Abstract
Transposable elements (TEs) are ubiquitous genomic parasites. The deleterious consequences of the presence and activity of TEs have fueled debate about the evolutionary forces countering their expansion. Purifying selection is thought to purge TE insertions from the genome, and TE sequences are targeted by hosts for epigenetic silencing. However, the interplay between epigenetic and evolutionary forces countering TE expansion remains unexplored. Here we analyze genomic, epigenetic, and population genetic data from Arabidopsis thaliana to yield three observations. First, gene expression is negatively correlated with the density of methylated TEs. Second, the signature of purifying selection is detectable for methylated TEs near genes but not for unmethylated TEs or for TEs far from genes. Third, TE insertions are distributed by age and methylation status, such that older, methylated TEs are farther from genes. Based on these observations, we present a model in which host silencing of TEs near genes has deleterious effects on neighboring gene expression, resulting in the preferential loss of methylated TEs from gene-rich chromosomal regions. This mechanism implies an evolutionary tradeoff in which the benefit of TE silencing imposes a fitness cost via deleterious effects on the expression of nearby genes.
Transposable elements (TEs) have been found in almost every eukaryotic genome surveyed to date (Wicker et al. 2007). The complement of TEs within any one genome can be quite diverse and typically includes both Class I retroelements and Class II DNA elements. The proliferation of these elements is largely responsible for differences in genome size among eukaryotes (Kidwell 2002). In some cases, particularly in plants, TE proliferation can fuel rapid shifts in genome size. For example, cotton, maize, and a relative of rice have at least doubled in genome size within the last ∼5 million yr (Myr) due to the proliferation of TEs (SanMiguel et al. 1998; Hawkins et al. 2006; Piegu et al. 2006).
Presumably host defenses against TEs counteract genome expansion, and three distinct mechanisms have been proposed to limit TE copy number. New TE insertions are hypothesized to be eliminated by purifying selection due to (1) the deleterious effect of insertion at a specific site (Finnegan 1992; McDonald et al. 1997), (2) ectopic recombination between TEs at different chromosomal positions (Langley et al. 1988), or (3) metabolic costs to the host (Badge and Brookfield 1997). Evolutionary genetic analyses have attempted to discriminate among these mechanisms. To our knowledge, all species studied to date exhibit a dearth of TE insertions within coding genes (Charlesworth and Langley 1989; Bartolome et al. 2002; Rizzon et al. 2003; Wright et al. 2003), signaling either a strong bias against insertion into protein coding regions or, more likely, that the “deleterious insertion” model applies fairly universally to insertions within genes (Yang et al. 2007).
The forces that limit the accumulation of TEs within noncoding regions are less clear, however, and may vary from species to species. In Drosophila melanogaster, for example, the accumulation of TEs is negatively correlated with recombination rate, suggesting that ectopic recombination acts to remove TEs (Bartolome et al. 2002; Rizzon et al. 2002). This view is bolstered by the finding that large TEs are generally found at lower population frequencies (Petrov et al. 2003), which is consistent with the notion that longer TEs are more effective templates for ectopic recombination.
In contrast to D. melanogaster, there is no consistent negative correlation between TE accumulation and recombination in the partial selfers Caenorhabditis elegans or Arabidopsis thaliana (Duret et al. 2000; Wright et al. 2003). In part, this may reflect the theoretical expectation that ectopic recombination among TEs occurs less frequently in inbred species, because ectopic recombination events are more likely in heterozygotes (Montgomery et al. 1991). Instead, in these species the genomic distribution of TEs seems to be governed by a combination of insertion biases, which vary among TE families, and selection against TE insertions near genes (Rizzon et al. 2003; Gaut et al. 2007). However, the mechanism of selection against TEs near genes has not been elucidated fully for any organism and particularly for these species in which ectopic recombination may be infrequent.
Epigenetic pathways also shape the proliferation and accumulation of TE sequences. In many eukaryotic lineages, TEs are targeted for DNA methylation by small RNA (sRNA)–mediated pathways (Almeida and Allshire 2005). In plants, DICER-LIKE RNase enzymes produce 24-bp sRNA that guides ARGONAUTE and other downstream proteins to complementary DNA sequences, thereby promoting and maintaining DNA and histone methylation (Zhang 2008; Teixeira et al. 2009). There is a strong correlation between methylation and sRNA targeting; silenced TE sequences are generally characterized by identity with 24-bp sRNAs and by dense, even DNA methylation (Lippman et al. 2004; Zilberman et al. 2007; Lister et al. 2008).
In the model plant A. thaliana, the overarching effect of TE methylation is to silence transposition (Zhang 2008), as evidenced by dramatically increased levels of TE transcription in met1 methylation mutants (Zilberman et al. 2007; Lister et al. 2008). In addition to preventing proliferation of new TE sequences, silencing of TEs near genes may also prevent the production of aberrant transcripts via read-through transcription beyond TE termini (Barkan and Martienssen 1991). However, methylated sequences may also affect the expression of nearby genes, typically reducing expression (Jahner and Jaenisch 1985; Lippman et al. 2004; Zhang et al. 2008). On occasion, the reduction of gene expression could prove adaptive. For example, Lippman et al. (2004) demonstrated that expression of the flowering time gene FWA is correlated with the methylation status of a nearby SINE-like TE. More generally, however, one might expect that alteration of gene expression due to methylation of nearby TEs may have deleterious effects on gene and genome function.
In this study, we examine the relationship among TE density, TE methylation, and gene expression in A. thaliana, building on the hypothesis that TE methylation may suppress gene expression and thus typically have negative consequences. To test this hypothesis, we focus on three predictions. First, if TE methylation suppresses neighboring gene expression, then gene expression should be a function of the genomic distribution of methylated (silenced) TEs. Therefore, we predict a negative correlation between gene expression and the density of silenced TEs. Second, if suppression of gene expression often has deleterious effects, then population genetic analysis should reveal that purifying selection acts more strongly on methylated TEs that are close to genes. Third, if silenced TEs near genes are often deleterious, they should be preferentially lost from gene-rich regions, resulting in an age distribution in which old, methylated TE insertions are rare or absent from genic regions. To test these predictions, we generated an extensive data set of TE population frequencies throughout the A. thaliana genome, and employed a combination of genomic, epigenetic, and population genetic analyses.
Results
TE genomic distribution and gene expression
We compiled a data set of 5986 TEs from the A. thaliana genome release version 8, including families of both Class I retrotransposons and Class II DNA transposons (Table 1). We employed a BLAST-based culling procedure to ensure that our data set contained only TEs belonging to well-defined, multicopy families (see Methods). Consistent with prior investigations (Wright et al. 2003), 85.7% of the TEs in our data set occupied intergenic regions; i.e., they were not within exons, introns, or untranslated regions (UTRs). To compare the observed distribution of TE insertions to random expectations, we generated 100 replicates of 5986 random insertion sites across the five A. thaliana chromosomes. Across replicates, between 47% and 51% of the simulated insertion sites were within genes compared with ∼15% of observed TE insertions (P < 0.01), suggesting an obvious bias against genic insertions. The underrepresentation of TEs in genes likely reflects strong selection against such insertions (Golding et al. 1986; Charlesworth and Langley 1989; Naito et al. 2006).
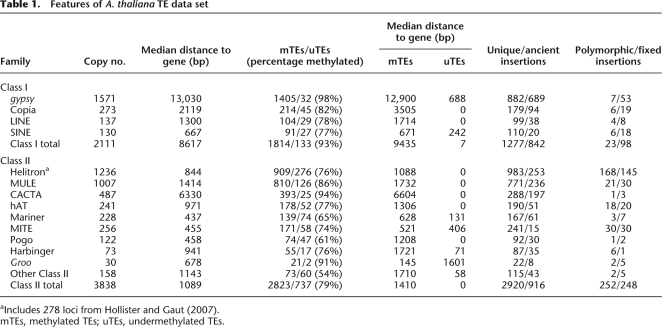
Table 1.
Features of A. thaliana TE data set
aIncludes 278 loci from Hollister and Gaut (2007).
mTEs, methylated TEs; uTEs, undermethylated TEs.
