Abstract
Differentiation of female murine ES cells triggers silencing of one X chromosome through X-chromosome inactivation (XCI). Immunofluorescence studies showed that soon after Xist RNA coating the inactive X (Xi) undergoes many heterochromatic changes, including the acquisition of H3K27me3. However, the mechanisms that lead to the establishment of heterochromatin remain unclear. We first analyze chromatin changes by ChIP-chip, as well as RNA expression, around the X-inactivation center (Xic) in female and male ES cells, and their day 4 and 10 differentiated derivatives. A dynamic epigenetic landscape is observed within the Xic locus. Tsix repression is accompanied by deposition of H3K27me3 at its promoter during differentiation of both female and male cells. However, only in female cells does an active epigenetic landscape emerge at the Xist locus, concomitant with high Xist expression. Several regions within and around the Xic show unsuspected chromatin changes, and we define a series of unusual loci containing highly enriched H3K27me3. Genome-wide ChIP-seq analyses show a female-specific quantitative increase of H3K27me3 across the X chromosome as XCI proceeds in differentiating female ES cells. Using female ES cells with nonrandom XCI and polymorphic X chromosomes, we demonstrate that this increase is specific to the Xi by allele-specific SNP mapping of the ChIP-seq tags. H3K27me3 becomes evenly associated with the Xi in a chromosome-wide fashion. A selective and robust increase of H3K27me3 and concomitant decrease in H3K4me3 is observed over active genes. This indicates that deposition of H3K27me3 during XCI is tightly associated with the act of silencing of individual genes across the Xi.
Gene dosage of X-chromosomal genes in mammals is equalized between sexes by inactivation of one of the two X chromosomes in female cells. This inactivation process (XCI) occurs in the embryonic lineage of female embryos (Lyon 1961), as well as during differentiation of female mouse embryonic stem (ES) cells, the latter providing a powerful model system.
The onset of XCI is controlled by the X-inactivation center (Xic), a region of ∼1 Mb on the X chromosome (Heard and Avner 1994). The Xic is comprised of several genetic elements that encode long noncoding RNAs (ncRNAs), which include Xist and Tsix. The Tsix transcribed region completely overlaps Xist, and the coordinated expression of these two genes determines XCI. On the future inactive X (Xi), down-regulation of Tsix is accompanied by up-regulation of Xist expression during differentiation. On the active X (Xa), persistence of Tsix expression keeps Xist silent preventing the Xa from being inactivated (Lee and Lu 1999). Various studies using knockout systems reported that Tsix regulates Xist transcription through chromatin remodeling at the Xist locus (Navarro et al. 2005; Sado et al. 2005; Sun et al. 2006). However, the chromatin changes in wild-type female and male ES cells have not been studied and compared in greater detail or at high resolution. In particular, the presence of a transient heterochromatic state characterized by H3K27me3 at the promoter of the active Xist gene has been reported in some studies (Sun et al. 2006), but not in others (Navarro et al. 2005; Sado et al. 2005). Furthermore, a large region of heterochromatin located 5′ to Xist has been reported to have a potential role in the initiation of X inactivation (Heard et al. 2001; Rougeulle et al. 2004; Augui et al. 2007), but the detailed chromatin structure and dynamics of this region have not been examined.
RNA fluorescent in situ hybridization (FISH) studies have shown that the Xist transcript appears to associate with the Xi and/or the nuclear matrix around it (Brown et al. 1992; Clemson et al. 1996). Very little is known about possible interactors of the Xist transcript that enable it to coat a chromosome in cis, or about the mechanisms by which heterochromatin is generated across the length of the X chromosome. It has been proposed that Xist RNA creates a silent, repetitive nuclear compartment (Chaumeil et al. 2006; Clemson et al. 2006) into which genes become recruited as they become silenced on the one hand (Chaumeil et al. 2006). Also, Xist RNA might bind to high-affinity sites present throughout the X chromosome inducing local heterochromatinization, which subsequently spreads in a cooperative manner. The L1 long interspersed elements (LINEs) have been suggested to function as such “relay elements” or way stations (McBurney 1988; Lyon 1998), although there is little direct experimental evidence for this (Popova et al. 2006). Other hypotheses have evoked specific regions of chromatin as nucleation centers, for example, a 250 kb region 5′ to the Xist gene, which consists of a stretch of chromatin enriched in H3K9me2 and H3K27me3 (termed “hotspot”) (Heard et al. 2001; Rougeulle et al. 2004).
Immunofluorescence (IF) studies have shown that chromatin changes are among the earliest events on the inactive X after Xist accumulation. This includes loss of histone modifications associated with active chromatin, such as H3K9 acetylation and H3K4 methylation (Heard et al. 2001; Goto et al. 2002; Okamoto et al. 2004), as well as enrichment of H3K9me2, H4K20me1, and H3K27me3 (Heard et al. 2001; Plath et al. 2003; Silva et al. 2003; Kohlmaier et al. 2004; Okamoto et al. 2004), which are associated with heterochromatin and silencing. Among these, H3K27me3 has been studied most extensively. The Polycomb group repressive complex 2 (PRC2), and in particular its enzymatic component Ezh2, appears to be responsible for the deposition of this mark (Plath et al. 2003; Silva et al. 2003). The PRC2 complex is recruited to the inactive X early in XCI, in what appears to be a Xist-dependent fashion (Plath et al. 2003; Silva et al. 2003; de la Cruz et al. 2005; Zhao et al. 2008). H3K27me3 increases the affinity for binding of chromodomain-containing Polycomb proteins such as CBX7, which is a component of PRC1 (Fischle et al. 2003; Bernstein et al. 2006). Based on this, PRC2-dependent H3K27me3 deposition has been proposed to act as a recruitment signal for PRC1, which in turn would catalyze H2AK119ub1 as a further step in the XCI (de Napoles et al. 2004). Later changes of the Xi during XCI include a shift to late replication timing, as well as macroH2A enrichment (Takagi et al. 1982; Costanzi and Pehrson 1998). A recent study by Mietton et al. (2009) used chromatin immunoprecipitation with microarray hybridization (ChIP-chip) to show that the increased macroH2A is uniformly distributed over the Xi.
Most studies on the chromatin and transcriptional changes on the Xi have been performed using IF and RNA FISH. This reveals the dynamics of various epigenetic factors and histone marks on the inactive X chromosome at microscopic resolution. However, such approaches do not allow the monitoring of local epigenetic changes, specifically those that might be important for initiation and propagation of XCI. Furthermore, the degree to which the condensation of the Xi might influence detection of fluorescent signals remains enigmatic in immunofluorescence studies (Brinkman et al. 2006; Mietton et al. 2009). In order to obtain a detailed molecular view of the chromatin changes on the X chromosome during the early stages in X inactivation, we perform ChIP-chip, as well as ChIP combined with massive parallel sequencing (ChIP-seq), to obtain high-resolution maps of various chromatin-associated proteins on the X chromosome. We determine the X-linked changes during female ES cell differentiation for four histone modifications (tri-methylation of H3K4, H3K27, and H3K36, as well as H3K9me2), H3-core, TATA binding protein (TBP), the RNA polymerase II complex (RNAPII), as well as RNA expression, on a 15-Mb region on the X chromosome using ChIP-chip. Male ES cells were profiled in parallel to distinguish between general differentiation-dependent changes on the X chromosome versus female-specific changes associated with XCI. The profiled region includes the Xic, thus providing the first epigenetic high-resolution profile for the various functional elements within the Xic. To investigate chromatin dynamics during XCI at a chromosome-wide level we generate quantitative, genome-wide H3K4me3 and H3K27me3 ChIP-seq profiles. The sequences obtained in ChIP-seq allow allele-specific mapping, and enable to monitor the H3K27me3 dynamics and heterochromatin formation specifically on the Xi.
Results
ChIP-chip profiling during X-chromosome inactivation
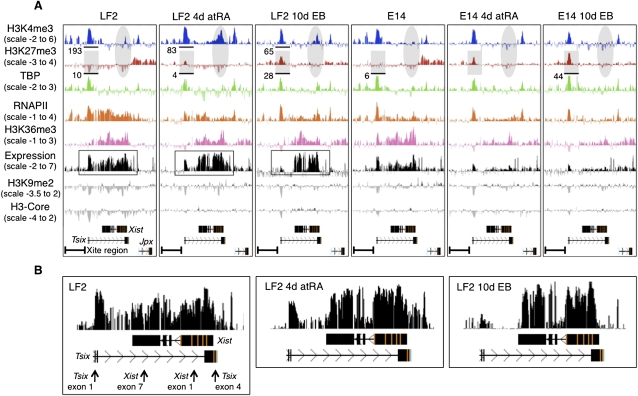
Various studies have focused on the epigenetic profile of genes on the inactive X (Xi) chromosome using targeted ChIP probing at a limited number of loci (Navarro et al. 2005, 2006; Sado et al. 2005; Sun et al. 2006; Shibata and Yokota 2008). As such, a spotlight view of the epigenetic landscape of the X chromosome was obtained. In order to assess the kinetics of the epigenetic changes associated with the early events of XCI at a higher resolution, we examined undifferentiated female (LF2) ES cells, as well as 4-d all-trans-retinoic acid (atRA) differentiated ES cells and 10-d embryoid bodies (EBs). Male (E14) ES cells were profiled in parallel to distinguish between general differentiation-dependent changes on the X chromosome versus female-specific changes associated with XCI. High-resolution ChIP-chip profiles for four histone modifications (trimethylation of H3K4, H3K27, and H3K36, as well as H3K9me2), H3-core, TBP, and RNAPII were generated. Furthermore, concomitant expression changes were analyzed using cDNAs generated from the same cell populations and hybridized to the same high-resolution microarrays. The genomic region analyzed was a ∼15 Mb segment centered around the X-inactivation center (Xic) (genome built MM9 95–109 Mb), which represents ∼10% of the X chromosome. Supplemental Figure S1 and the supporting Supplemental material document the differentiation of the E14 and LF2 cells, which in the female LF2 cells is accompanied by XCI.
