Abstract
Faithful transmission of the genome through sexual reproduction requires reduction of genome copy number during meiosis to produce haploid sperm and eggs. Meiosis entails steps absent from mitosis to achieve this goal. When meiosis begins, sisters are held together by sister chromatid cohesion (SCC), mediated by the cohesin complex. Homologs then become linked through crossover recombination. SCC subsequently holds both sisters and homologs together. Separation of homologs and then sisters requires two successive rounds of chromosome segregation and the stepwise removal of Rec8, a meiosis-specific cohesin subunit. We show that HTP-3, a known component of the C. elegans axial element (AE), molecularly links these meiotic innovations. We identified HTP-3 in a genetic screen for factors necessary to maintain SCC until meiosis II. Our data show that interdependent loading of HTP-3 and cohesin is a principal step in assembling the meiotic chromosomal axis and in establishing SCC. HTP-3 recruits all known AE components to meiotic chromosomes and promotes cohesin loading, the first known involvement of an AE protein in this process. Furthermore, REC-8 and two paralogs, called COH-3 and COH-4, together mediate meiotic SCC, but they perform specialized functions. REC-8 alone is necessary and sufficient for the persistence of SCC after meiosis I. In htp-3 and rec-8 mutants, sister chromatids segregate away from one another in meiosis I (equational division), rather than segregating randomly, as expected if SCC were completely eliminated. AE assembly fails only when REC-8, COH-3, and COH-4 are simultaneously disrupted. Premature equational sister separation in rec8 mutants of other organisms suggests the involvement of multiple REC-8 paralogs, which may have masked a conserved requirement for cohesin in AE assembly.
Keywords: Meiosis, cohesin, sister chromatid cohesion, sister chromatid co-orientation, chromosome axis, kinetochore co-orientation, equational division
Meiosis is a specialized cell division program that reduces genome copy number from four in germline stem cells after premeiotic DNA replication to one in haploid gametes. Two key innovations distinguish meiosis from mitosis and underlie the reduction in chromosome number. First, DNA is exchanged between homologous chromosomes (homologs) through meiotic crossover (CO) recombination. The CO, along with sister chromatid cohesion (SCC), tethers homologs together. A structure called the axial element (AE) promotes multiple events required for interhomolog recombination. Second, two successive rounds of meiotic chromosome segregation reduce chromosome copy number to one. Homologs separate in meiosis I (reductional division). Sister chromatids disjoin in meiosis II (equational division). The consecutive separation of homologs and sisters requires that SCC, mediated by the cohesin complex, be removed in two steps. Our work demonstrates a molecular link between these meiotic innovations. We report the role of HTP-3 in AE assembly and meiotic cohesin loading, and provide new insights into assembly of the meiotic chromosomal axis.
The distinct patterns of chromosome segregation in meiosis and mitosis result in part from the different compositions of meiotic and mitotic cohesin complexes. Cohesin is composed of four proteins—two Structural Maintenance of Chromosomes (SMC) proteins called Smc1 and Smc3, a non-SMC subunit called Scc3, and a kleisin subunit that differs between meiotic and mitotic cohesin complexes (for review, see Onn et al. 2008). Scc1 is the mitotic kleisin, and Rec8 is the meiotic kleisin. The kleisin is proteolytically cleaved to allow chromosomes to separate, and substitution of Rec8 for Scc1 is necessary for stepwise release of meiotic SCC (Klein et al. 1999; Watanabe and Nurse 1999; Buonomo et al. 2000; Toth et al. 2000). Cohesin complexes associate with chromosomes prior to S-phase and link sister chromatids together following replication (for review, see Onn et al. 2008). In meiotic prophase, Rec8 is present in linear tracks along the length of chromosomes in most organisms.
AE assembly is one of the earliest known events of meiosis; the AE forms shortly after premeiotic DNA replication (Padmore et al. 1991). The AE was initially defined by electron microscopy as a structure that forms between sister chromatids, along their entire length, prior to synapsis. Genetic and biochemical studies identified AE components and showed that the AE is required for several events that culminate in the reciprocal exchange of DNA between homologs, including homolog pairing and synapsis (for review, see Zickler and Kleckner 1999).