To explore the interaction between TE insertions and host gene expression, we used massively parallel signature sequencing (MPSS) data to obtain mRNA expression profiles of 20,756 A. thaliana genes (Meyers et al. 2004). The nearest neighboring genes to the TE insertions in our data set (n = 4606 genes; see Methods) were expressed at lower levels compared with the genome-wide distribution of gene expression (median expression for nearest gene to a TE 2.00 transcripts per million [TPM] vs. 7.25 TPM genome-wide; Mann-Whitney U test [MWU] P < 10−15). To minimize potential effects of interactions between particular genes and TEs, we calculated the number of TEs in a 10-kb window around the 20,756 genes for which MPSS expression data were available. Genome wide, gene expression was negatively correlated with the number of TEs within 10 kb (Spearman's ρ = −0.23; P < 10−15) (Fig. 1). This trend was also apparent considering only the distal portions of chromosome arms (i.e., >5 Mb from centromeres; Spearman's ρ = −0.11; P < 10−15). Taken together, these results indicate either that TEs do not accumulate around highly expressed genes or that the accumulation of TEs dampens gene expression.
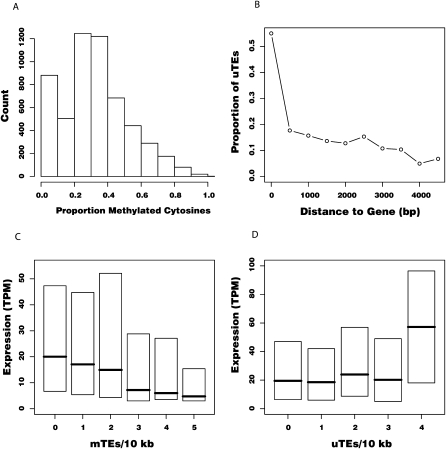
Figure 1.
(A) Histogram showing the proportion of methylated cytosines for n = 4665 TEs, based on bisulfite-treated genomic DNA sequencing (BS-seq) data. Note the large proportion of TEs with <10% methylation, which we deemed as “undermethylated TEs” (uTEs). TEs with >10% methylation are referred to as “methylated TEs” (mTEs). (B) Proportion of uTEs in 500-bp windows of increasing distance from genes. “0 bp” represents TEs within genes. Half of the TEs within genes are unmethylated, but this proportion drops below 20% within 1.5 kb. (C,D) Box plots showing normalized expression for genes as a function of density of mTEs (C) or uTEs (D) within a 10-kb window centered on the gene. Gene expression is measured in transcripts per million (TPQ). Box heights represent inter-quartile distance; horizontal lines, median value for each category.
Methylated TEs are associated with reduced neighboring gene expression
The negative correlation between expression and TE density may be caused by chromatin modifications associated with epigenetic silencing. To address this hypothesis, we used previously published genome-wide data for A. thaliana to assess the level of cytosine methylation of TE sequences in our data set. The data were produced by Illumina sequencing of bisulfite-treated genomic DNA (BS-seq) (Lister et al. 2008). Using these data, we were able to assess the level of methylation of 93% of the TEs in our data set (see Methods). A substantial proportion of TE sequences had low proportions of methylated cytosines (≤10%) (Fig. 1A). Hereafter, we refer to these TEs as undermethylated TEs (uTEs; n = 881), and to TEs with >10% methylation of cytosines as methylated (mTEs; n = 4665). We cross-validated assignment of methylation status using another A. thaliana DNA methylation data set (Zilberman et al. 2007). There was high correspondence between data sets with regard to TE methylation status (see Methods), and we procured qualitatively identical results with both data sets. Hereafter we report results based on the BS-seq data.
There were striking differences in the distribution of mTEs and uTEs. mTEs had a significantly lower proportion of insertions into genes than uTEs (mTEs: 7.8% of 4665, uTEs: 50.6% of 881; Fisher's exact test [FET] P < 10−20). In addition, intergenic mTEs were farther from genes than intergenic uTEs (median distances 3277 and 934 bp, respectively; MWU P < 10−15). To evaluate these differences based on a random expectation, we compared the distribution of mTEs and uTEs to the 100 simulated random TE distributions (see above). Considering intergenic insertions only, mTEs were significantly farther from genes than the random expectation (median distance to neighboring gene of 3277 bp versus 1094–1330 bp for simulated distributions; P < 0.05 for 100 MWU) and uTEs were significantly nearer (median 934 bp; P < 0.05 for 100 MWU). These results suggest that uTEs are preferentially clustered around genes. To further untangle the relationship between TE methylation and proximity to genes, we calculated the proportion of uTEs in 500-bp windows of increasing distance to the nearest gene. Over half of TE insertions within genes were uTEs, but this proportion rapidly decreased until, at a distance of 1.5–2 kb from genes, the proportion of uTEs was similar to the genome-wide proportion (15%) (Fig. 1B).
Notably, genes nearest to mTEs were expressed at lower levels than genes nearest to uTEs (2.00 TPM vs. 8.0 TPM; MWU P < 10−15). We implemented a multiple regression model to assess the independent associations of mTE and uTE density on genome-wide gene expression. In A. thaliana, expression is highly correlated with chromosomal location (Yamada et al. 2003), so we also controlled for the normalized distance between genes and centromeric regions in the linear model. Under this model, the density of mTEs was significantly correlated with reduced gene expression (P < 10−4) (Fig. 1C) while the density of uTEs was not (P = 0.88) (Fig. 1D). These patterns suggest that abundance of mTEs, not simply abundance of TEs, is associated with low expression of nearby genes and that this effect is independent of chromosomal location.
Population frequency of TEs
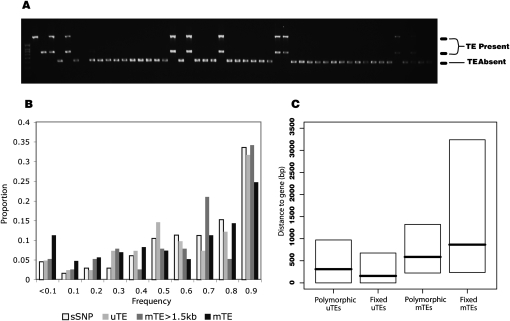
Population genetic analyses are ultimately required to evaluate the strength of selection acting on TEs (Golding et al. 1986; Wright et al. 2001; Petrov et al. 2003; Lockton et al. 2008). We screened a panel of 48 A. thaliana individuals for presence/absence of 621 TE insertions using a PCR-based assay (Fig. 2A), representing the largest data set of TE polymorphisms ever assembled for a plant species. Insertions were surveyed randomly with respect to genomic location. Our assays covered TEs in intergenic locations (n = 530), exons (n = 44), introns (n = 22), and UTR (n = 25). Overall, 44% of the 621 TE insertions were polymorphic in our panel of 48 individuals.
Figure 2.
(A) An example of a gel representing one of 621 TE frequency assays in a panel of 48 individuals. The first three lanes are Col-0 positive controls with combinations of primers; subsequent lanes represent one individual in the panel. Large, paired bands (∼700 and ∼1500 bp) indicate presence of TE in one individual; the smaller, single band (∼400 bp) represents absence of the TE. (B) Comparison of site-frequency spectra (SFS) of sSNPs, unmethylated TEs, methylated TEs >1.5 kb from genes, and all methylated TEs. The SFS of methylated TEs contains an excess of the low-frequency TEs, consistent with a deleterious effect of TE silencing near genes. (C) Box plots showing distance to the nearest gene for polymorphic and fixed uTEs and mTEs.
If silencing of TEs has deleterious effects on the expression of nearby genes, a key prediction is that the site frequency spectrum (SFS) of mTEs near genes should be skewed toward rare variants due to the effects of purifying selection (Williamson et al. 2004). To test this prediction, we divided the TE frequency data set (n = 621) into mTE (n = 527) and uTE (n = 78) subsets. There was a higher percentage of polymorphic insertions in the uTE sample (53%) than the mTE sample (44%), but the excess was not significant (FET P = 0.15).