Xist/Tsix
We first focused on the Tsix–Xist locus within the Xic as this region is involved in controlling the onset of XCI. Thus, we expected to see changes occurring between undifferentiated and differentiated female ES cells and between male and female differentiated ES cells. In undifferentiated female LF2 and male E14 cells, a clear accumulation of H3K4me3, TBP, and RNAPII could be observed at the promoter of Tsix, while H3-core and H3K9me2 appear to be depleted (Fig. 1A). The Tsix transcribed region is decorated with H3K36me3 and RNAPII, whereas H3K27me3 is absent. As expected from this profile, the (unspliced) Tsix transcript is highly expressed as is particularly evident at the 5′ end of Tsix, which best distinguishes Tsix from Xist transcripts (Fig. 1B). The Xist expression levels are very low (Fig. 1A,B; Supplemental Fig. S1b), and indeed active histone modifications or TBP/RNAPII are absent or very low at the Xist promoter in undifferentiated ES cells. Upstream of the Tsix promoter, within the so-called Xite region (Ogawa and Lee 2003), both male and female undifferentiated cells show an active epigenetic landscape of H3K4me3, TBP, and RNAPII accumulation. The peaks correlate with the two major transcription start sites in the Xite region identified by Ogawa and Lee (2003).
Figure 1.
Overview of the epigenetic landscape, as well as gene expression within and centered around the Xic. A screen shot from the UCSC Genome Browser showing the distribution of the ChIP-chip profiles across a 100 kb region of mouse chromosome X in mouse embryonic cells (UCSC Mouse [mm9], July 2007; chromosome X genomic coordinates 100.615–100.715 Mb). Expression was determined by hybridizing polyA RNA to the tiling array. Y-axis, log2 ChIP/input ratio or log2 cDNA/input ratio; x-axis, 50 bp oligonucleotides from sequence included on the NimbleGen mouse chromosome X tiling array. The quantification, as shown below the profiles, was obtained from the XT67E1 ChIP-seq experiments as described later. For quantifications, ChIP-seq tags were counted within the indicated regions. For the E14 cells, the tags are obtained from one X chromosome, while in XT67E1 cells they are obtained from both X chromosomes. (A) Overview of the Tsix–Xist interplay. Tsix and Xist noncoding RNAs, and direction of transcription, are indicated below all profiles. (B) Enlargement of the expression profiles of the LF2 cells, showing a clear correlation between the expression signal and the Xist exons after 4-d atRA treatment or 10-d EB formation. The arrows shown for the undifferentiated LF2 cells indicate PCR amplicons used for RT-qPCR validation (Supplemental Fig. S1b).
Upon differentiation, H3K27me3 appears at the Tsix promoter in both male and female cells, concomitant with a clear decrease in Tsix expression (Fig. 1B). RT-qPCR confirms the decreased Tsix expression (∼10-fold) after 4-d atRA treatment (Supplemental Fig. S1b). As after 4-d atRA treatment Tsix expression is almost completely lost from both the inactive and active X chromosomes (Xi and Xa, respectively; Lee and Lu 1999), the observed H3K27me3 at the Tsix promoter is probably present on both alleles. This is in line with the H3K27me3 emerging at the Tsix promoter in the male E14 cells (Fig. 1A), in which the single X chromosome is thought to behave similar to the Xa in the female LF2 cells. As H3K4me3 persists at the Tsix promoter throughout differentiation of the male cells, the deposition of H3K27me3 might result in a bivalent H3K4me3 and H3K27me3 mark at the Tsix promoter in these cells. The low expression level of Tsix at the differentiation stages of the E14 cells is in line with the low expression levels reported for bivalent marks (Bernstein et al. 2006). Otherwise, the H3K27me3 on the Tsix promoter might be present in a different subset of cells as compared to the H3K4me3.
In differentiating female but not male cells, the decrease in Tsix expression is accompanied by increased expression of Xist at both differentiation stages, as expected for the onset of XCI (Fig. 1A,B). The shift from Tsix to Xist expression at 4 d and 10 d of XCI in the LF2 cells can be concluded from the clear decrease in expression of Tsix (best seen over its first two exons), as well as from the concordance between the exon pattern of Xist and the expression signals at 4 d and 10 d of XCI (Fig. 1B). RT-qPCR on the 4-d atRA treated LF2 cells confirmed Xist expression (Supplemental Fig. S1b), while RNA FISH analysis showed the accumulation of Xist RNA over one X chromosome in 81% of the LF2 10-d EBs. H3K4me3 emerges at the Xist promoter at these stages concomitant with an accumulation of TBP and RNAPII. This presumably occurs on the X chromosome that is undergoing inactivation as Xist expression has been shown to be monoallelic from the future Xi (Lee and Lu 1999). In line with the monoallelic activation of Xist on the Xi in female cells, H3K4me3 and H3K36me3 deposition or recruitment of TBP/RNAPII are not observed at the Xist locus in male cells during differentiation (Fig. 1A).
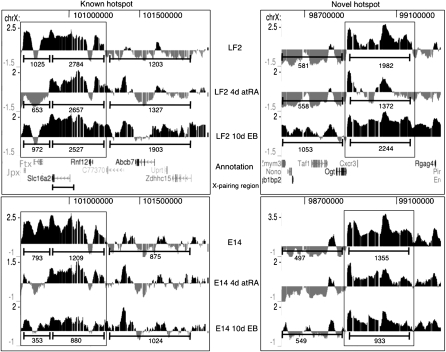
H3K27me3 hotspots around the Xic
Previous studies using targeted ChIP in undifferentiated ES cells have documented a domain upstream of Xist and including the 2010000I03Rik (also known as Jpx) locus that is enriched in H3K9me2 and H3K27me3 (“hotspot”) (Heard et al. 2001; Rougeulle et al. 2004). Part of this hotspot, called the X-pairing region (Xpr), has been implicated in the initiation of XCI and, more specifically, in a pairing event between the two X chromosomes in female ES cells (Augui et al. 2007).
Inspection of our profiles indeed reveals the presence of an ∼0.5 Mb genomic region almost completely covered with H3K27me3 in female, as well as in male, undifferentiated cells (Fig. 2, “Known hotspot”). The H3K27me3 enrichment over the proximal half of the hotspot (spanning 100.680–100.875 Mb and including the 2010000I03Rik and B230206F22Rik [also known as Ftx] locus) disappears after 4-d atRA treatment in both female and male cells. After 10-d EB differentiation some H3K27me3 reappears in female and male cells albeit to a lower extent than in undifferentiated cells. The distal part of the hotspot, containing the Xpr (Fig. 2), is invariantly marked with H3K27me3 in male and female cells although it decreases slightly in overall enrichment in differentiated cells. The Slc16a2 (or Xpct) gene, which overlaps with most of the Xpr region (Fig. 2), shows an unusual pattern of expression in female LF2 and male E14 cells (Supplemental Fig. S2a,b). In male E14 cells, we invariantly detect Slc16a2 mRNA at all stages, in line with the H3K4me3, TBP, and RNAPII occupancies at the Slc16a2 promoter. In female LF2 cells, Slc16a2 is very low or absent in undifferentiated cells or 10-d EB cells. However at 4-d atRA treatment, we detect Slc16a2 expression and increased H3K4me3 at the promoter concomitant with decreased H3K27me3 over the gene body. As X chromosome pairing only occurs at early XCI stages (between 0 and 4 d of differentiation), it is tempting to speculate that the temporal activation of Slc16a2 in LF2 cells, cells is related to the Xpr pairing event (Augui et al. 2007).
Figure 2.
Kinetics of the two H3K27me3 hotspots (which are boxed) surrounding the Xic in LF2 and E14 ES cells as obtained by H3K27me3 ChIP-chip. Screen shots from the UCSC Genome Browser showing the distribution of the H3K27me3 ChIP-chip profiles across a ∼0.5 Mb region of mouse chromosome X in mouse embryonic stem cells (UCSC Mouse [mm9], July 2007; chromosome X genomic coordinates 100.68–101.25 and 98.88–99.31 Mb for the known hotspot and the novel hotspot, respectively). The MM9 coordinates of the Xic are ∼100.3–101 Mb. Annotation is provided below the profiles. For further details, see Figure 1. For quantification using the ChIP-Seq tags of the XT67E1 cells, the “known” hotspot was split into two parts based on their differential kinetics. For comparison, tags were also counted in equally sized neighboring regions.
Remarkably, we identified a second region rich in H3K27me3 proximal of the Xic, which showed similar H3K27me3 kinetics as the hotspot described above (Fig. 2, “Novel hotspot”). In contrast to the “Known hotspot,” in this second region the H3K27me3 levels are higher in the 10-d EBs than in the undifferentiated LF2 cells (Fig. 2). Within this novel hotspot, the epigenetic and expression profiles of the genes across this region were similar in undifferentiated E14 and LF2 cells, as well as during differentiation of both.