Based on the colocalization of AE components and cohesin, it has been proposed that cohesin is either the foundation upon which the AE is built or an integral part of the AE itself (Klein et al. 1999; Eijpe et al. 2003). Indeed, Rec8 is essential for AE formation in budding yeast (Klein et al. 1999). However, the relationship between cohesin and AE assembly is controversial in other organisms. In Caenorhabditis elegans, SCC-3 depletion disrupts the chromosomal association of some AE proteins (Pasierbek et al. 2003; Goodyer et al. 2008). However, AE proteins associate with meiotic chromosomes in rec8 mutants of C. elegans, mouse, Arabidopsis, and maize (Bhatt et al. 1999; Colaiácovo et al. 2003; Bannister et al. 2004; Xu et al. 2005; Golubovskaya et al. 2006; Martinez-Perez et al. 2008). Thus, an intact meiotic cohesin complex may not be essential for AE assembly, the AE and bound cohesin may be structurally separate entities (Kleckner 2006), or cohesin is integral to the AE but REC-8 depletion is insufficient to disrupt SCC.
The AE and meiotic recombination are important for reductional division. Inviable, aneuploid spores result from incorrect homolog segregation in meiosis I of budding yeast mutant for the AE protein Hop1 or the endonuclease Spo11, which creates the double-strand DNA breaks (DSBs) that initiate meiotic recombination (Hollingsworth and Byers 1989; Bergerat et al. 1997; Keeney et al. 1997; Klein et al. 1999). However, successful recombination does not guarantee the successive separation of homologs and sisters. Rather, cohesin removal must be regulated to ensure that sisters remain together until anaphase II. Also, sister chromatids must attach to microtubules (MTs) from the same spindle pole (or co-orient) to ensure their cosegregation in meiosis I, but later must attach to MTs from opposite spindle poles (or biorient) to allow their separation in meiosis II. MTs attach to chromosomes through protein structures called kinetochores, which assemble onto discrete chromosomal regions called centromeres.
The chromosomes of all widely studied model organisms except C. elegans are monocentric; each has a single discrete centromere. Centromere-associated factors including Spo13 and Mei-S332/Shugoshin prevent destruction of centromeric SCC at the metaphase to anaphase transition of meiosis I (for review, see Marston and Amon 2004; Dunleavy et al. 2005; Vagnarelli et al. 2008). Cohesin complexes along the chromosome arms lack these protective factors and are therefore removed, allowing homologs to separate.
On holocentric chromosomes of C. elegans, centromeric proteins reside along the entirety of meiotic chromosomes (Albertson et al. 1997; Moore et al. 1999). Therefore, protecting centromeric SCC cannot promote the successive separation of homologs and sisters. Instead, the single CO establishes the regions where SCC is protected. The CO can form anywhere along the chromosome but usually divides the homologs asymmetrically into a long arm and a short arm (Albertson et al. 1997). Following pachytene exit, homolog pairs are reorganized around the CO by the condensin II complex to form the compact, cruciform bivalents present in diakinesis, the final stage of prophase I (Chan et al. 2004; Nabeshima et al. 2005). Concomitantly, cohesin subunits and the paralogous AE components HIM-3 and HTP-3 appear as a cruciform pattern between sister chromatids along both arms (Zetka et al. 1999; Pasierbek et al. 2001, 2003; Chan et al. 2003; Goodyer et al. 2008). In contrast, the partially redundant HIM-3 paralogs HTP-1 and HTP-2 (HTP-1/2 henceforth) persist only on the long arm, where they help protect SCC until anaphase II (Martinez-Perez et al. 2008). However, much remains unknown about the mechanisms that promote meiotic cohesin loading and AE assembly to ensure the production of haploid gametes.
Our work reveals unexpected roles of cohesin and the AE during meiosis. We show that multiple kleisins act in the context of cohesin to mediate meiotic SCC in C. elegans. REC-8, COH-3, and the previously unknown kleisin COH-4 are each sufficient for association of AE proteins with meiotic chromosomes. AE assembly fails only in animals lacking all three kleisins, demonstrating that cohesin is required for AE formation in C. elegans, as in yeast. REC-8, COH-3, and COH-4 are all important for meiotic SCC, but REC-8 alone is necessary for sister chromatid co-orientation and the persistence of SCC until meiosis II. Thus, COH-3/COH-4 and REC-8 perform specialized meiotic functions. In a screen to identify proteins that promote REC-8•cohesin loading or protect SCC at the bivalent long arm, we identified the AE component HTP-3. We show that, in addition to its known roles in meiosis (Goodyer et al. 2008), HTP-3 is required for AE assembly and the association of REC-8•cohesin with meiotic chromosomes. In htp-3 and rec-8 mutants, premature, equational sister separation occurs in anaphase I. Thus, the interdependent loading of HTP-3 and meiotic cohesin is a key step in the genesis of the meiotic chromosomal axis. Moreover, our work reveals a molecular link between multiple meiotic innovations that together maintain ploidy during sexual reproduction.