The presence and direction of selection acting on a class of mutations is commonly inferred by comparing the SFS to a (presumably) neutral class of polymorphisms, typically synonymous single nucleotide polymorphisms (sSNPs). We compared the TE SFS with the SFS for A. thaliana sSNPs, utilizing a genome-wide nucleotide sequence data set consisting of 876 fragments of 500–800 bp sequenced in the same 48 individuals that made up our TE frequency panel (Nordborg et al. 2005). After using information from the 8× draft version of the Arabidopsis lyrata genome to correct for ascertainment biases (see Methods), we tested whether the SFS for polymorphic insertions in either mTEs or uTEs was similar to the sSNP SFS. The SFS of mTEs was significantly skewed toward low frequencies, compared with sSNPs (MWU test P < 10−7) (Fig. 2B). Conversely, the SFS of uTEs was indistinguishable from sSNPs (P = 0.56) (Fig. 2B).
Under the hypothesis proposed here, mTEs are deleterious because of their effect on nearby genes. Therefore, mTEs that are relatively distant from genes should have an SFS similar to sSNPs and uTEs. We noted that the proportion of uTEs decreased sharply until insertions were >1.5 kb from genes, suggesting a convenient demarcation for designation of proximity to genes. Consistent with our hypothesis, polymorphic mTEs >1.5 kb (n = 38) from genes were statistically indistinguishable from sSNPs (MWU P = 0.67) (Fig. 2B). However, considering only polymorphic mTEs <1.5 kb from genes (n = 180), there was a significant skew toward low frequencies compared with sSNPs. The failure to detect a difference in polymorphic mTEs >1.5 kb from genes could be due to the small sample size of only 38 insertions. To investigate statistical power, we generated 10,000 random subsamples of size 38 from polymorphic mTEs <1.5 kb from genes and calculated the number of times a subsample generated a significant MWU relative to the sSNP SFS. Over 90% of the tests showed a significant difference, indicating that sample size alone is unlikely to be responsible for the similarity between the mTEs >1.5 kb from genes and the sSNP data set. These SFS comparisons suggest that purifying selection is detectable predominantly for mTEs near genes.
The age distribution of TE insertions
Our hypothesis further predicts that mTEs are preferentially removed from genic regions over time, due to their deleterious effects on gene expression. To investigate this directly one must have some notion of the age distribution of TE insertions. We made a rough determination of ages of TEs in two ways. We first made use of the polymorphism data set, separating polymorphic from fixed TE insertions. Under neutrality, fixed TE insertions should be older than polymorphic insertions, on average (Kimura 1983). This interpretation is subject to caveats about selection coefficients on TE insertions (see Discussion), but the contrast between fixed and polymorphic insertions should roughly approximate “older” versus “newer” insertions. In our sample of 621 insertions, polymorphic mTEs were significantly closer to genes than fixed mTEs (median distance of 587 bp for polymorphic TEs, 865 bp for mTEs; MWU P < 0.001) (Fig. 2C). In contrast, polymorphic and fixed uTEs were distributed at similar distances from genes (median of 310 bp for polymorphic, 157 bp for fixed; MWU P = 0.5) (Fig. 2C).
As an alternative approach to examining the age distribution of TEs, we determined which insertions were unique to A. thaliana and which were shared (orthologous) between A. thaliana and its closest relative A. lyrata. The shared TEs must have been present in the common ancestor of the two species and thus inserted >5 Myr ago (Mya) (Koch et al. 2000). Hereafter, we refer to these elements as “ancient.” We identified ancient insertions by computational comparison of A. thaliana TEs and flanking regions to the draft A. lyrata genome (see Methods). For a subset of 66 TEs, the computational results were confirmed by comparison to a data set of resequenced TE flanking regions in A. lyrata (DeRose-Wilson and Gaut 2007), with 97% agreement between methods.
Based on this computational approach, we estimated that 30% of 5986 TEs are ancient and presumably inserted >5 Mya. Conversely, 4218 TEs (70%) are present only in the A. thaliana genome, representing “unique” insertions to A. thaliana. These unique TEs are roughly evenly distributed near (≤1.5 kb) and distant (>1.5 kb) from genes. The distribution has shifted over time, however, such that a much higher proportion of ancient TEs are >1.5 kb from genes (FET P < 10−17); as a consequence, ancient TEs are distributed farther from genes than unique TEs (MWU P < 10−15). Ancient and unique TEs can be further discriminated as mTEs or uTEs. Ancient mTEs are farther from genes than unique mTEs (median distance of 6980 vs. 1862 bp; MWU P < 10−15), while ancient uTEs are nearer to genes than unique uTEs (median distance of 142 vs. 0 bp; MWU P < 0.005), due to a higher proportion of ancient uTE insertions into genes (FET P < 0.03). Considering intergenic uTEs only, ancient and recent insertions are similarly distributed (median distance of 988 vs. 851 bp; MWU P = 0.4). Overall, the analysis of ancient (i.e., either fixed or shared) and more recent (i.e., either polymorphic or unique) TEs is consistent with our prediction that mTEs are preferentially removed from gene-rich regions over time.
Using sRNA targeting as a proxy for methylation
Both de novo DNA methylation and restoration of methylation patterns are dependent upon sRNA, which targets homologous sequences for epigenetic modification (Chan et al. 2004; Teixeira et al. 2009). Although it is clear that sRNA targeting and methylation status do not share a one-to-one correspondence (Lister et al. 2008), methylation of TE sequences and presence of sRNA mapping to TEs are highly correlated (Zhang et al. 2006). Therefore, as an independent test of the robustness of the silencing-associated patterns reported above, we repeated all analyses using sRNA mapping to our TE data set as a proxy for methylation.
We quantified sRNA matching the TEs in our data set using the MPSS library of sRNA signatures (Lu et al. 2005). For each TE, we measured sRNA targeting both in the number of sRNA hits per base pair (hbp) and also at the sRNA expression level (abundance; see Methods). We counted both sRNA that were unique matches to a given TE and those that mapped to multiple TEs, because multiply mapping sRNA sequences have been shown to be necessary to explain DNA methylation patterns in the A. thaliana genome (Lister et al. 2008).
Targeting was pervasive among TEs: For the full TE data set, median hbp was 0.003 and mean abundance was 1.6 transcripts per quarter-million (TPQ) per TE. As expected (Zhai et al. 2008), methylated TEs matched significantly more sRNA than unmethylated TEs: Median hbp and abundance for methylated TEs were 0.004 and 2.0 TPQ, respectively, while median hbp and abundance for unmethylated TEs were both 0 (MWU P < 10−15 for both measures).
In contrast to our methylation-based analysis, from which we had to exclude 8% of TEs due to poor coverage of BS-seq data (see Methods), we were able to use 100% of our data set of TEs with this analysis of sRNA targeting. Using several quantitative thresholds for sRNA targeting (see Methods), we reanalyzed our genomic and population genetic data. The simplest threshold labeled TEs matching at least one sRNA as “sRNA+,” and TEs with no matches as “sRNA−” (see Methods). Assuming sRNA targeting is a proxy for epigenetic silencing, we were able to replicate our main results: (1) the density of sRNA+ TEs was correlated negatively with gene expression (P < 2 × 10−14) while the density of sRNA− TEs was not (P = 0.134); (2) only sRNA+ TEs within 1.5 kb of neighboring genes have a frequency spectrum skewed toward low frequency insertions compared with synonymous sites (MWU P < 9 × 10−8) (Supplemental Fig. S1) consistent with purifying selection acting on these insertions; and (3) ancient sRNA+ TEs are distributed farther from genes than the simulated random distribution (permutation test P < 0.01), unique sRNA+ TEs (MWU P < 10−15), and sRNA− TEs (both unique and ancient; MWU P < 10−15) (Supplemental Fig. S2). That we uncover the same basic patterns using sRNA data strongly, and independently, supports the preceding analyses based on BS-seq data.