Epigenetic changes at X-linked genes during female and male ES cell differentiation
To investigate the nature of the chromatin dynamics affecting genes on the Xi during X inactivation, we inspected the epigenetic profile of three genes that are representative for different kinetic classes of XCI. Pgk1, a highly expressed housekeeping gene, is considered a classical example of a gene that undergoes inactivation on the Xi (Penny et al. 1996). Atrx is an active gene that is inactivated on the Xi late after the onset of XCI, i.e., not at day 4, but by day 8 (Supplemental Fig. S3). Gdpd2, an osteoblast differentiation-promoting factor, is shown as control for a constitutive silent gene (GNF Expression Atlas 2). The 15 Mb profiled region did not contain known genes that escape XCI.
For Atrx and Pgk1, we clearly detect mRNA in undifferentiated LF2 and E14 cells (Supplemental Fig. S4a,b). This is in good agreement with the epigenetic profile for both genes in undifferentiated cells, characterized by high occupancies of H3K4me3, TBP, and RNAPII at the promoter, as well as H3K36me3 and RNAPII over the coding body. However, during differentiation of LF2 cells we do not detect any major changes occurring in mRNA levels or in the epigenetic landscape of Atrx and Pgk1 (Supplemental Fig. S4a,b). The patterns between female, with supposedly one allele inactivated, and male are virtually indistinguishable. We do, however, observe subtle quantitative differences. The most notable is a slight overall increase in H3K27me3 signal at 10-d EB formation that appears to be restricted to the female LF2 cells (Supplemental Fig. S4a,b). This increase is present for 30 of the 33 highest expressed genes present on the X-chromosomal region examined, while it was not seen over inactive genes, such as Gdpd2 (Supplemental Fig. S4c; 27 of the 36 inactive genes did not show any apparent increase in H3K27me3).
Kinetics of H3K4me3/H3K27me3 during XCI
X inactivation clearly occurs in differentiating LF2 cells as shown by Xist activation (Fig. 1; Supplemental Fig. S1) and Xist RNA coating of the Xi (Supplemental material). This is apparently not accompanied by qualitative or quantitative changes in the combined epigenetic profiles of X-linked genes unambiguously measurable by ChIP-chip (Supplemental Fig. S4a–c). However, ChIP-chip, a hybridization-based assay, does not allow robust quantification. Therefore, we performed ChIP-seq for H3K4me3 and H3K27me3. ChIP-seq is quantitative and can be used to analyze repeat regions (see below). Moreover, the complete genome is profiled allowing direct comparisons of the X chromosome with autosomes. We performed H3K27me3 ChIP-seq profiling for E14 and LF2 undifferentiated cells, as well as 10-d EBs from the same batches of chromatin as used for ChIP-chip. Moreover, H3K27me3 and H3K4me3 ChIP-seq profiles were generated for female undifferentiated XT67E1 ES cells, as well as for two differentiation stages of these cells, 4-d atRA treatment or cells grown as EBs for 10 d. The XT67E1 cells originate from mouse crossbreeds of two inbred strains (129 and C3H/He; both Mus musculus domesticus [Frazer et al. 2007]) with the distant strain PGK-1a/Ws (Mus musculus musculus). The 129 derived X chromosome of the XT67E1 cells, furthermore, contains a deletion in the Xist gene (Penny et al. 1996), resulting in nonrandom inactivation of the PGK derived X chromosome. Thus, the XT67E1 cells harbor two genetically distinct X chromosomes that can be distinguished in deep sequencing. Therefore, the XT67E1 cells combined with ChIP-seq provide an invaluable system to study the epigenetic marking of the Xi specifically. RNA FISH for Xist and immunofluorescent (IF) staining of H3K27me3 show that most of the XT67E1 cells undergo XCI during differentiation (Supplemental Fig. S5).
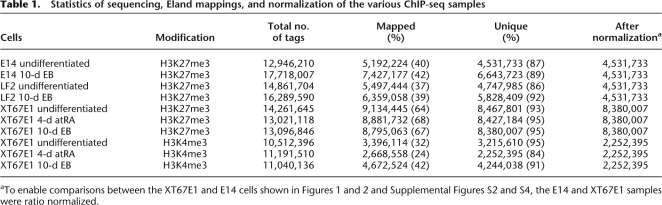
To achieve comprehensive genome coverage, we sequenced around 15 million tags per sample (Table 1). After aligning the unique tags onto the reference MM9 genome allowing one mismatch, the samples were normalized to enable direct and quantitative comparative analyses. Supplemental Figure S6a–c illustrates the quality of our data. Despite originating from different female ES cell lines, the ChIP-chip profiles for LF2 cells are very similar to the ChIP-seq results obtained for the XT67E1 cells (Supplemental Fig. S6b,c; see also the numbers in Figs. 1, 2 and Supplemental Figs. S2a, S4a–c for ChIP-seq quantification of the ChIP-chip data). As the female XT67E1 and LF2 cells gave very similar results, the ChIP-seq analyses performed for the LF2 cells are shown in the Supplemental material.
Table 1.
Statistics of sequencing, Eland mappings, and normalization of the various ChIP-seq samples
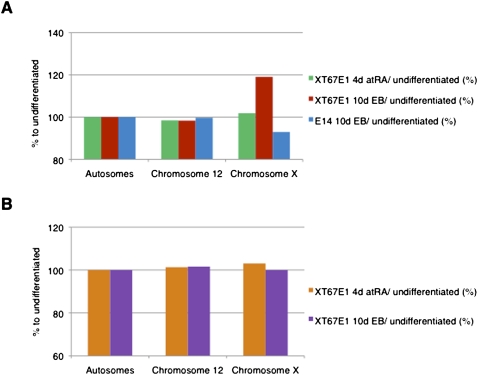
We determined the ratios of the number of H3K27me3 tags of differentiated (4-d atRA-treated cells and 10-d EBs) versus undifferentiated cells per chromosome. Plotting these ratios shows a gradual increase of H3K27me3 on the X chromosome during differentiation only in female ES cells (Fig. 3A). The XT67E1 cells show an increase of 2% after 4-d atRA treatment and 19% after EB formation. A similar increase after EB formation is observed for LF2 cells (Supplemental Fig. S7a). The increase in H3K27me3 does not occur on the X chromosome in the male E14 cells (Fig. 3A); on the contrary, its H3K27me3 level decreases. Assuming that the Xa in the female cells will have a similar epigenetic profile as the single, active X chromosome in the male E14 cells, we conclude that the increase in H3K27me3 is occurring on the Xi (see also below). The degree of H3K27me3 on the autosomes remained similar, showing that the increase in H3K27me3 observed in the female cells is specific for the X chromosomes. In contrast to H3K27me3, the distribution of H3K4me3 during XCI in the female XT67E1 remained largely identical over all chromosomes, including the X chromosome (Fig. 3B; Supplemental Fig. S7b), reinforcing the notion that the increase is specific for H3K27me3.
Figure 3.
Specific enrichment of H3K27me3 on the X chromosome after 10-d EB formation in mouse female XT67E1 ES cells, but not in male ES cells. (A) H3K27me3 tag distribution (representing amounts of H3K27me3) over autosomes or the X chromosome in XT67E1 or E14 cells (ratio of 4-d atRA or 10-d EB versus undifferentiated cells in percent). See Supplemental material and Supplemental Figure S7 for more details. (B) H3K4me3 tag distribution (representing amounts of H3K4me3) over autosomes or the X chromosome in XT67E1 cells (ratio of 4-d atRA or 10-d EB versus undifferentiated cells in percent). See Supplemental material and Supplemental Figure S7 for more details.
Allele-specific single nucleotide polymorphism (SNP) assignment
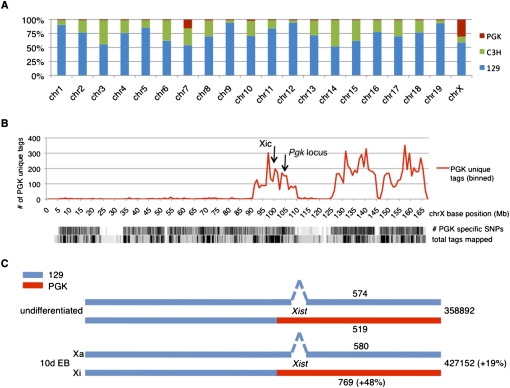
To investigate whether the increased H3K27me3 is indeed occurring on the Xi we exploited the presence of SNPs on the different alleles (129/C3H versus PGK) in the female XT67E1 cells. The strain identity of the X chromosomes of the XT67E1 cells was originally validated only by Southern blot screening of the Pgk marker locus (Penny et al. 1996). Based on this single measurement it was concluded that the X chromosome in the XT67E1 cells was heterozygous 129/PGK. To validate the origin of the chromosomes in detail we first determined the chromosomal contribution of each of the parental strains to the XT67E1 cells using the known SNPs among the 129, C3H, and PGK mouse strains. SNP containing tags were extracted from all mapped XT67E1 sequence tags and assigned to either the 129, C3H, or PGK strains. Subsequently, the parental contribution to each of the chromosomes was calculated. A picture emerges of a mosaic genome (Fig. 4A). The PGK strain contributes significantly to chromosome 7 and surprisingly only to about 25% of the X chromosomes (Fig. 4A). The overall contributions for each of the strains are calculated to be 3.27% (PGK), 23.19% (C3H), and 73.54% (129), which is in good agreement with the theoretical contributions of ∼6.25% (PGK), 18.75% (C3H), and 75% (129) based on the number of mouse crossbreeds (Penny et al. 1996).