Results
SCC is established in rec-8 mutants but sisters separate equationally in anaphase I
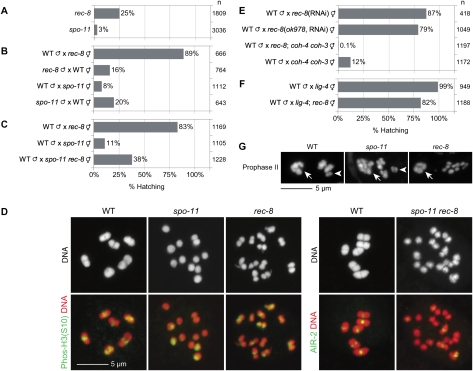
While characterizing C. elegans hermaphrodites mutant for rec-8(ok978), a deletion allele predicted to severely reduce or eliminate REC-8 function (Hayashi et al. 2007), we noticed phenotypes contradicting the view that REC-8 depletion eliminates SCC and causes separation of all 24 sister chromatids (Pasierbek et al. 2001). A surprisingly high number of progeny (25%) from rec-8(ok978) mutants hatched, inconsistent with the random segregation of chromatids expected if SCC were eliminated (Fig. 1A). Many hatched rec-8 embryos developed into fertile adults (Supplemental Table S1). In contrast, only 3% of progeny hatched from spo-11(me44) mutants, in which homologs remain apart as univalents (Dernburg et al. 1998) and segregate randomly in meiosis I (see below).
Figure 1.
Reducing REC-8 function rescues the embryonic lethality of spo-11 mutants. (A) The self progeny of spo-11(me44) mutant hermaphrodites exhibit much higher levels of embryonic lethality than the self progeny of rec-8(ok978) animals. (B) Mating crosses demonstrate that most of the lethality of rec-8 embryos results from sperm abnormalities (likely aneuploidy), while spo-11 embryos die from defects in oogenesis and spermatogenesis. Data shown hereafter represent analyses of oocyte meiosis (mutant mothers mated with wild-type males). Analyses of the self progeny of mutant hermaphrodites are included in Supplemental Figure S4. (C) Reducing REC-8 function suppresses the embryonic lethality of spo-11 mutants. (D) DAPI staining and S10-phosphorylated histone H3 or AIR-2 staining of meiotic chromosomes in prometaphase I. The six homolog pairs of wild-type animals are held together by SCC and the CO. AIR-2 localizes at the midbivalent in wild-type worms, where it phosphorylates histone H3 on Ser 10. PhosH3(S10) is present between the discrete sister chromatids of rec-8(ok978) worms. In contrast, discrete sisters are rarely observed following NEBD in spo-11(me44) worms, and a patch of phosH3(S10) is present on a subset of univalents. Discrete sisters can be resolved in spo-11 rec-8 animals; however, AIR-2 is only present between sisters in some univalents. (E) Synergistic embryonic lethality occurs in rec-8(ok978); coh-4(tm1857) coh-3(gk112) mutants. (F) Disrupting NHEJ with a deletion allele of lig-4 does not substantially increase the lethality of rec-8 mutants, suggesting that DSBs made in rec-8 mutants are repaired by a different mechanism, likely homologous recombination. (G) SCC persists until anaphase II in spo-11 mutants. After anaphase I and extrusion of the first polar body (arrows), sister chromatids remain together in wild-type and spo-11 zygotes (arrowheads), while only detached sisters are visible in rec-8 mutants.
The differential viability of rec-8 and spo-11 embryos was more pronounced in matings between mutant hermaphrodites and wild-type males. Eighty-nine percent of embryos laid by mated rec-8 mutants hatched compared with 8% of embryos laid by mated spo-11 mutants, suggesting that sperm abnormalities cause substantial lethality among self progeny of rec-8 mutants (Fig. 1B). Supporting this view, only 16% of progeny from wild-type hermaphrodites mated with rec-8 males were viable (Fig. 1B).