TE family dynamics
Thus far, we have not carefully considered heterogeneity in characteristics among TE classes and TE families. Although each TE family follows the three general trends we have documented, i.e., gene expression is a function of TE density, mTEs are farther from genes than uTEs (Table 1); and older mTEs are farther from genes—there are differences among them. For example, one longstanding view has been that some Class I TEs tend to insert into gene-poor heterochromatic regions (Pereira 2004), while some Class II TEs insert preferentially into or near genes (Cresse et al. 1995; SanMiguel et al. 1996). Consistent with this view, we also find that Class I TEs are distributed farther from genes than Class II TEs (P < 10−15). In addition, a higher proportion of Class I insertions are methylated (93%) and fixed (81%) compared with Class II elements (79% and 50%, respectively). Overall, these patterns suggest that Class I TEs have been less active recently, on average, in A. thaliana (see Discussion).
There is also considerable heterogeneity among families within the same class. For example, CACTA elements and MULEs, both of which are families of class II DNA transposons, have strikingly different distributions with respect to genes (MWU P < 10−15) (Table 1). Similarly, among Class I families, gypsy-like LTR elements are distributed very far from genes (median distance, 13,030 bp), while LINE elements tend to be much nearer (median, 1300 bp; MWU P < 10−15). Such within-class differences may reflect insertion preferences; MULEs are known to insert preferentially near transcription start sites (Cresse et al. 1995), while gypsy-like elements prefer insertion into heterochromatin (Pereira 2004). However, these patterns may also reflect a combination of the proportion of mTEs (presumably silenced) in a family and purifying selection against mTEs near genes. For example, CACTA and gypsy-like TEs are nearly all methylated (94% and 98%, respectively), while MULE and LINE elements have proportionally fewer methylated insertions (86% and 78%, respectively).
Discussion
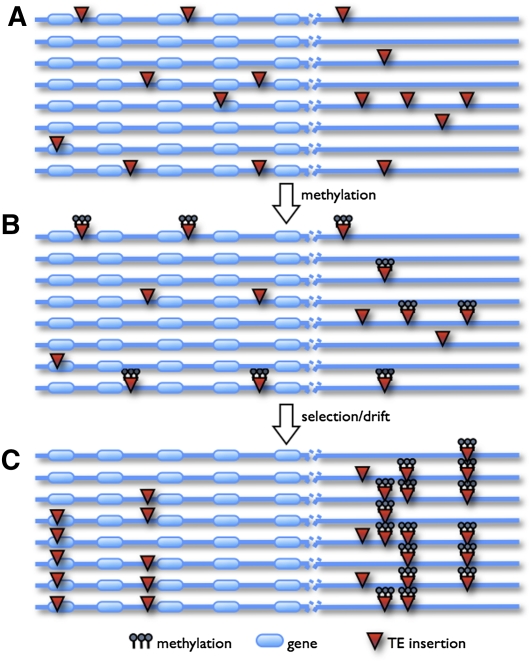
Our investigations of gene expression, DNA methylation, and TE population genetics suggest a model for the removal of TEs from the A. thaliana genome (Fig. 3). It is undoubtedly true that a subset of new insertions interrupts crucial genes and is rapidly removed by strong natural selection (Naito et al. 2006). This strong selection likely accounts for much of the under representation of TE insertions in protein coding genes. Many of the remaining TE insertions are silenced via DNA methylation (Slotkin et al. 2003; Teixeira et al. 2009). In turn, silencing has effects on the expression of nearby genes (Iida et al. 2004; Lippman et al. 2004; Slotkin and Martienssen 2007; Zhang 2008). We hypothesize that these are largely—although not exclusively—deleterious effects, promoting preferential removal of methylated TEs from gene-dense regions over time.
Figure 3.
Model of the effect of silencing on the distribution of a TE family. Horizontal lines represent chromosomes in a population sample of eight individuals and are divided by dashed lines into regions of high (left) and low (right) gene density. (A) New TE insertions are distributed randomly along chromosomes. (B) Some insertions become silenced by DNA methylation. mTEs near genes interfere with gene expression, and are removed by purifying selection, while the evolutionary trajectory of other TEs are governed largely by genetic drift. (C) Eventually these processes result in a TE distribution in which most insertions are methylated and sequestered in regions of low gene density.
Four observations are consistent with this model: (1) gene expression is negatively correlated with the density of methylated, but not unmethylated, TEs (Fig. 1C,D); (2) the signature of purifying selection, a skewed SFS relative to sSNPs, was detectable only for mTEs within 1.5 kb of the nearest gene (Fig. 2B); (3) mTEs are farther from genes than uTEs, even on a per family basis (Table 1; Fig. 1B); and (4) the distribution of mTEs, but not uTEs, migrates away from genes over time (Fig. 2C). Among Class II elements, ancient families, such as CACTA-like elements, tend to be farther from genes (P < 10−15), while more recently active families, such as MITEs and Helitrons, tend to be closer to genes (P < 4 × 10−11 and P < 10−15, respectively). The net result of this model of selection against TEs is the now familiar distribution of TEs concentrated in gene-poor regions (Charlesworth et al. 1994; Wright et al. 2003).
We propose this model while recognizing at least three caveats to our analysis. First, we cannot discriminate cause from effect. For example, we find an association between gene expression and the density of mTEs but do not know whether TEs tend to preferentially insert near lowly expressed genes or whether the insertion of TEs causes low expression. While both scenarios may be true to some degree, our model relies on the latter interpretation, which is reasonable in light of substantial molecular evidence that methylation of nearby sequences lowers gene expression (Jacobsen and Meyerowitz 1997; Lippman et al. 2004; Zilberman et al. 2007; Zhang et al. 2008). Moreover, our interpretation of cause and effect is more consistent with our population genetic evidence that suggests stronger selection against mTEs near genes. Experimental confirmation of the effect of TE silencing on gene expression could be obtained by studies of allele-specific expression in F1 progeny of parents that differ in the presence of methylated TE insertions near genes.
Second, our study is limited by the fact that the A. thaliana methylome has been fully characterized in only the Col-0 accession to date. If there is substantial variation in methylation status of individual TEs among accessions, we may incorrectly have assigned the predominant (population-wide) methylation status. However, a recent examination of methylation patterns on chromosome 4 of the Col-0 and Landsberg erecta accessions revealed that fewer than 10% of TEs differed in methylation between accessions (Vaughn et al. 2007), indicating a high degree of stability of TE methylation patterns. Also note that all singleton insertions identified in this study, which contribute predominantly to the inference of purifying selection, are present only in Col-0. The methylation status for these singletons is known.
Third, we cannot be certain to what extent differences in insertion bias among TE families affect the genomic patterns we report. Our analysis suggests that differences in the genomic distribution and activity of Class I and Class II TEs, as well as among TE families, may be a function of the proportion of methylated elements. Overall, TE families that have a higher proportion of methylated insertions are distributed farther from genes (Table 1). This suggests a causal relationship between TE methylation and genomic distribution, but we cannot be certain to what extent insertion biases, transposition rate, and selection against methylated TEs near genes influence this relationship. It is likely that all these factors interact to determine the genomic distribution of TEs, but further study will be necessary to untangle the interactions between these factors.
Finally, our age analyses are limited by our assumptions about the relative ages of polymorphic versus fixed TEs and of unique versus ancient TEs, respectively. If a substantial proportion of fixed TE insertions were driven to fixation by positive selection (or if many polymorphic insertions have been subject to balancing selection), it is plausible that our assumptions regarding the relationship between population frequency and age are incorrect. There is also uncertainty associated with our analysis of TEs shared between A. thaliana and A. lyrata. We cannot be sure whether TEs that are unique to the A. thaliana genome have inserted subsequent to divergence from A. lyrata or if they were lost (or unrecognizable) in the sequenced A. lyrata strain. Importantly, both analyses of age-dependent TE distribution reveal the same pattern, so our conclusion that the distribution of methylated TEs only migrates away from genes over time appears to be robust.
TE methylation: An evolutionary trade-off
Despite caveats, our analysis is consistent with the hypothesis that silencing of TEs may increase the deleterious effect of insertions near genes. This implies an evolutionary tradeoff between decreased activity of TEs and increased deleterious effects of some insertions on nearby genes. At least two lines of evidence support the plausibility of this tradeoff by highlighting the catastrophic effect of unfettered transposition. First, P-element activity in Drosophila is associated with sterility and lethality (Kidwell 1985). Second, transposition rates estimated from TEs in natural populations (nearly neutral rates, presumably) are one to two orders of magnitude below rates estimated from mutation accumulation studies (where purifying selection is relaxed), indicating that most new TE insertions are subject to strong purifying selection (Nuzhdin and Mackay 1995; Maside et al. 2001; Bergman and Bensasson 2007). These findings indicate that the benefit of silencing TE families may often outweigh the deleterious effect of silenced TEs on nearby genes.