Figure 4.
Allele-specific mapping of tags obtained during the ChIP-seq experiments. (A) Genetic make-up of XT67E1 mouse ES cells, showing the contribution of the parental PGK, C3H, or 129 mouse strains to the individual chromosomes of the XT67E1 cells. For individual chromosomes, the contribution of each strain was calculated by dividing the number of uniquely identified SNPs in the ChIP-seq experiments by the total number of SNPs, as determined from the SNP database (mouse.perlegen.com/mouse/index.html). (B) Distribution of the uniquely identified PGK SNPs within the total pool of XT67E1 ChIP-seq tags in 1 Mb bins over the X chromosome (represented by the x-axis). Density plots of the total number of PGK-specific SNPs (114,781 for the complete X chromosome), as well as of the total number of mapped tags, are plotted below the graph. The genomic locations of the Xic and the Pgk locus are indicated. (C) Schematic representation of the X chromosomes in the undifferentiated and the 10-d EB XT67E1 cells, with the Xist deletion in the 129 derived X chromosome (which will become the Xa during XCI). The number of total H3K27me3 tags mapped over both chromosomes, as well as the number of tags that could be mapped allele-specific, is indicated for both stages.
As only one-quarter of the X chromosomes in the female XT67E1 cells originates from the PGK strain (Fig. 4A), we performed further analysis to obtain a more precise strain origin. Chromosome X was subdivided in bins and the number of the tags specifically assigned to the PGK strain was counted within each bin (Fig. 4B). This analysis shows that only the distal part of chromosome X (MM9 location ∼90–167 Mb) contains PGK strain-specific tags, implying that only this part of chromosome X is heterozygous PGK. The proximal part of chromosome X (∼3–90 Mb) is mainly 129 (data not shown). The genetic mosaic composition was not detected before because the Pgk locus that was used as marker is located around 103 Mb on the X chromosome (Fig. 4B,C). A similar analysis for chromosome 7 shows that the 36–77 Mb region on this chromosome originates from the PGK mouse strain (data not shown).
Next, we assigned the tags obtained in the H3K27me3 ChIP-seq experiments for the undifferentiated XT67E1 cells and the 10-d EBs to the heterozygous 129/PGK part of the X chromosome (Fig. 4C). The number of tags containing a 129 specific SNP remains constant during EB formation (574 versus 580). In contrast, the number of tags containing a PGK SNP increased with 48% (519 versus 769). Thus, the increase in H3K27me3 during XCI occurs on the Xi and not on the Xa. Although the 48% increase is based on a small subset of sequence tags, it is in good agreement with the 38% increase on the Xi as predicted from the total number of mapped tags (two times the 19% increase shown in Fig. 3A; note that the tags in Fig. 3A are mapped over both X chromosomes of the XT67E1 cells).
Distribution of the H3K27me3 increase across the inactive X chromosome during XCI
Several models have been proposed to explain Xist induced silencing across the X chromosome and propagation of heterochromatin formation during XCI. Heterochromatin may spread evenly from the Xic across the chromosome or may propagate in two phases: first to Xist “docking stations” inducing local heterochromatinization from which it spreads secondarily (McBurney 1988; for review, see Ng et al. 2007). We set out to distinguish between these models using the deposition of H3K27me3 as a hallmark of heterochromatin formation.
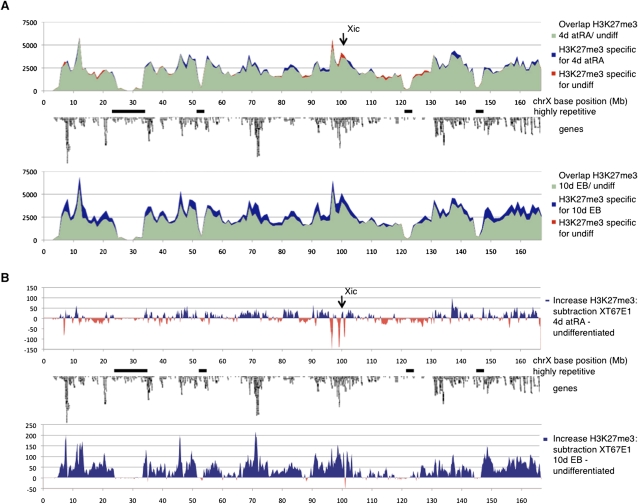
We computed the H3K27me3 tag densities in bins over chromosome X and plotted the change in the number of tags for the different times of differentiation as compared to undifferentiated cells (Fig. 5A,B). At the early stages of XCI (4-d atRA) the profile on the X chromosome is highly similar to that of undifferentiated XT67E1 cells (Fig. 5A,B). The 2% increase in H3K27me3 (Fig. 3A) does not appear to be localized to specific loci, but rather dispersed over the entire chromosome (Fig. 5B). Three loci near the Xic show a dramatic drop in H3K27me3 (Fig. 5B). The two distal troughs, centered around the Xic, correspond very well to the “hotspot” profiles observed in ChIP-chip, i.e., high levels of H3K27me3 that disappear after 4-d atRA treatment (Fig. 2). The most proximal of the three H3K27me3 troughs (MM9 coordinates 96.3–96.8 Mb) appears to be specific for the XT67E1 cells, as it is not present in the LF2 cells. The XT67E1 EB profile displays a prominent chromosome-wide increase of H3K27me3 as compared to undifferentiated cells (Fig. 5A,B). The loci showing the strongest increase are dispersed over chromosome X and overlap with gene-dense regions (Fig. 5B). We do not observe clear differences in the distribution of the increased H3K27me3 between 4-d and 10-d differentiated LF2 cells (Fig. 5B), which could have been indicative for nucleation centers from which H3K27me3 spreads. Very similar results were obtained in the female LF2 cells (Supplemental material; Supplemental Fig. S8). Taken together, these data suggest that the deposition of H3K27me3 and thus heterochromatin formation occurs evenly over the Xi.
Figure 5.
Kinetics of H3K27me3 over chromosome X during XCI in XT67E1 cells, showing a chromosome-wide increase of H3K27me3 during XCI. (A) H3K27me3 profiles over chromosome X in XT67E1 cells. Chromosome X is subdivided in bins of 1 Mb, followed by counting of the number of tags per bin. undiff, undifferentiated. (B) Subtraction tracks of the H3K27me3 profiles shown in A at a higher resolution (bins of 100 kb). Increase of H3K27me3 as compared to undifferentiated cells is indicated in blue, decrease in red.
H3K27me3 over repetitive elements
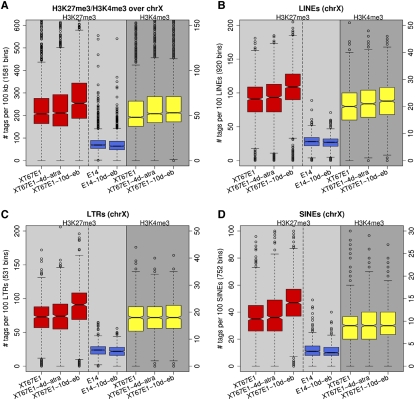
It has been reported that repetitive genomic elements, in particular LINEs, may serve as relay elements that may help in the propagation or spread of inactivation and heterochromatinization across the X chromosome (Ke and Collins 2003; Lyon 2006; Popova et al. 2006). Therefore, we calculated the H3K27me3 distribution on the X chromosome over various repeat classes including LINEs (Fig. 6A–D). Figure 6A shows the spread in the number of H3K27me3 tags per 100 kb of the X chromosome in female XT67E1 (red boxplots) and male E14 cells (blue boxplots). The amount of H3K27me3 clearly increases during differentiation in female cells and remains the same in male cells (Fig. 6A). Note that the number of tags in male E14 cells is derived from one X chromosome, while in the female XT67E1 it is derived from two X chromosomes.
Figure 6.
Boxplots of the occupancies of H3K27me3 and H3K4me3 over various repeats of chromosome X in female XT67E1 and male E14 cells. (A) All tags on chromosome X; a boxplot representation of the graphs shown in Fig. 5A. (B–D) Distributions over various repeat classes (LINE, LTR, and SINE repeats).
Analyses of the LINE, LTR, and SINE repeats show that none of these repeat classes seems to be specifically enriched in H3K27me3 as compared to the entire chromosome X either at 4-d or at 10-d differentiation of the XT67E1 cells (Fig. 6B–D). The increase in H3K27me3 over Simple repeats, Low Complexity repeats, and Satellite repeats is also similar to the increase calculated for the entire X chromosome (data not shown). Moreover, full-length LINEs, which have been shown to be particularly enriched on the X chromosome and have therefore been proposed as likely relay elements, do also show a similar increase in H3K27me3 as observed for the entire chromosome X (data not shown). No sign of an above-average increase in H3K27me3 levels could be detected around these full-length L1 regions in day 4 differentiated cells, as might be expected for relay elements from which heterochromatin spreads. Thus, our data do not provide support that LINEs, SINEs, or any other repeat class, act as relay elements at least at the level of H3K27me3 spread. Intriguingly, we did observe a slight increase of H3K4me3 over LINE repeats during X inactivation (Fig. 6B), which might be indicative for a small increase in expression from these repeats.
