–5656. doi: 10.1128/MCB.24.13.5650-5656.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis A, Mitsuya K, Umlauf D, Smith P, Dean W, Walter J, Higgins M, Feil R, Reik W. Imprinting on distal chromosome 7 in the placenta involves repressive histone methylation independent of DNA methylation. Nat Genet. 2004b;36:1291–1295. doi: 10.1038/ng1468. [DOI] [PubMed] [Google Scholar]
- Lewis A, Green K, Dawson C, Redrup L, Huynh KD, Lee JT, Hemberger M, Reik W. Epigenetic dynamics of the Kcnq1 imprinted domain in the early embryo. Development. 2006;133:4203–4210. doi: 10.1242/dev.02612. [DOI] [PubMed] [Google Scholar]
- Li E, Beard C, Jaenisch R. Role for DNA methylation in genomic imprinting. Nature. 1993;366:362–365. doi: 10.1038/366362a0. [DOI] [PubMed] [Google Scholar]
- Mikkelsen TS, Ku M, Jaffe DB, Issac B, Lieberman E, Giannoukos G, Alvarez P, Brockman W, Kim TK, Koche RP, et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448:553–560. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monk M, Boubelik M, Lehnert S. Temporal and regional changes in DNA methylation in the embryonic, extraembryonic and germ cell lineages during mouse embryo development. Development. 1987;99:371–382. doi: 10.1242/dev.99.3.371. [DOI] [PubMed] [Google Scholar]
- Moore T, Constancia M, Zubair M, Bailleul B, Feil R, Sasaki H, Reik W. Multiple imprinted sense and antisense transcripts, differential methylation and tandem repeats in a putative imprinting control region upstream of mouse Igf2. Proc Natl Acad Sci. 1997;94:12509–12514. doi: 10.1073/pnas.94.23.12509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann B, Kubicka P, Barlow DP. Characteristics of imprinted genes. Nat Genet. 1995;9:12–13. doi: 10.1038/ng0195-12. [DOI] [PubMed] [Google Scholar]
- Oakes CC, La Salle S, Smiraglia DJ, Robaire B, Trasler JM. A unique configuration of genome-wide DNA methylation patterns in the testis. Proc Natl Acad Sci. 2007;104:228–233. doi: 10.1073/pnas.0607521104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Geen H, Nicolet CM, Blahnik K, Green R, Farnham PJ. Comparison of sample preparation methods for ChIP-chip assays. Biotechniques. 2006;41:577–580. doi: 10.2144/000112268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oswald J, Engemann S, Lane N, Mayer W, Olek A, Fundele R, Dean W, Reik W, Walter J. Active demethylation of the paternal genome in the mouse zygote. Curr Biol. 2000;10:475–478. doi: 10.1016/s0960-9822(00)00448-6. [DOI] [PubMed] [Google Scholar]
- Regha K, Sloane MA, Huang R, Pauler FM, Warczok KE, Melikant B, Radolf M, Martens JH, Schotta G, Jenuwein T, et al. Active and repressive chromatin are interspersed without spreading in an imprinted gene cluster in the mammalian genome. Mol Cell. 2007;27:353–366. doi: 10.1016/j.molcel.2007.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reik W, Walter J. Genomic imprinting: Parental influence on the genome. Nat Rev Genet. 2001;2:21–32. doi: 10.1038/35047554. [DOI] [PubMed] [Google Scholar]
- Reinhart B, Paoloni-Giacobino A, Chaillet JR. Specific differentially methylated domain sequences direct the maintenance of methylation at imprinted genes. Mol Cell Biol. 2006;26:8347–8356. doi: 10.1128/MCB.00981-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto A, Liu J, Greene A, Chen M, Weinstein LS. Tissue-specific imprinting of the G protein Gsα is associated with tissue-specific differences in histone methylation. Hum Mol Genet. 2004;13:819–828. doi: 10.1093/hmg/ddh098. [DOI] [PubMed] [Google Scholar]
- Santos F, Hendrich B, Reik W, Dean W. Dynamic reprogramming of DNA methylation in the early mouse embryo. Dev Biol. 2002;241:172–182. doi: 10.1006/dbio.2001.0501. [DOI] [PubMed] [Google Scholar]
- Schilling E, Rehli M. Global, comparative analysis of tissue-specific promoter CpG methylation. Genomics. 2007;90:314–323. doi: 10.1016/j.ygeno.2007.04.011. [DOI] [PubMed] [Google Scholar]
- Shiota K, Kogo Y, Ohgane J, Imamura T, Urano A, Nishino K, Tanaka S, Hattori N. Epigenetic marks by DNA methylation specific to stem, germ and somatic cells in mice. Genes Cells. 2002;7:961–969. doi: 10.1046/j.1365-2443.2002.00574.x. [DOI] [PubMed] [Google Scholar]
- Suzuki M, Sato S, Arai Y, Shinohara T, Tanaka S, Greally JM, Hattori N, Shiota K. A new class of tissue-specifically methylated regions involving entire CpG islands in the mouse. Genes Cells. 2007;12:1305–1314. doi: 10.1111/j.1365-2443.2007.01136.x. [DOI] [PubMed] [Google Scholar]
- Vu TH, Li T, Hoffman AR. Promoter-restricted histone code, not the differentially methylated DNA regions or antisense transcripts, marks the imprinting status of IGF2R in human and mouse. Hum Mol Genet. 2004;13:2233–2245. doi: 10.1093/hmg/ddh244. [DOI] [PubMed] [Google Scholar]
- Wang Z, Zang C, Rosenfeld JA, Schones DE, Barski A, Cuddapah S, Cui K, Roh TY, Peng W, Zhang MQ, et al. Combinatorial patterns of histone acetylations and methylations in the human genome. Nat Genet. 2008;40:897–903. doi: 10.1038/ng.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber M, Davies JJ, Wittig D, Oakeley EJ, Haase M, Lam WL, Schubeler D. Chromosome-wide and promoter-specific analyses identify sites of differential DNA methylation in normal and transformed human cells. Nat Genet. 2005;37:853–862. doi: 10.1038/ng1598. [DOI] [PubMed] [Google Scholar]
- Weber M, Hellmann I, Stadler MB, Ramos L, Paabo S, Rebhan M, Schubeler D. Distribution, silencing potential and evolutionary impact of promoter DNA methylation in the human genome. Nat Genet. 2007;39:457–466. doi: 10.1038/ng1990. [DOI] [PubMed] [Google Scholar]
- Wen B, Wu H, Bjornsson H, Green RD, Irizarry R, Feinberg AP. Overlapping euchromatin/heterochromatin-associated marks are enriched in imprinted gene regions and predict allele-specific modification. Genome Res. 2008;18:1806–1813. doi: 10.1101/gr.067587.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Li T, Vu TH, Ulaner GA, Hu JF, Hoffman AR. The histone code regulating expression of the imprinted mouse Igf2r gene. Endocrinology. 2003;144:5658–5670. doi: 10.1210/en.2003-0798. [DOI] [PubMed] [Google Scholar]