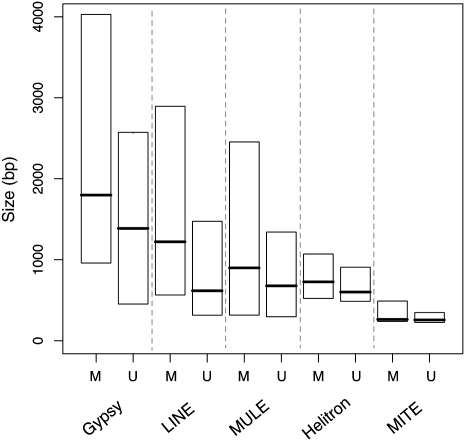
The evolutionary tradeoff of silencing we propose provides new explanations for some well-known patterns of TE distribution. For example, longer TE sequences show evidence of being subject to stronger purifying selection than short TE sequences, and this observation has been interpreted as evidence for the ectopic recombination model (Petrov et al. 2003; Hollister and Gaut 2007). However, it is possible that longer TEs may be more deleterious not because of more frequent ectopic recombination but rather because they are methylated more often. Consistent with this conjecture, methylated TEs were significantly larger than their unmethylated counterparts in most families of both Class I and II TEs (Fig. 4). Even among methylated TEs, longer TEs represent larger regions of modified chromatin, which could have a greater impact on expression of surrounding genes.
Figure 4.
Box plot showing sizes of methylated and unmethylated TEs from five representative families. Horizontal lines display medians; box heights, interquartile range. M, methylated insertions; U, unmethylated insertions. Unmethylated elements are significantly smaller than methylated elements in each family (MWU P < 0.01).
These observations provide an additional reason for the success of small nonautonomous TE families. It has already been suggested that nonautonomous TEs may attain high copy-number by recruiting the transposition machinery of distantly related TE families, thereby remaining active even after their full-length progenitor elements have been silenced (Feschotte et al. 2003). Their small size may also enable nonautonomous TEs to avoid silencing. In Neurospora crassa, sequences <300 bp are not efficiently methylated (Lewis et al. 2009). If a similar size limit exists in plants, it helps explain the abundance of nonautonomous TEs, such as MITEs, SINEs, and nonautonomous Helitrons, in diverse plant lineages (Feschotte et al. 2003; Hedges et al. 2004). In support of these ideas, MITEs, SINEs, and Helitrons have higher proportions of unmethylated insertions than most other TE families (FET P < 0.004, P < 10−4, P < 10−18, respectively).
One interesting product of our analysis is the high proportion of uTEs that are located within genes (50.5%) (Table 1). An important feature of our model is that methylation of some subset of TEs is deleterious to organismal function. It follows that natural selection could prevent the methylation (or, alternatively, promote demethylation) of a small subset of TEs that are located near critical genes. A recent study of genome-wide patterns of DNA methylation in A. thaliana supports this possibility (Lister et al. 2008). This study revealed a subset of TE insertions near genes that are unmethylated in the wild-type Col-0 background but are hypermethylated in the DNA demethylase triple mutant rdd. This finding shows that these TEs are actively demethylated in wild-type plants, allowing transcription of transposase enzymes (Lister et al. 2008). DNA demethylation is thought to ameliorate deleterious methylation events (Penterman et al. 2007), so the observation that a small subset of TEs near genes is actively demethylated is consistent with the notion that methylation of such TEs can be deleterious, supporting the evolutionary trade-off we propose.
Remaining questions and broader implications
At least three crucial questions remain. First, how does the methylation of TEs in noncoding regions affect gene expression? Thus far, most experimental examples of mTEs that influence gene expression are quite close to genes. For example, Lippman et al. (2004) showed TE–gene coregulation only when TEs were inserted into or within a few hundred bp of genes. Here we seem to uncover effects that are more general and perhaps extend to 1.5 kb, based on genomic correlations (Fig. 1B). One possibility is that methylation extends from distant mTEs into flanking regions, thus affecting the chromatin conformation of promoter regions. Extension of methylated regions has been observed in both mice and plants (Jahner and Jaenisch 1985; Arnaud et al. 2000), but de novo methylation and remethylation appear to be targeted accurately in A. thaliana (Teixeira et al. 2009). It thus seems unlikely that extension of methylation from TEs to genes could cause the effects on gene expression predicted by our model. A second possibility is that the methylated regions are targets for RNA polymerase IV (Pol IV) complexes, which interferes with regular gene transcription by Pol II (Erhard et al. 2009). To our knowledge, there is no information about the physical extent over which Pol IV interference could occur, however. Finally, TE silencing is associated with changes in heterochromatin formation (Lippman et al. 2004), which might have a decreasing effect on gene expression with distance. In this last case, the effect of distant mTEs on gene expression could be very subtle and perhaps only observable with evolutionary analyses such as those described here, which integrate over both genomic location and time.
Second, is this a general phenomenon, or one that is limited to A. thaliana? The highly selfing nature of A. thaliana may have several effects that are pertinent to the model we propose. On the one hand, selfing lowers the effective population size, which increases the proportion of “nearly neutral” alleles that might be purged from populations with greater effective size (Charlesworth and Wright 2001). All other things being equal, selfing should allow TEs to attain higher copy numbers due to decreased efficacy of purifying selection. On the other hand, selfing greatly decreases heterozygosity, which “unmasks” deleterious recessive alleles (Barrett and Charlesworth 1991). This process may increase purifying selection on TEs near genes, leading to more efficient purging of TEs from the genome. A. thaliana has a small, streamlined genome and small TE complement compared with closely related species, indicating that the latter feature of a selfing habit may be important in the stabilization of TE copy number. Because of this, other species with higher TE copy numbers may show a proportionally greater effect of TE silencing on the epigenetic regulation of genes, as has been suggested in a recent review (Zhang 2008). Additional studies in plant and animal species with larger genomes (and TE copy numbers) will likely provide ample tests of the model we propose.
Finally, what is the general picture of selection on TEs? Some insertions may be adaptive; the frequency of such events has not been well established. Some insertions, particularly those far from genes and also those that are not methylated, are probably neutral, or nearly so. However, most insertions are likely deleterious, for various reasons. Some may have strongly deleterious consequences by interrupting gene function directly. Others may affect gene expression after methylation, as our results suggest, representing a tradeoff between the costs and benefits of silencing. In this scenario, the cost of silencing is a function of the size of the TE, its distance from neighboring genes, and the function and/or expression of the neighboring genes. The benefit of silencing is reduction in TE proliferation with a lower probability of new, highly deleterious insertions, and perhaps reduced metabolic costs to the host.
Methods
Annotation of A . thaliana TEs
The annotated A. thaliana genome sequence version 8 was downloaded from TAIR (http://www.arabidopsis.org). TE sequences were annotated using RepeatMasker, with a query set of sequences from Repbase. We performed a BLAST-based culling analysis of RepeatMasker output in which putative TE sequences were removed from the database if they did not match at least one terminal sequence of the most complete TE sequence (E-value cutoff, 10−5). We did this to prevent non-TE sequences matching sequence fragments captured by Helitrons, MULEs, or other TEs from being included in our data set. Our data set of n = 5986 TEs is greater than that of a previous genome-wide analysis of TEs (n = 5069) (Wright et al. 2003).
We compared the position of TE sequences to the TAIR 8 gene annotations, excluding pseudogenes and TE-associated genes. The nearest gene adjacent to each TE insertion, and the distance from gene to TE, was recorded. When multiple TEs shared the same “nearest gene,” the gene was annotated as nearest to each TE, but was only considered once in statistical analyses (i.e., sample size was n = 5986 for TEs, n = 4606 for nearest neighboring genes). TEs whose positions were within, or overlapped with, genes were used as a BLAST query (E-value = 10−10) against a separate TAIR 8 gene sequence database to confirm their position. TEs that overlapped with exons were also used as a BLAST query against the TAIR 8 full-length cDNA database. TEs that overlapped with the exon annotations but did not match cDNA sequences were annotated as intronic or intergenic based on the correspondence of the TE position with the cDNA sequence and gene annotation.