H3K27me3 deposition on the Xi is tightly associated with active transcription
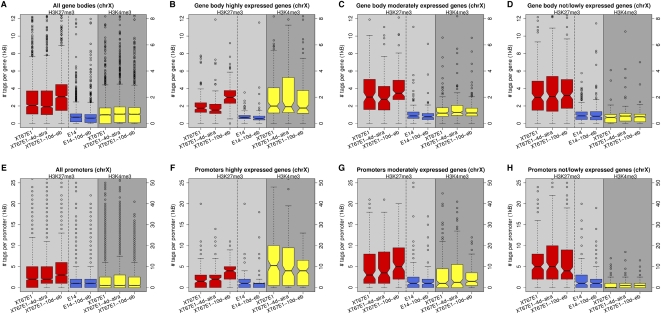
We next investigated whether the increased deposition of H3K27me3 across the Xi during XCI coincides with transcribed regions, as suggested by our ChIP-chip analysis (Supplemental Fig. S4a–c). The X-linked genes were categorized into three groups according to their expression level in undifferentiated ES cells, after which the number of H3K27me3 tags over the gene bodies and promoters was determined at the various stages of differentiation (Fig. 7). H3K27me3 over the gene bodies of all genes on chromosome X during XCI is increased to a similar degree as calculated over the entire chromosome (cf. Fig. 6A with Fig. 7A). An above-average increase of H3K27me3 is observed during XCI at highly expressed genes (55%; Fig. 7B), whereas limited and no H3K27me3 increase are detected on moderately or not expressed genes, respectively (Fig. 7C,D). At EB day 10, the level of H3K27me3 over genes highly expressed in undifferentiated ES, but inactivated during XCI reaches the same distribution range as measured for moderately or not/lowly expressed genes throughout differentiation. The increase in H3K27me3 levels over highly expressed genes is not observed in the male E14 cells and hence specific for XCI (Fig. 7B).
Figure 7.
Boxplots of the occupancies of H3K27me3 and H3K4me3 over promoters and gene bodies of chromosome X. Genes were binned in three equally sized groups of 212 genes according to expression level, representing not/lowly-, moderately-, and highly-expressed genes, respectively. The red boxplots represent H3K27me3 occupancies for the female XT67E1 cells, while the yellow boxplots represent H3K4me3 occupancies. The blue boxplots represent H3K27me3 occupancies for the male E14 cells. (A–D) Distribution of H3K27me3 and H3K4me3 over gene bodies (from +500 to the end of Ensemble genes). (E–H) Distributions of H3K27me3 and H3K4me3 over gene promoters (from −700 to +300 of Ensemble genes).
A similar specific deposition of H3K27me3, as seen over gene bodies, is observed for promoter regions (Fig. 7E–H). Interestingly, the above-average prominent increase of H3K27me3 over the promoters of genes highly active in undifferentiated cells, but inactivated during XCI is accompanied by a correspondingly strong decrease of H3K4me3 (Fig. 7F; yellow boxplots). Taken together, our results suggest that the observed deposition of H3K27me3 on the Xi occurs on genes that are being inactivated during XCI.
Discussion
In this study we describe the first comprehensive approach for the investigation of epigenetic changes correlated with XCI. We profile H3K4me3, H3K27me3, H3K36me3, H3K9me2, H3-core, TATA binding protein, and the RNA polymerase II complex in wild-type (wt) female ES cells and male ES cells in order to discriminate between differentiation and XCI events at two stages of differentiation: 4-d atRA treatment and 10-d EB formation. Using this approach, we show that Tsix is down-regulated in female, as well as male, ES cells during differentiation (Fig. 1A,B). The accumulation of H3K27me3 at the Tsix promoter suggests that the mechanism of Tsix repression might be very similar in female and male ES cells. The down-regulation of Tsix is concomitant with a female-specific up-regulation of Xist during XCI and the deposition of H3K4me3 at the Xist promoter on the Xi (Fig. 1A,B). We do not observe a transient heterochromatic state represented by an accumulation of H3K27me3 at the Xist promoter during XCI as observed by Sun et al. (2006), although this could be due to the fact that we use a different female ES cell line and slightly different time points for monitoring. Tsix down-regulation has been shown to permit up-regulation of Xist on the future Xi in female cells, but it neither triggers expression of Xist in male cells (Fig. 1A,B; Lee and Lu 1999), nor remodels chromatin marking at the Xist promoter in male cells as observed in female cells (Fig. 1A). Our data corroborates and extends previous findings that the transcription status of Tsix is not the only factor determining Xist expression at the onset of XCI (Lee and Lu 1999; Luikenhuis et al. 2001). Recently it was reported that, next to the loss of Tsix, the absence of the Polycomb repressive complex 2 (PRC2) component Eed can lead to Xist expression in male cells (Shibata et al. 2008). This suggested that Eed might be involved in keeping Xist silent by deposition of H3K27me3 at the Xist promoter (Shibata et al. 2008). However, we do not observe H3K27me3 at the Xist promoter in either wt male or female ES cells or in their differentiated progeny (Fig. 1A). This suggests that during 4-d atRA treatment or 10-d EB formation additional factors might be required to keep Xist silent in male ES cells and on the Xa in female ES cells. The deposition of H3K27me3 may occur at later stages, playing a role in Xist silencing during maintenance of XCI rather than during the XCI initiation phase (4-d or 10-d differentiation) examined in this study.
We show that the up-regulation of the Xist RNA is accompanied by a disappearance of H3K27me3 “hotspots” surrounding the Xic only during the early stages of XCI (4-d atRA treatment; Figs. 2, 5B). Originally one of these H3K27me3 hotspots was identified as a long stretch (>300 kb) of DNA containing high levels of H3K27me3 as observed by targeted ChIP using local probing on chromosome X (Rougeulle et al. 2004). A recent study using genome-wide ChIP-chip and ChIP-seq profiling identified similar sized regions containing high levels of H3K27me3 as a general phenomenon over all chromosomes, referred to as blocks (Pauler et al. 2009). Close inspection of our ChIP-seq profiles indeed identified multiple large genomic regions containing high levels of H3K27me3, both in female and male ES cells. Therefore, the presence of H3K27me3 “hotspots” per se does not necessarily implicate the hotspots surrounding the Xic in X-chromosomal inactivation. However, the loss of H3K27me3 at the early stages of XCI (4-d atRA treatment) is highly specific for the hotspots surrounding the Xic. The remainder of the genome only contains five loci showing a similar disappearance of H3K27me3 dispersed over different chromosomes. In contrast to the hotspots surrounding the Xic, all of these correlate with single genes (Supplemental Fig. S11). Also, the fact that one of the H3K27me3 hotspots surrounding the Xic coincides with a pairing region of the two X chromosomes at the onset of XCI (Augui et al. 2007) argues in support of a functional role. Whether this would involve chromosome choice, X chromosome pairing, heterochromatin formation or propagation of XCI, as previously suggested (Heard et al. 2001; Rougeulle et al. 2004; Augui et al. 2007) is still not clear. Possibly, the disappearance of H3K27me3 early in XCI allows relaxation of higher-order chromatin structure along these regions, allowing spread of Xist from the Xic across the X chromosome. Further functional characterization of the hotspots is needed to shed more light on their role in the XCI process.
Using H3K4me3 and H3K27me3 ChIP-seq profiling in female and male cells, we investigate the initiation and/or propagation of heterochromatin across the X chromosome during XCI. For this purpose, we perform allele-specific mapping of ChIP-seq tags to distinguish between the inactive and the active X chromosome in the XT67E1 ES cells. This confirms for the first time the feasibility of monitoring allele-specific chromatin dynamics on a genome-wide scale using ChIP-seq. By assigning H3K27me3 ChIP-seq tags to either the active or inactive X chromosome, we are able to quantify the relative amounts of H3K27me3 over the X chromosomes. The analysis indisputably shows that the level of H3K27me3 increases over the Xi, while it remains constant over the Xa (Fig. 4C). Following 4-d atRA treatment, the overall H3K27me3 increase is small and importantly dispersed over the chromosome (Fig. 5A,B). We do not observe an elevated level of H3K27me3 at or surrounding the Xic, nor do we observe increased levels of H3K27me3 from the Xic on. We would have expected this if heterochromatin formation starts from the Xic and gradually spreads across the Xi. After 10 d we detect a chromosome-wide increase of H3K27me3 (Fig. 5A,B). Our observations seem to be in contrast to classical studies by Russell and Montgomery (1970). Analyzing X chromosome:autosome translocations, Russell and Montgomery conclude that inactivation of genes spreads from the Xic toward both ends of the chromosome because the physical distance from a gene to the Xic appeared to correlate with its chance to be inactivated as monitored by phenotypes of mice. One explanation for this discrepancy could be that the heterogeneity in H3K27me3 deposition on the Xi, caused by the asynchronous onset of XCI in the ES cell population used for our ChIP-seq profiling, prevents the detection of H3K27me3 spread. Another possibility is that “spread” is in fact a three-dimensional process occurring across the X-chromosome territory and that multiple regions, chromosome wide, are targeted simultaneously for H3K27 methylation due to the nature of chromosome folding in nuclear space. Alternatively, H3K27me3 deposition or heterochromatin formation on the Xi might follow different kinetics, as compared to gene silencing as monitored on autosomes.