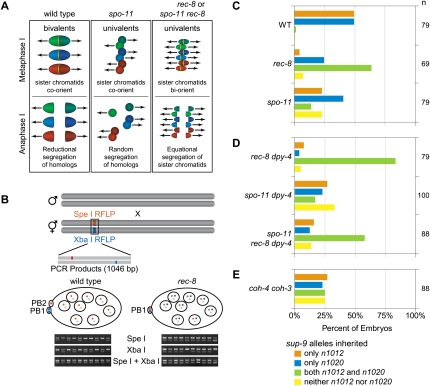
The high viability of rec-8 mutant embryos was not caused by residual REC-8 function in rec-8(ok978) mutants: Similar viability occurred in broods of rec-8(RNAi) animals and rec-8(ok978) mutants treated with rec-8(RNAi) (Fig. 1E). Instead, we found the high viability of rec-8 mutants to be the consequence of highly penetrant defects in both meiosis I and II. Premature equational separation of sisters in meiosis I coupled with failed polar body extrusion in meiosis II resulted in the production of viable triploids by rec-8 mutants mated with wild-type males (Fig. 2A). In contrast, random homolog segregation during meiosis I of mated spo-11 mutants resulted in aneuploidy and embryonic lethality.
Figure 2.
Sister chromatids separate equationally during meiosis I of rec-8 and spo-11 rec-8 mutants, while homologs segregate randomly in spo-11 mutants. (A) Model of meiotic chromosome segregation in wild-type, spo-11, and rec-8 mutant worms as shown by RFLP analysis in B. Three chromosomes are shown (green, blue, and red); homologs are indicated by different shades of the same color. The Aurora B kinase AIR-2, thought to mark REC-8 for destruction, is depicted in yellow. Arrows indicate the direction of MT-dependent forces exerted on chromosomes. In wild-type zygotes, sister chromatids co-orient. AIR-2 accumulates between homologs, which separate during meiosis I. Sister chromatids also co-orient in spo-11 mutants, SCC is protected during meiosis I, and sisters remain together while homologs segregate randomly in meiosis I. spo-11 zygotes inherit zero, one, or two copies of each chromosome, resulting in aneuploidy. In rec-8 mutants, sister chromatids biorient. AIR-2 localizes between sisters, which separate equationally when SCC is removed prematurely during anaphase I. Zygotes inherit a chromatid from each homolog. Because extrusion of the second polar body fails (Fig. 3), fertilization by a wild-type sperm would produce a viable triploid. (B) Strategy for examining meiotic chromosome segregation with ChrII-RFLPs. This assay, together with knowledge of the number of polar bodies extruded (Fig. 3) and whether or not sister chromatids remain together in prophase II (Fig. 1G), allows unambiguous determination of the pattern of chromosome segregation in meiosis I and II, as explained in Supplemental Table S2. Trans-heterozygous mothers [genotype sup-9(n1012)/sup-9(n1020)] are mated with wild-type males. n1012 and n1020 introduce a SpeI RFLP and an XbaI RFLP, respectively (de la Cruz et al. 2003). The lesions are amplified in a single PCR product. The cross progeny of an n1012/n1020 hermaphrodite and a wild-type male inherit a single sup-9 allele from their mother and a wild-type copy from their father. The progeny of n1012/n1020; rec-8 mutant worms mated with wild-type males inherit both ChrII-RFLPs and the wild-type sup-9 allele. ChrII-RFLPs are revealed by a doublet in SpeI or XbaI digests. An uncut product in SpeI/XbaI double digests reveals the wild-type allele. The alleles partitioned into polar bodies are not detected. (C) During wild-type meiosis, zygotes inherit a single ChrII-RFLP. In contrast, most rec-8 mutant embryos inherit both ChrII-RFLPs. The four possible outcomes of meiosis (inheritance of an n1012 allele, an n1020 allele, both alleles, or neither allele) occur at similar levels in spo-11 embryos. (D) Most progeny of spo-11 rec-8 mutants inherit both ChrII-RFLPs. The dpy-4 mutation was used in the construction of the spo-11 rec-8 double mutant. (E) Chromosome segregation in coh-4(tm1857) coh-3(gk112) embryos resembles that of spo-11 mutants.