Gene expression, methylation, and sRNA analysis
We obtained a library of 17-bp signatures and their abundances from the MPSS Gene Expression database (Meyers et al. 2004). Gene expression was quantified as the average normalized abundance of signatures uniquely matching a single gene from the TAIR 8 gene annotations. We also measured gene expression as the maximum abundance of a signature uniquely matching a gene, and obtained qualitatively similar results.
BS-seq data were downloaded from the NCBI Short Read Archive (SRA000284). These data consisted of 39,113,599 approximately 36-bp reads produced by Illumina sequencing of bisulfite-treated genomic DNA from inflorescence tissue and mapped to the A. thaliana reference genome (Lister et al. 2008). We determined the percentage of methylated cytosines relative to genomic DNA for each read that overlapped with a TE sequence, and averaged over reads to determine the percentage of methylated cytosines for each TE. Using this method, individual cytosines that had conflicting evidence of methylation among reads were given less weight in the calculation of overall methylation levels. We considered the methylation status of a TE “ambiguous” if <50% of its length was covered by one or more reads. Using this cutoff, we were able to determine the percentage of methylated cytosines for 93% of our TE data set (n = 5546). TEs with <10% methylated cytosines were considered uTEs.
To independently verify the BS-seq data, tiling array DNA methylation data were obtained from the NCBI Gene Expression Omnibus (accession no. GSE5974). We assigned methylation status to TEs following the method described by Zilberman et al. (2007). Ninety-five percent of TEs labeled as mTEs (≥10% methylated cytosines) were also methylated in the tiling array data, and 80% of TEs labeled uTEs were unmethylated in the tiling array. We expected a higher proportion of uTEs to show evidence of methylation on the tiling arrays due to cross hybridization, because most TEs are methylated. We were unable to confirm the methylation status of 18% of the TEs with tiling array data due to lack of coverage on the array. Because the tiling array data set was used for quality-control purposes and the correspondence between the two data sets was so high, we included all TEs with adequate coverage in the BS-seq data set in our analysis (n = 5546 or 93%). However, all results were fully reproducible using only the tiling array data to determine methylation of TEs (data not shown).
sRNA data were downloaded from the Arabidopsis MPSS Plus website (http://mpss.udel.edu/at/) (Lu et al. 2005). These data include the sequenced 17-bp sRNA signatures generated by MPSS and full-length approximately 24-bp sRNA generated by 454 Life Sciences (Roche) sequencing, as well as the normalized abundances of all sRNA in number of TPQ in both seedling and inflorescence tissues. We analyzed the FLR inflorescence and SDC seedling libraries because they had the highest read counts of any available sRNA library.
We assigned sRNA sequences to TEs based on a perfect match between the sRNA and the TE. sRNA signatures that had more than one matching sequence on a particular TE (i.e., the sRNA motif was repeated) were counted only once. Overlapping sRNA hits were counted separately. To obtain a measure of sRNA targeting density independent of the size of TEs, we divided the total number of sRNA signatures matching a TE by the length of the TE, in bp, to calculate the hbp. To quantify the abundance of sRNAs matching TEs, we took the average TPQ for all sRNA signatures matching a TE. We tried three cutoffs for labeling a TE sRNA+, arranged in order of stringency: (1) presence of at least one sRNA signature that matched the TE, (2) presence of at least one sRNA signature at abundance higher than one TPQ that matched the TE, or (3) a mean TPQ higher than one for all sRNA matching the TE. We used cutoff 1 for the reported analysis, but our results were qualitatively consistent using any of the cutoffs (data not shown).
Population genetic data
TEs were screened for presence/absence in a panel of 48 A. thaliana individuals, as described by Hollister and Gaut (2007). In addition to the 278 insertion frequencies previously published, we screened another 343 insertions for the present work (Supplemental Table S1). We downloaded alignments of 876 sequenced fragments in 96 A. thaliana accessions, and divided each into the subset of 48 individuals that made up our TE population frequency screen panel. For each of these alignments, we identified the A. lyrata orthologous sequence by reciprocal BLASTN, using the 8× version of the A. lyrata genome (http://genome.jgi-psf.org/Araly1/Araly1.download.ftp.html; D. Weigel, pers. comm.). The A. lyrata outgroup sequence was aligned using the Profile Alignment option in ClustalW. We continued analyses only on the subset of alignments in which the A. lyrata sequence and the A. thaliana alignment shared >90% identity over >80% of the alignment length.
Because we assayed the frequency of TEs first identified in the Col-0 sequenced genome, our TE polymorphism data have a strong ascertainment bias that ultimately results in underrepresentation of low-frequency TE insertions. We created similar biases in the SNP data set by including only derived sSNPs in Col-0, based on comparison to the A. lyrata outgroup. We compared this corrected sSNP data to the TE frequency data of unique—i.e., derived—TE insertions. The ascertained SNP data set contains 1134 synonymous sites. Polymorphism information was extracted from sequence alignments using the Libsequence evolutionary genetics software package (Thornton 2003).
Identification of ancient and unique TEs
Each TE and 2 kb of flanking sequence at its 3′ an 5′ ends was extracted from the A. thaliana genome. These sequences were used as a BLAST query against the 8× draft A. lyrata genome sequence (E-value cutoff, 10−10). A TE insertion was considered ancient if it fulfilled one or both of two complementary conditions (Supplemental Fig. S3). The first condition required a continuous stretch of A. lyrata sequence matching the A. thaliana sequence from 50 bp flanking the sequence to 50 bp inside the TE sequence. The second criterion attempted to control for divergence between shared insertions. We found the best A. lyrata matches to the flanking sequences nearest to both ends of the TE in A. thaliana. We then subtracted the distance from the end (for the 5′ match) or the start (for the 3′ match) of the matches to the beginning (5′) or end (3′) of the TE in A. thaliana. If the distance between the A. lyrata matches to the flanks, minus the distance from the matches to the TE in A. thaliana, was greater than half the TE length but less than two times its length, we considered the TE ancient.
Acknowledgments
We thank R. Gaut for laboratory work. S. Lockton, J. Ross-Ibarra, and L. DeRose-Wilson provided helpful comments and discussion. The comments of three anonymous reviewers greatly enhanced the presentation. We thank the Joint Genome Institute and the Arabidopsis lyrata sequencing consortium for the use of unpublished data. This work was supported by National Science Foundation Grant DEB-0426166 and DEB-0723860 (to B.S.G.).
Footnotes
[Supplemental material is available online at www.genome.org.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.091678.109.