Bioinformatic and genetic analysis has predicted a role for LINE repeats as relay elements that facilitate the propagation of XCI over the X chromosome (the so-called “LINE hypothesis” reviewed in Lyon 1998, 2003, 2006). We do not find any support for this hypothesis in our analysis using H3K27me3. The measured H3K27me3 level was not found to be increased above average on LINE repeats (Fig. 6) or any subclasses of LINE repeats, such as young LINEs or full-length LINEs. We also do not find a correlation between the increased H3K27me3 and other genomic elements that previously have been implicated in the XCI process and might have served as relay elements, such as SINEs or LTRs (Fig. 6). Thus, chromatin marks other than H3K27me3 may participate in the role of LINEs in spreading. Instead, we observe that during XCI, H3K27me3 is increased above-average over active genes, as compared to inactive genes (Fig. 7). This deposition of H3K27me3 over active genes is specific for XCI and not the result of differentiation of female ES cells, as it is not observed in male ES cells. The increase of H3K27me3 over (active) genes fits very well with previous studies considering overall H3K27me3 localization (Mak et al. 2002; Chadwick 2007; Pauler et al. 2009). Immunofluorescent stainings on metaphase chromosomes indicated that H3K27me3 (or PRC2, which deposits H3K27me3) occurs in a banded pattern, correlating with gene density and Giemsa light bands (Mak et al. 2002; Chadwick 2007). In line with these immunofluorescent studies, we observe H3K27me3 mainly in gene rich regions (Fig. 5A).
H3K27me3 is generally linked to gene silencing, and therefore the observed deposition of H3K27me3 over active genes during XCI (Fig. 7) seems to be associated with the inactivation of these genes on the Xi. Decoration of the Xi with H3K27me3 alone has been suggested to be insufficient for X-chromosome silencing (Plath et al. 2003). Recently the PRC2 complex, mediating tri-methylation of H3K27, was shown to recruit the H3K4me3 demethylase KDM5A (also known as JARID1A) in mouse ES cells during silencing of developmental genes (Pasini et al. 2008). We observe that the increase of H3K27me3 during XCI over active genes is accompanied by a decrease of H3K4me3 at the promoters of these genes (Fig. 7). Therefore, the recruitment of KDM5A by PRC2 might explain how gene silencing on the Xi proceeds after the PRC2 complex decorates the Xi.
Together, our data provide supporting evidence that Tsix down-regulation is not the only factor regulating Xist expression, but that multiple dynamic chromatin changes across the Xic are likely to participate in Xist regulation. After monoallelic up-regulation of Xist and Xist RNA coating of the Xi, we provide unequivocal, quantitative proof that H3K27me3 increases on the Xi during XCI. The level of H3K27me3 on the Xa remains constant. Our results further indicate that the chromatin changes of H3K4me3 and H3K27me3 on the Xi seem to be tightly linked with the process of chromosome-wide silencing of active genes on the Xi. Our study represents the first chromosome-wide and allele-specific analysis of X inactivation and provides new evidence that the loss of active and gain of repressive chromatin marks are tightly integrated during the onset of this process.
Methods
E14, LF2, and XT67E1 cell culture
The LF2 and XT67E1 female ES cells were cultured as described previously for the LF2 cells (Rougeulle et al. 2004; Augui et al. 2007). Male E14 ES cells were cultured according to standard protocols of the SIGTR (www.sanger.ac.uk/PostGenomics/genetrap/protocols/GeneralInformation.pdf).
Differentiation of ES cells using all-trans-retinoic acid (for 4 d) was performed as described by Rougeulle et al. (2004). For embryoid body (EB) differentiations, cells in an 80% confluent T25 flask were lightly trypsinized with 0.05% Trypsin (2 min at 37°C), then split into two low-adherence 10-cm dishes and maintained in DMEM media containing 10% FBS and 0.1 μM 2-mercaptoethanol (Smith 1991). For RNA FISH and immunofluorescence analyses, the EBs were plated onto gelatinized coverslips after 4 d in suspension and then allowed to grow on coverslips for an additional 6 d.
RT-qPCR, expression microarray analysis
For RNA isolation, ES cells were homogenized in TRIzol (Invitrogen) and subjected to chloroform extraction, isopropanol precipitation and ethanol washing according to the manufacturer's recommendations. For RT-qPCR, 2–5 μg of isolated RNA was treated with DNase I (Invitrogen). The DNase treated RNA was subject to an RT reaction primed by a mix of hexamers and oligo(dT)-primer, performed using SuperScript III Reverse Transcriptase (Invitrogen) according to the protocol of the manufacturer. qPCR was performed using iQ SYBR Green Supermix (Bio-Rad) with a Bio-Rad MyIQ Cycler. Primer sequences used for RT-qPCR are shown in Supplemental Table S1.
The cDNA for expression microarray analysis was prepared similarly as for RT-qPCR, with the following adjustments: the starting material was 40–100 μg RNA. Following DNase treatment, mRNA was isolated using the Oligotex mRNA Mini Kit (Qiagen) obtaining around 1 μg mRNA. After reverse transcription, the cDNA was subject to RNase H (Ambion) treatment and column purified (Qiaquick PCR purification kit, Qiagen). One microgram of this cDNA was used in the Cy5 labeling reaction prior to microarray hybridization. Microarray hybridization was the same as for the ChIP-chip. In short, Cy5 labeled cDNA was hybridized together with Cy3 labeled input material on NimbleGen MM8 385K whole-genome tiling arrays.
ChIP and ChIP-chip
ChIP experiments were performed according to standard protocols as described in Brinkman et al. (2006). The antibodies used are summarized in Supplemental Table S2.
For ChIP-chip, the T7-based amplification procedure (Liu et al. 2003) was used to amplify both ChIP and total (input) material. Input DNA (labeled Cy3) and ChIP DNA (labeled Cy5) were hybridized to MM8 385K whole genome tiling arrays manufactured by NimbleGen Systems, Inc. (NimbleGen MM8 Tiling Set 38).
ChIP-chip hybridizations were performed by NimbleGen according to standard protocols of the full ChIP-chip microarray service. After Tukey bi-weight normalization, the log2-ratio values of ChIP over input were used for further analysis. For visualization purposes, the output data were converted to Wiggle (WIG) files for viewing the data in the UCSC Genome Browser (except for the expression, all ChIP-seq profiles are shown with a smoothing window of 4; the H3K27me3 hotspots are shown with a smoothing window of 8). Mouse genome built MM9 coordinates were acquired from the MM8 coordinates using the Batch Coordinate Conversion (liftOver) of the UCSC Genome Browser. For identification of H3K27me3 rich regions (hotspots), an algorithm was developed that scored genomic regions of at least 75 kb in which at least 80% of the probe values was above 0 in the H3K27me3 ChIP-chip profiles of undifferentiated LF2 cells.
ChIP-seq
ChIP-seq sample preparation for sequencing was performed according to standard protocols of the manufacturer (Illumina). In short, ChIP-seq was carried out by conventional ChIP followed by end repair of 15 ng enriched DNA (Pico Green measurements, Quant-iTTM PicoGreen dsDNA kit, Invitrogen). Adaptors were ligated to DNA fragments, which were subsequently size selected (∼200 base pair [bp]). The adapter-modified DNA fragments were subjected to limited PCR amplification (18 cycles) and quality control was made by qPCR (primers sequences are available upon request). Finally, cluster generation and sequencing-by-synthesis (32 bp) was performed using the Illumina 1G Analyzer according to standard protocols of the manufacturer (Illumina).
The image files generated by the Genome Analyzer were processed to extract DNA sequence data. Sequences were aligned to the mouse MM9 reference genome using the Illumina Analysis Pipeline allowing one mismatch. The total number of sequenced fragments and mapped fragments are shown in Table 1. For all analysis performed, only the unique tags (Table 1) were considered. The 32 bp sequence reads were subsequently directionally extended to 200 bp, corresponding to the length of the original fragments used for sequencing. The output data were converted to Browser Extensible Data (BED) files for downstream analysis and Wiggle (WIG) files for viewing the data in the UCSC Genome Browser. All ChIP-seq sequence analyses were conducted based on the Mus musculus NCBI m37 genome assembly (MM9) accessed from the Ensembl (release 49) or the UCSC (assembly July 2007) Genome Browsers.
Data normalization ChIP-seq
To compensate for differences in sequencing depth and mapping efficiency among ChIP-seq samples, the total number of unique tags of each sample was uniformly equalized relative to the sample with the lowest number of tags, allowing quantitative comparisons (Table 1). The H3K27me3 samples of the XT67E1 cells, the H3K27me3 samples of the LF2/E14 cells, and the H3K4me3 samples of the XT67E1 cells were normalized as independent groups. To enable comparisons between the XT67E1 and E14 cells in the ChIP-seq countings shown in Figures 1 and 2 and Supplemental Figures S2 and S4, the E14 and XT67E1 samples were ratio normalized. All ChIP-chip, RNA-chip, and RNA-seq data are present in the NCBI GEO SuperSeries GSE15884.
ChIP-seq tag counts
For the normalized H3K27me3 tracks, we counted the number of unique sequence tags within Ensembl genes (from +500 bp to the end of gene; genes ≤500 bp were filtered out), promoters (from −700 to +300 bp around the gene start) and intergenic regions (700 bp downstream of a gene end to 700 bp upstream of the next gene start). Gene body and intergenic counts were normalized by the length of the region (number of tags per 1 kb). Sequence coordinates of various repeat classes were downloaded from the UCSC Table Browser. ChIP-seq tags were considered to represent a genomic element in case of an overlap of 50% (100 bp) or more, or if the tag completely overlapped the genomic element. For categorizing genes according to expression levels, we used the expression data generated for the female LF2 undifferentiated cells by RNA-ChIP (using all genes located between MM9 95 and 167 Mb). The expression level for a gene was determined by averaging the probes located within gene exons. Next, genes were binned in three equally sized groups of 212 genes according to expression level, representing not/lowly, moderately, and highly expressed genes, respectively.