Evidence for premature equational sister separation in rec-8 mutants came from an assay we devised to assess chromosome segregation. Transmission of chromosome II is followed through meiosis in strains for which each chromosome II homolog is uniquely tagged with a single-nucleotide polymorphism that introduces a restriction fragment length polymorphism (ChrII-RFLPs) (Fig. 2B). This assay, together with knowledge of the number of polar bodies extruded and whether precocious sister separation occurs in anaphase I, permits unambiguous determination of the pattern of meiotic chromosome segregation (Supplemental Table S2). In the assay, hermaphrodites trans-heterozygous for two ChrII-RFLPs are mated with wild-type males and the cross progeny scored for each ChrII-RFLP and the sperm-derived wild-type allele. In wild-type meiosis, zygotes inherit a chromatid from one homolog of each chromosome, resulting in embryos with a wild-type allele and a single ChrII-RFLP (Fig. 2B,C). In contrast, 64% of rec-8 embryos and 85% of rec-8 dpy-4 embryos tested positive for both ChrII-RFLPs and the wild-type allele. This pattern would occur if sister chromatids separate equationally during a single round of meiotic chromosome segregation. Furthermore, spo-11 and spo-11 dpy-4 mutant embryos inherited only the matroclinous ChrII-RFLP, only the patroclinous ChrII-RFLP, both ChrII-RFLPs, or neither ChrII-RFLP. This pattern is expected if homologs segregate randomly in meiosis I and sisters separate equationally in meiosis II.
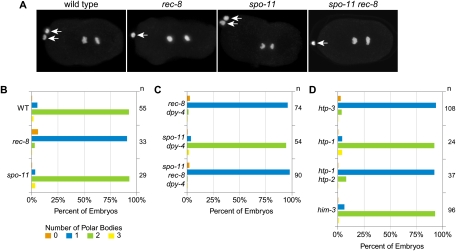
Cytological observations confirmed these conclusions (Fig. 3A,B). Fluorescence in situ hybridization of the 5S rDNA locus demonstrated that rec-8 zygotes inherited two copies of chromosome V during oocyte meiosis (Supplemental Fig. S1). In living rec-8 embryos, we found that polar body extrusion occurred during meiosis I, but extrusion of the second polar body failed (Supplemental Fig. S2). Thus, all chromosomes inherited during meiosis I were trapped in the embryo and detected by the ChrII-RFLP assay. Furthermore, detached sister chromatids were observed after anaphase I in DAPI-stained rec-8 mutants, indicating precocious loss of SCC (Fig. 1G). Together, these results demonstrate the frequent occurrence of equational sister separation in rec-8 mutants. In contrast, spo-11 mutant embryos had two polar bodies, and sisters remained together in anaphase I, indicating the persistence of SCC (Figs. 1G, 3A,B). Together with the ChrII-RFLP assay, these results indicate that homologs segregate randomly in meiosis I of spo-11 mutants.
Figure 3.
Extrusion of the second polar body fails in rec-8 mutants. (A) Polar bodies in DAPI-stained embryos. Arrows indicate the two polar bodies of wild-type and spo-11 mutants and the single polar body of rec-8 and spo-11 rec-8 mutants. (B–D) A single polar body is extruded in most rec-8, spo-11 rec-8 dpy-4, htp-1 htp-2 (see also Martinez-Perez et al. 2008), and htp-3 mutant embryos. In contrast, two polar bodies are present in most wild-type, spo-11, him-3, and htp-1 mutant embryos.
The equational sister separation observed in rec-8 mutants indicates that SCC persists until anaphase I in the absence of REC-8, but sister chromatids prematurely biorient on the meiosis I spindle, and SCC is destroyed to allow sister separation. Indeed, diakinesis nuclei of rec-8(ok978), rec-8(RNAi), and rec-8(ok978, RNAi) animals contained 12 univalents, not 24 sister chromatids (Figs. 1D, 4A; Supplemental Fig. S3A,B). Each univalent was bilobed, and the two sister chromatids were visible as discrete structures, consistent with their biorientation.
Figure 4.