References
- Almeida R, Allshire RC. RNA silencing and genome regulation. Trends Cell Biol. 2005;15:251–258. doi: 10.1016/j.tcb.2005.03.006. [DOI] [PubMed] [Google Scholar]
- Arnaud P, Goubely C, Pelissier T, Deragon JM. SINE retroposons can be used in vivo as nucleation centers for de novo methylation. Mol Cell Biol. 2000;20:3434–3441. doi: 10.1128/mcb.20.10.3434-3441.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badge RM, Brookfield JF. The role of host factors in the population dynamics of selfish transposable elements. J Theor Biol. 1997;187:261–271. doi: 10.1006/jtbi.1997.0432. [DOI] [PubMed] [Google Scholar]
- Barkan A, Martienssen RA. Inactivation of maize transposon Mu suppresses a mutant phenotype by activating an outward-reading promoter near the end of Mu1. Proc Natl Acad Sci. 1991;88:3502–3506. doi: 10.1073/pnas.88.8.3502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett SC, Charlesworth D. Effects of a change in the level of inbreeding on the genetic load. Nature. 1991;352:522–524. doi: 10.1038/352522a0. [DOI] [PubMed] [Google Scholar]
- Bartolome C, Maside X, Charlesworth B. On the abundance and distribution of transposable elements in the genome of Drosophila melanogaster. Mol Biol Evol. 2002;19:926–937. doi: 10.1093/oxfordjournals.molbev.a004150. [DOI] [PubMed] [Google Scholar]
- Bergman CM, Bensasson D. Recent LTR retrotransposon insertion contrasts with waves of non-LTR insertion since speciation in Drosophila melanogaster. Proc Natl Acad Sci. 2007;104:11340–11345. doi: 10.1073/pnas.0702552104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan SW, Zilberman D, Xie Z, Johansen LK, Carrington JC, Jacobsen SE. RNA silencing genes control de novo DNA methylation. Science. 2004;303:1336. doi: 10.1126/science.1095989. [DOI] [PubMed] [Google Scholar]
- Charlesworth B, Langley CH. The population genetics of Drosophila transposable elements. Annu Rev Genet. 1989;23:251–287. doi: 10.1146/annurev.ge.23.120189.001343. [DOI] [PubMed] [Google Scholar]
- Charlesworth D, Wright SI. Breeding systems and genome evolution. Curr Opin Genet Dev. 2001;11:685–690. doi: 10.1016/s0959-437x(00)00254-9. [DOI] [PubMed] [Google Scholar]
- Charlesworth B, Jarne P, Assimacopoulos S. The distribution of transposable elements within and between chromosomes in a population of Drosophila melanogaster. III. Element abundances in heterochromatin. Genet Res. 1994;64:183–197. doi: 10.1017/s0016672300032845. [DOI] [PubMed] [Google Scholar]
- Cresse AD, Hulbert SH, Brown WE, Lucas JR, Bennetzen JL. Mu1-related transposable elements of maize preferentially insert into low copy number DNA. Genetics. 1995;140:315–324. doi: 10.1093/genetics/140.1.315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeRose-Wilson LJ, Gaut BS. Transcription-related mutations and GC content drive variation in nucleotide substitution rates across the genomes of Arabidopsis thaliana and Arabidopsis lyrata. BMC Evol Biol. 2007;7:66. doi: 10.1186/1471-2148-7-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duret L, Marais G, Biemont C. Transposons but not retrotransposons are located preferentially in regions of high recombination rate in Caenorhabditis elegans. Genetics. 2000;156:1661–1669. doi: 10.1093/genetics/156.4.1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erhard KFJ, Stonaker JL, Parkinson SE, Lim JP, Hale CJ, Hollick JB. RNA polymerase IV functions in paramutation in Zea mays. Science. 2009;323:1201–1205. doi: 10.1126/science.1164508. [DOI] [PubMed] [Google Scholar]
- Feschotte C, Swamy L, Wessler SR. Genome-wide analysis of mariner-like transposable elements in rice reveals complex relationships with stowaway miniature inverted repeat transposable elements (MITEs) Genetics. 2003;163:747–758. doi: 10.1093/genetics/163.2.747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finnegan DJ. Transposable elements. The genome of Drosophila melanogaster. In: Lindsey DL, Zimm G, editors. Academic Press; New York: 1992. pp. 1096–1107. [Google Scholar]
- Gaut BS, Wright SI, Rizzon C, Dvorak J, Anderson LK. Recombination: An underappreciated factor in the evolution of plant genomes. Nat Rev Genet. 2007;8:77–84. doi: 10.1038/nrg1970. [DOI] [PubMed] [Google Scholar]
- Golding GB, Aquadro CF, Langley CH. Sequence evolution within populations under multiple types of mutation. Proc Natl Acad Sci. 1986;83:427–431. doi: 10.1073/pnas.83.2.427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawkins JS, Kim H, Nason JD, Wing RA, Wendel JF. Differential lineage-specific amplification of transposable elements is responsible for genome size variation in Gossypium. Genome Res. 2006;16:1252–1261. doi: 10.1101/gr.5282906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedges DJ, Callinan PA, Cordaux R, Xing J, Barnes E, Batzer MA. Differential Alu mobilization and polymorphism among the human and chimpanzee lineages. Genome Res. 2004;14:1068–1075. doi: 10.1101/gr.2530404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollister JD, Gaut BS. Population and evolutionary dynamics of Helitron transposable elements in Arabidopsis thaliana. Mol Biol Evol. 2007;24:2515–2524. doi: 10.1093/molbev/msm197. [DOI] [PubMed] [Google Scholar]
- Iida S, Morita Y, Choi JD, Park KI, Hoshino A. Genetics and epigenetics in flower pigmentation associated with transposable elements in morning glories. Adv Biophys. 2004;38:141–159. [PubMed] [Google Scholar]
- Jacobsen SE, Meyerowitz EM. Hypermethylated SUPERMAN epigenetic alleles in Arabidopsis. Science. 1997;277:1100–1103. doi: 10.1126/science.277.5329.1100. [DOI] [PubMed] [Google Scholar]
- Jahner D, Jaenisch R. Retrovirus-induced de novo methylation of flanking host sequences correlates with gene inactivity. Nature. 1985;315:594–597. doi: 10.1038/315594a0. [DOI] [PubMed] [Google Scholar]
- Kidwell MG. Hybrid dysgenesis in Drosophila melanogaster: Nature and inheritance of P element regulation. Genetics. 1985;111:337–350. doi: 10.1093/genetics/111.2.337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidwell MG. Transposable elements and the evolution of genome size in eukaryotes. Genetica. 2002;115:49–63. doi: 10.1023/a:1016072014259. [DOI] [PubMed] [Google Scholar]
- Kimura M. The neutral theory of molecular evolution. Cambridge University Press; Cambridge: 1983. [Google Scholar]
- Koch MA, Haubold B, Mitchell-Olds T. Comparative evolutionary analysis of chalcone synthase and alcohol dehydrogenase loci in Arabidopsis, Arabis, and related genera (Brassicaceae) Mol Biol Evol. 2000;17:1483–1498. doi: 10.1093/oxfordjournals.molbev.a026248. [DOI] [PubMed] [Google Scholar]
- Langley CH, Montgomery E, Hudson R, Kaplan N, Charlesworth B. On the role of unequal exchange in the containment of transposable element copy number. Genet Res. 1988;52:223–235. doi: 10.1017/s0016672300027695. [DOI] [PubMed] [Google Scholar]
- Lewis ZA, Honda S, Khlafallah TK, Jeffress JK, Freitag M, Mohn F, Schubeler D, Selker EU. Relics of repeat-induced point mutation direct heterochromatin formation in Neurospora crassa. Genome Res. 2009;19:427–437. doi: 10.1101/gr.086231.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippman Z, Gendrel AV, Black M, Vaughn MW, Dedhia N, McCombie WR, Lavine K, Mittal V, May B, Kasschau KD, et al. Role of transposable elements in heterochromatin and epigenetic control. Nature. 2004;430:471–476. doi: 10.1038/nature02651. [DOI] [PubMed] [Google Scholar]
- Lister R, O'Malley RC, Tonti-Filippini J, Gregory BD, Berry CC, Millar AH, Ecker JR. Highly integrated single-base resolution maps of the epigenome in Arabidopsis. Cell. 2008;133:523–536. doi: 10.1016/j.cell.2008.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lockton S, Ross-Ibarra J, Gaut BS. Demography and weak selection drive patterns of transposable element diversity in natural populations of Arabidopsis lyrata. Proc Natl Acad Sci. 2008;105:13965–13970. doi: 10.1073/pnas.0804671105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu C, Tej SS, Luo S, Haudenschild CD, Meyers BC, Green PJ. Elucidation of the small RNA component of the transcriptome. Science. 2005;309:1567–1569. doi: 10.1126/science.1114112. [DOI] [PubMed] [Google Scholar]