Chromosome assignment XT67E1
The XT67E1 ES cells originate from a mouse crossbreed of C3H/He with Pgk-1a/Ws, which after a backcross to C3H/He was twice crossbred into a 129 background. The parental Pgk1a/Ws mouse strain belongs to the subspecies Mus musculus musculus and is distantly related to the classical C3H and 129 inbred strains, which are Mus musculus domesticus (Green 1989; Frazer et al. 2007). For obtaining SNPs between C3H, 129, and PGK we made use of a database (accessible from mouse.perlegen.com/mouse/index.html) containing the nucleotide variation (SNPs) between 15 mouse strains, including the 129S1/SvlmJ, C3H/HeJ, and PWD/PhJ (Frazer et al. 2007). The Pgk-1a/Ws strain is very closely related to the PWD/PhJ strain. Therefore, the known PWD/PhJ SNPs were used as a reference to identify SNPs within the XT67E1 cells. From the SNP database, we selected the SNPs that could be used to discriminate between 129, C3H, and PGK (“strain-specific SNPs”). From the ChIP-seq experiments, the collective mapped XT67E1 sequence tags from all H3K27me3 and H3K4me3 ChIP-seq experiments were gathered to obtain a large pool of XT67E1 tags. Next we counted per chromosome the number of strain-specific SNPs for which the genotype was present in our pool of XT67E1 tags (“assigned SNPs”). The contribution of each of the three strains to the individual chromosomes was determined by calculating the ratio between the “assigned SNPs” and the strain-specific SNPs for each of the strains. Per chromosome, the total sum of the contributions of the three strains was set to 100%, after which the relative contributions were determined (Fig. 4A).
Allele-specific assignment tags
For allele-specific assignment of the tags of the individual H3K27me3 ChIP-seq experiments of the XT67E1 cells, we only considered SNPs for which both alleles were sequenced in any of the tags generated (and thereby confirmed to be different between both alleles). In this way we make sure that the SNPs that are reported in the database (mouse.perlegen.com/mouse/index.html) are indeed present in the XT67E1 cells that we used. In total we identified 2154 SNPs in the XT67E1 cells on the X chromosome. Almost all of these (2099; 98%) were present in the distal part as expected based on our genotyping (Fig. 4B).
RNA FISH, combined RNA/DNA FISH, and immunofluorescent stainings
Cells used for immunofluorescence (IF) and FISH were grown on gelatinized coverslips. The cells were permeabilized on ice for 4 min in cytoskeletal buffer containing 0.5% Triton X-100 and 2 mM vanadyl ribonucleoside complex (New England Biolabs), and then fixed in 4% paraformaldehyde. Subsequently the coverslips were rinsed and kept in 70% ethanol at −20°C. The coverslips were progressively dehydrated through an ethanol series and air-dried. The probes used for RNA FISH and DNA FISH include a 19 kb genomic lambda clone (510) to detect Xist and a BAC probe (RP23-265D6) to detect Atrx. Probes were fluorescently labeled according to the Vysis nick translation kit using Spectrum-Red dUTP or Spectrum-Green dUTP. The Atrx gene probe was denatured, competed with Cot-1 DNA (3 μg/coverslip) for 30 min at 37°C and combined with the Xist RNA FISH probe just prior to hybridization. Preparation of the chromosome paint probes was performed according to the supplier's instructions (Cambio). For RNA FISH, hybridization was performed overnight at 37°C in a dark and humid chamber. For RNA/DNA FISH experiments, cells were additionally denatured in 50% formamide/2× SSC at 80°C for 35 min and rinsed several times in ice-cold 2× SSC prior to overnight hybridization at 42°C. After hybridization, the coverslips were washed three times in 50% formamide/2× SSC and three times in 2× SSC at 42°C, and then stained with DAPI. IF stainings were performed as described previously (Chaumeil et al. 2002). The antibody for H3K27me3 used for IF was provided by Danny Reinberg (Howard Hughes Medical Institute, New York) and was used at a dilution of 1/200.
Microscopy
Images were obtained using the Delta Vision system (Applied Precision) with a 100×/1.35NA objective (Olympus) and deconvolved using the Softworx software algorithm (Applied Precision, enhanced ratio method, 10 iterations).
Acknowledgments
We thank Eva Janssen-Megens and Arjen Brinkman for their help with the ChIP-seq experiments; Simon van Heeringen, Willem Welboren, Stefan Gräf, and Paul Flicek for bioinformatic help; Joost Martens for general recommendations and discussion; Thomas Jenuwein for sharing H3K27me3 and H3K9me2 antibodies for ChIP; Danny Reinberg for sharing the H3K27me3 antibody for immunofluorescence; Konstantinos Anastassiadis for providing LIF; SIGTR for providing the E14 cells. J.C.C. and E.H. were funded by the Fondation pour Recherche Medicale and the Agence Nationale de la Recherche. This work is supported by HEROIC, an Integrated Project funded by the European Union under the 6th Framework Programme (LSHG-CT-2005-018883).
Footnotes
[Supplemental material is available online at www.genome.org. ChIP-chip, RNA-chip, and RNA-seq data from this study have been submitted to NCBI Gene Expression Omnibus (GEO) (http://www.ncbi.nlm.nih.gov/geo/) under series accession no. GSE15884.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.092643.109.
References
- Augui S, Filion GJ, Huart S, Nora E, Guggiari M, Maresca M, Stewart AF, Heard E. Sensing X chromosome pairs before X inactivation via a novel X-pairing region of the Xic. Science. 2007;318:1632–1636. doi: 10.1126/science.1149420. [DOI] [PubMed] [Google Scholar]
- Bernstein E, Duncan EM, Masui O, Gil J, Heard E, Allis CD. Mouse Polycomb proteins bind differentially to methylated histone H3 and RNA and are enriched in facultative heterochromatin. Mol Cell Biol. 2006;26:2560–2569. doi: 10.1128/MCB.26.7.2560-2569.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brinkman AB, Roelofsen T, Pennings SW, Martens JH, Jenuwein T, Stunnenberg HG. Histone modification patterns associated with the human X chromosome. EMBO Rep. 2006;7:628–634. doi: 10.1038/sj.embor.7400686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown CJ, Hendrich BD, Rupert JL, Lafreniere RG, Xing Y, Lawrence J, Willard HF. The human XIST gene: Analysis of a 17 kb inactive X-specific RNA that contains conserved repeats and is highly localized within the nucleus. Cell. 1992;71:527–542. doi: 10.1016/0092-8674(92)90520-m. [DOI] [PubMed] [Google Scholar]
- Chadwick BP. Variation in Xi chromatin organization and correlation of the H3K27me3 chromatin territories to transcribed sequences by microarray analysis. Chromosoma. 2007;116:147–157. doi: 10.1007/s00412-006-0085-1. [DOI] [PubMed] [Google Scholar]
- Chaumeil J, Okamoto I, Guggiari M, Heard E. Integrated kinetics of X chromosome inactivation in differentiating embryonic stem cells. Cytogenet Genome Res. 2002;99:75–84. doi: 10.1159/000071577. [DOI] [PubMed] [Google Scholar]
- Chaumeil J, Le Baccon P, Wutz A, Heard E. A novel role for Xist RNA in the formation of a repressive nuclear compartment into which genes are recruited when silenced. Genes & Dev. 2006;20:2223–2237. doi: 10.1101/gad.380906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clemson CM, McNeil JA, Willard HF, Lawrence JB. XIST RNA paints the inactive X chromosome at interphase: Evidence for a novel RNA involved in nuclear/chromosome structure. J Cell Biol. 1996;132:259–275. doi: 10.1083/jcb.132.3.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clemson CM, Hall LL, Byron M, McNeil J, Lawrence JB. The X chromosome is organized into a gene-rich outer rim and an internal core containing silenced nongenic sequences. Proc Natl Acad Sci. 2006;103:7688–7693. doi: 10.1073/pnas.0601069103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costanzi C, Pehrson JR. Histone macroH2A1 is concentrated in the inactive X chromosome of female mammals. Nature. 1998;393:599–601. doi: 10.1038/31275. [DOI] [PubMed] [Google Scholar]
- de la Cruz CC, Fang J, Plath K, Worringer KA, Nusinow DA, Zhang Y, Panning B. Developmental regulation of Suz 12 localization. Chromosoma. 2005;114:183–192. doi: 10.1007/s00412-005-0008-6. [DOI] [PubMed] [Google Scholar]
- de Napoles M, Mermoud JE, Wakao R, Tang YA, Endoh M, Appanah R, Nesterova TB, Silva J, Otte AP, Vidal M, et al. Polycomb group proteins Ring1A/B link ubiquitylation of histone H2A to heritable gene silencing and X inactivation. Dev Cell. 2004;7:663–676. doi: 10.1016/j.devcel.2004.10.005. [DOI] [PubMed] [Google Scholar]
- Fischle W, Wang Y, Jacobs SA, Kim Y, Allis CD, Khorasanizadeh S. Molecular basis for the discrimination of repressive methyl-lysine marks in histone H3 by Polycomb and HP1 chromodomains. Genes & Dev. 2003;17:1870–1881. doi: 10.1101/gad.1110503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frazer KA, Eskin E, Kang HM, Bogue MA, Hinds DA, Beilharz EJ, Gupta RV, Montgomery J, Morenzoni MM, Nilsen GB, et al. A sequence-based variation map of 8.27 million SNPs in inbred mouse strains. Nature. 2007;448:1050–1053. doi: 10.1038/nature06067. [DOI] [PubMed] [Google Scholar]
- Goto Y, Gomez M, Brockdorff N, Feil R. Differential patterns of histone methylation and acetylation distinguish active and repressed alleles at X-linked genes. Cytogenet Genome Res. 2002;99:66–74. doi: 10.1159/000071576. [DOI] [PubMed] [Google Scholar]
- Green MC. Genetic variants and strains of the laboratory mouse. Oxford University Press; Oxford, UK: 1989. [Google Scholar]
- Heard E, Avner P. Role play in X-inactivation. Hum Mol Genet. 1994;3:1481–1485. doi: 10.1093/hmg/3.suppl_1.1481. [DOI] [PubMed] [Google Scholar]
- Heard E, Rougeulle C, Arnaud D, Avner P, Allis CD, Spector DL. Methylation of histone H3 at Lys-9 is an early mark on the X chromosome during X inactivation. Cell. 2001;107:727–738. doi: 10.1016/s0092-8674(01)00598-0. [DOI] [PubMed] [Google Scholar]
- Ke X, Collins A. CpG islands in human X-inactivation. Ann Hum Genet. 2003;67:242–249. doi: 10.1046/j.1469-1809.2003.00038.x. [DOI] [PubMed] [Google Scholar]
- Kohlmaier A, Savarese F, Lachner M, Martens J, Jenuwein T, Wutz A. A chromosomal memory triggered by Xist regulates histone methylation in X inactivation. PLoS Biol. 2004;2:E171. doi: 10.1371/journal.pbio.0020171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JT, Lu N. Targeted mutagenesis of Tsix leads to nonrandom X inactivation. Cell. 1999;99:47–57. doi: 10.1016/s0092-8674(00)80061-6. [DOI] [PubMed] [Google Scholar]
- Liu CL, Schreiber SL, Bernstein BE. Development and validation of a T7 based linear amplification for genomic DNA. BMC Genomics. 2003;4:19. doi: 10.1186/1471-2164-4-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luikenhuis S, Wutz A, Jaenisch R. Antisense transcription through the Xist locus mediates Tsix function in embryonic stem cells. Mol Cell Biol. 2001;21:8512–8520. doi: 10.1128/MCB.21.24.8512-8520.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyon MF. Gene action in the X-chromosome of the mouse (Mus musculus L.) Nature. 1961;190:372–373. doi: 10.1038/190372a0. [DOI] [PubMed] [Google Scholar]
- Lyon MF. X-chromosome inactivation: A real hypothesis. Cytogenet Cell Genet. 1998;80:133–137. doi: 10.1159/000014969. [DOI] [PubMed] [Google Scholar]
- Lyon MF. The Lyon and the LINE hypothesis. Semin Cell Dev Biol. 2003;14:313–318. doi: 10.1016/j.semcdb.2003.09.015. [DOI] [PubMed] [Google Scholar]
- Lyon MF. Do LINEs have a role in X-chromosome inactivation? J Biomed Biotechnol. 2006;2006:59746. doi: 10.1155/JBB/2006/59746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mak W, Baxter J, Silva J, Newall AE, Otte AP, Brockdorff N. Mitotically stable association of polycomb group proteins Eed and Enx1 with the inactive X chromosome in trophoblast stem cells. Curr Biol. 2002;12:1016–1020. doi: 10.1016/s0960-9822(02)00892-8. [DOI] [PubMed] [Google Scholar]
- McBurney MW. X chromosome inactivation: A hypothesis. BioEssays. 1988;9:85–88. doi: 10.1002/bies.950090211. [DOI] [PubMed] [Google Scholar]
- Mietton F, Sengupta AK, Molla A, Picchi G, Barral S, Heliot L, Grange T, Wutz A, Dimitrov S. Weak but uniform enrichment of the histone variant macroH2A1 along the inactive X chromosome. Mol Cell Biol. 2009;29:150–156. doi: 10.1128/MCB.00997-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro P, Pichard S, Ciaudo C, Avner P, Rougeulle C. Tsix transcription across the Xist gene alters chromatin conformation without affecting Xist transcription: implications for X-chromosome inactivation. Genes & Dev. 2005;19:1474–1484. doi: 10.1101/gad.341105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro P, Page DR, Avner P, Rougeulle C. Tsix-mediated epigenetic switch of a CTCF-flanked region of the Xist promoter determines the Xist transcription program. Genes & Dev. 2006;20:2787–2792. doi: 10.1101/gad.389006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng K, Pullirsch D, Leeb M, Wutz A. Xist and the order of silencing. EMBO Rep. 2007;8:34–39. doi: 10.1038/sj.embor.7400871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa Y, Lee JT. Xite, X-inactivation intergenic transcription elements that regulate the probability of choice. Mol Cell. 2003;11:731–743. doi: 10.1016/s1097-2765(03)00063-7. [DOI] [PubMed] [Google Scholar]
- Okamoto I, Otte AP, Allis CD, Reinberg D, Heard E. Epigenetic dynamics of imprinted X inactivation during early mouse development. Science. 2004;303:644–649. doi: 10.1126/science.1092727. [DOI] [PubMed] [Google Scholar]
- Pasini D, Hansen KH, Christensen J, Agger K, Cloos PA, Helin K. Coordinated regulation of transcriptional repression by the RBP2 H3K4 demethylase and Polycomb-Repressive Complex 2. Genes & Dev. 2008;22:1345–1355. doi: 10.1101/gad.470008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauler FM, Sloane MA, Huang R, Regha K, Koerner MV, Tamir I, Sommer A, Aszodi A, Jenuwein T, Barlow DP. H3K27me3 forms BLOCs over silent genes and intergenic regions and specifies a histone banding pattern on a mouse autosomal chromosome. Genome Res. 2009;19:221–233. doi: 10.1101/gr.080861.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penny GD, Kay GF, Sheardown SA, Rastan S, Brockdorff N. Requirement for Xist in X chromosome inactivation. Nature. 1996;379:131–137. doi: 10.1038/379131a0. [DOI] [PubMed] [Google Scholar]
- Plath K, Fang J, Mlynarczyk-Evans SK, Cao R, Worringer KA, Wang H, de la Cruz CC, Otte AP, Panning B, Zhang Y. Role of histone H3 lysine 27 methylation in X inactivation. Science. 2003;300:131–135. doi: 10.1126/science.1084274. [DOI] [PubMed] [Google Scholar]
- Popova BC, Tada T, Takagi N, Brockdorff N, Nesterova TB. Attenuated spread of X-inactivation in an X;autosome translocation. Proc Natl Acad Sci. 2006;103:7706–7711. doi: 10.1073/pnas.0602021103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rougeulle C, Chaumeil J, Sarma K, Allis CD, Reinberg D, Avner P, Heard E. Differential histone H3 Lys-9 and Lys-27 methylation profiles on the X chromosome. Mol Cell Biol. 2004;24:5475–5484. doi: 10.1128/MCB.24.12.5475-5484.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell LB, Montgomery CS. Comparative studies on X-autosome translocations in the mouse. II. Inactivation of autosomal loci, segregation, and mapping of autosomal breakpoints in five T (X;1) S. Genetics. 1970;64:281–312. doi: 10.1093/genetics/64.2.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sado T, Hoki Y, Sasaki H. Tsix silences Xist through modification of chromatin structure. Dev Cell. 2005;9:159–165. doi: 10.1016/j.devcel.2005.05.015. [DOI] [PubMed] [Google Scholar]
- Shibata S, Yokota T. Alteration of histone tail modifications in the Xist locus in wild-type and Tsix-mutant male embryonic stem cells during differentiation. Exp Anim. 2008;57:153–157. doi: 10.1538/expanim.57.153. [DOI] [PubMed] [Google Scholar]
- Shibata S, Yokota T, Wutz A. Synergy of Eed and Tsix in the repression of Xist gene and X-chromosome inactivation. EMBO J. 2008;27:1816–1826. doi: 10.1038/emboj.2008.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva J, Mak W, Zvetkova I, Appanah R, Nesterova TB, Webster Z, Peters AH, Jenuwein T, Otte AP, Brockdorff N. Establishment of histone h3 methylation on the inactive X chromosome requires transient recruitment of Eed-Enx1 polycomb group complexes. Dev Cell. 2003;4:481–495. doi: 10.1016/s1534-5807(03)00068-6. [DOI] [PubMed] [Google Scholar]
- Smith AG. Culture and differentiation of embryonic stem cells. J Tissue Cult Methods. 1991;13:89–94. [Google Scholar]
- Sun BK, Deaton AM, Lee JT. A transient heterochromatic state in Xist preempts X inactivation choice without RNA stabilization. Mol Cell. 2006;21:617–628. doi: 10.1016/j.molcel.2006.01.028. [DOI] [PubMed] [Google Scholar]
- Takagi N, Sugawara O, Sasaki M. Regional and temporal changes in the pattern of X-chromosome replication during the early post-implantation development of the female mouse. Chromosoma. 1982;85:275–286. doi: 10.1007/BF00294971. [DOI] [PubMed] [Google Scholar]
- Zhao J, Sun BK, Erwin JA, Song JJ, Lee JT. Polycomb proteins targeted by a short repeat RNA to the mouse X chromosome. Science. 2008;322:750–756. doi: 10.1126/science.1163045. [DOI] [PMC free article] [PubMed] [Google Scholar]