- Maside X, Bartolome C, Assimacopoulos S, Charlesworth B. Rates of movement and distribution of transposable elements in Drosophila melanogaster: In situ hybridization vs Southern blotting data. Genet Res. 2001;78:121–136. doi: 10.1017/s0016672301005201. [DOI] [PubMed] [Google Scholar]
- McDonald JF, Matyunina LV, Wilson S, Jordan IK, Bowen NJ, Miller W. LTR retrotransposons and the evolution of eukaryotic enhancers. Genetica. 1997;100:111–115. [PubMed] [Google Scholar]
- Meyers BC, Tej SS, Vu TH, Haudenschild CD, Agrawal V, Edberg SB, Ghazal H, Decola S. The use of MPSS for whole-genome transcriptional analysis in Arabidopsis. Genome Res. 2004;14:1641–1653. doi: 10.1101/gr.2275604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montgomery EA, Huang SM, Langley CH, Judd BH. Chromosome rearrangement by ectopic recombination in Drosophila melanogaster: Genome structure and evolution. Genetics. 1991;129:1085–1098. doi: 10.1093/genetics/129.4.1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naito K, Cho E, Yang G, Campbell MA, Yano K, Okumoto Y, Tanisaka T, Wessler SR. Dramatic amplification of a rice transposable element during recent domestication. Proc Natl Acad Sci. 2006;103:17620–17625. doi: 10.1073/pnas.0605421103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordborg M, Hu TT, Ishino Y, Jhaveri J, Toomajian C, Zheng H, Bakker E, Calabrese P, Gladstone J, Goyal R, et al. The pattern of polymorphism in Arabidopsis thaliana. PLoS Biol. 2005;3:e196. doi: 10.1371/journal.pbio.0030196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nuzhdin SV, Mackay TF. The genomic rate of transposable element movement in Drosophila melanogaster. Mol Biol Evol. 1995;12:180–181. doi: 10.1093/oxfordjournals.molbev.a040188. [DOI] [PubMed] [Google Scholar]
- Penterman J, Zilberman D, Huh JH, Ballinger T, Henikoff S, Fischer RL. DNA demethylation in the Arabidopsis genome. Proc Natl Acad Sci. 2007;104:6752–6757. doi: 10.1073/pnas.0701861104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira V. Insertion bias and purifying selection of retrotransposons in the Arabidopsis thaliana genome. Genome Biol. 2004;5:R79. doi: 10.1186/gb-2004-5-10-r79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrov DA, Aminetzach YT, Davis JC, Bensasson D, Hirsh AE. Size matters: Non-LTR retrotransposable elements and ectopic recombination in Drosophila. Mol Biol Evol. 2003;20:880–892. doi: 10.1093/molbev/msg102. [DOI] [PubMed] [Google Scholar]
- Piegu B, Guyot R, Picault N, Roulin A, Saniyal A, Kim H, Collura K, Brar DS, Jackson S, Wing RA, et al. Doubling genome size without polyploidization: Dynamics of retrotransposition-driven genomic expansions in Oryza australiensis, a wild relative of rice. Genome Res. 2006;16:1262–1269. doi: 10.1101/gr.5290206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizzon C, Marais G, Gouy M, Biemont C. Recombination rate and the distribution of transposable elements in the Drosophila melanogaster genome. Genome Res. 2002;12:400–407. doi: 10.1101/gr.210802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizzon C, Martin E, Marais G, Duret L, Segalat L, Biemont C. Patterns of selection against transposons inferred from the distribution of Tc1, Tc3 and Tc5 insertions in the mut-7 line of the nematode Caenorhabditis elegans. Genetics. 2003;165:1127–1135. doi: 10.1093/genetics/165.3.1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SanMiguel P, Tikhonov A, Jin YK, Motchoulskaia N, Zakharov D, Melake-Berhan A, Springer PS, Edwards KJ, Lee M, Avramova Z, et al. Nested retrotransposons in the intergenic regions of the maize genome. Science. 1996;274:765–768. doi: 10.1126/science.274.5288.765. [DOI] [PubMed] [Google Scholar]
- SanMiguel P, Gaut BS, Tikhonov A, Nakajima Y, Bennetzen JL. The paleontology of intergene retrotransposons of maize. Nat Genet. 1998;20:43–45. doi: 10.1038/1695. [DOI] [PubMed] [Google Scholar]
- Slotkin RK, Martienssen R. Transposable elements and the epigenetic regulation of the genome. Nat Rev Genet. 2007;8:272–285. doi: 10.1038/nrg2072. [DOI] [PubMed] [Google Scholar]
- Slotkin RK, Freeling M, Lisch D. Mu killer causes the heritable inactivation of the Mutator family of transposable elements in Zea mays. Genetics. 2003;165:781–797. doi: 10.1093/genetics/165.2.781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teixeira FK, Heredia F, Sarazin A, Roudier F, Boccara M, Ciaudo C, Cruaud C, Poulain J, Berdasco M, Fraga MF, et al. A role for RNAi in the selective correction of DNA methylation defects. Science. 2009;323:1600–1604. doi: 10.1126/science.1165313. [DOI] [PubMed] [Google Scholar]
- Thornton K. Libsequence: A C++ class library for evolutionary genetic analysis. Bioinformatics. 2003;19:2325–2327. doi: 10.1093/bioinformatics/btg316. [DOI] [PubMed] [Google Scholar]
- Vaughn MW, Tanurd Ic M, Lippman Z, Jiang H, Carrasquillo R, Rabinowicz PD, Dedhia N, McCombie WR, Agier N, Bulski A, et al. Epigenetic natural variation in Arabidopsis thaliana. PLoS Biol. 2007;5:e174. doi: 10.1371/journal.pbio.0050174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wicker T, Sabot F, Hua-Van A, Bennetzen JL, Capy P, Chalhoub B, Flavell A, Leroy P, Morgante M, Panaud O, et al. A unified classification system for eukaryotic transposable elements. Nat Rev Genet. 2007;8:973–982. doi: 10.1038/nrg2165. [DOI] [PubMed] [Google Scholar]
- Williamson S, Fledel-Alon A, Bustamante CD. Population genetics of polymorphism and divergence for diploid selection models with arbitrary dominance. Genetics. 2004;168:463–475. doi: 10.1534/genetics.103.024745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright SI, Le QH, Schoen DJ, Bureau TE. Population dynamics of an Ac-like transposable element in self- and cross-pollinating Arabidopsis. Genetics. 2001;158:1279–1288. doi: 10.1093/genetics/158.3.1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright SI, Agrawal N, Bureau TE. Effects of recombination rate and gene density on transposable element distributions in Arabidopsis thaliana. Genome Res. 2003;13:1897–1903. doi: 10.1101/gr.1281503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada K, Lim J, Dale JM, Chen H, Shinn P, et al. Empirical analysis of transcriptional activity in the Arabidopsis genome. Science. 2003;302:842–846. doi: 10.1126/science.1088305. [DOI] [PubMed] [Google Scholar]
- Yang G, Zhang F, Hancock CN, Wessler SR. Transposition of the rice miniature inverted repeat transposable element mPing in Arabidopsis thaliana. Proc Natl Acad Sci. 2007;104:10962–10967. doi: 10.1073/pnas.0702080104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai J, Liu J, Liu B, Li P, Meyers BC, Chen X, Cao X. Small RNA-directed epigenetic natural variation in Arabidopsis thaliana. PLoS Genet. 2008;4:e1000056. doi: 10.1371/journal.pgen.1000056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X. The epigenetic landscape of plants. Science. 2008;320:489–492. doi: 10.1126/science.1153996. [DOI] [PubMed] [Google Scholar]
- Zhang X, Yazaki J, Sundaresan A, Cokus S, Chan SW, Chen H, Henderson IR, Shinn P, Pellegrini M, Jacobsen SE, et al. Genome-wide high-resolution mapping and functional analysis of DNA methylation in Arabidopsis. Cell. 2006;126:1189–1201. doi: 10.1016/j.cell.2006.08.003. [DOI] [PubMed] [Google Scholar]
- Zhang X, Shiu S, Cal A, Borevitz JO. Global analysis of genetic, epigenetic and transcriptional polymorphisms in Arabidopsis thaliana using whole genome tiling arrays. PLoS Genet. 2008;4:e1000032. doi: 10.1371/journal.pgen.1000032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zilberman D, Gehring M, Tran RK, Ballinger T, Henikoff S. Genome-wide analysis of Arabidopsis thaliana DNA methylation uncovers an interdependence between methylation and transcription. Nat Genet. 2007;39:61–69. doi: 10.1038/ng1929. [DOI] [PubMed] [Google Scholar]