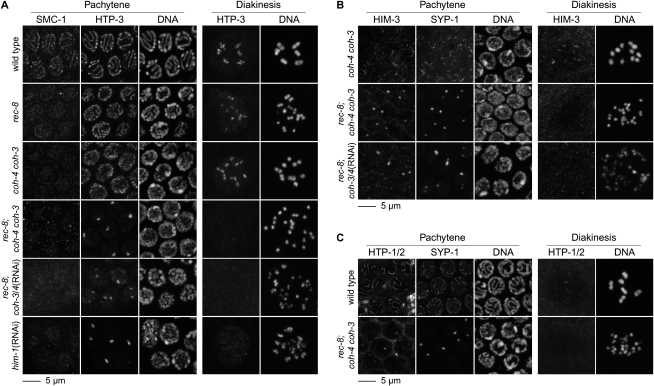
The kleisins COH-3/4 function in AE assembly and SCC during C. elegans meiosis. (A) In wild-type animals, the cohesin subunit SMC-1 and the AE protein HTP-3 colocalize along the length of synapsed pachytene chromosomes. Cruciform HTP-3 staining is observed in diakinesis nuclei. The diakinesis nuclei shown in this and subsequent images are from the oldest unfertilized oocyte (the −1 oocyte). SMC-1 is present at reduced levels in rec-8 and coh-4 coh-3 mutants, suggesting that these kleisins associate with SMC-1 in a cohesin complex. HTP-3 associates with pachytene chromosomes and diakinesis univalents of rec-8 and coh-4 coh-3 mutants. SMC-1 is undetectable in rec-8; coh-4 coh-3 triple mutants, HTP-3 associates with polycomplexes, and many detached sister chromatids are present in diakinesis nuclei, indicating severe disruption of SCC. Numerous chromosomal fragments are seen in diakinesis nuclei of rec-8; coh-3/4(RNAi) animals. Depletion of the cohesin subunit SMC-1, encoded by the him-1 gene, also disrupts AE assembly and causes chromosome fragments. (B) Short stretches of HIM-3 and SYP-1 associate with pachytene chromosomes of coh-4 coh-3 mutants. Both proteins are present in polycomplex in rec-8; coh-4 coh-3 triple mutants and rec-8; coh-3/4(RNAi) animals. (C) HTP-1/2 are not detected on meiotic chromosomes in rec-8; coh-4 coh-3 mutants.
Detection of Ser 10-phosphorylated histone H3 [phosH3(S10)] between sisters in rec-8 mutants confirmed that sister chromatids were not detached and suggested that the proteolytic machinery that normally triggers homolog separation in anaphase I instead triggers premature sister separation (Fig. 1D; Kaitna et al. 2002). PhosH3(S10) marks the short arms of wild-type bivalents in meiosis I and the interface between sister chromatids in meiosis II; it demarks activity of the Aurora B kinase AIR-2 (Hsu et al. 2000). Phosphorylation of REC-8 by AIR-2 is thought to tag it for destruction in wild-type animals (Rogers et al. 2002). Thus, the presence of PhosH3(S10) between sisters is consistent with their equational separation in rec-8 mutants.
The univalents of spo-11 mutants appeared different from those of rec-8 mutants. Sister chromatids were tightly associated in each univalent and could rarely be resolved following nuclear envelope breakdown (NEBD) (Fig. 1D). PhosH3(S10) was only detectable on some spo-11 univalents (Fig. 1D; Kaitna et al. 2002; Rogers et al. 2002), where it marked the surface of the univalent, not the region between sisters, consistent with our find-ing that sisters remain together until meiosis II of spo-11 mutants.
rec-8 mutations increase the viability of spo-11 mutants
Our data show that sister chromatids biorient in meiosis I of rec-8 mutants, and SCC is destroyed precociously, allowing equational sister separation, while sister chromatids co-orient in spo-11 single mutants, SCC persists until meiosis II, and most embryos die with aneuploid genomes resulting from random homolog segregation (Fig. 2A). Thus, disrupting rec-8 in a spo-11 mutant should increase viability by permitting equational sister separation. Indeed, rec-8(ok978) substantially suppressed the lethality of spo-11(me44) embryos (Fig. 1C; Supplemental Fig. S4A). Discrete sister chromatids were visible in diakinesis univalents of spo-11 rec-8 double mutants. AIR-2 was present between sisters in some, but not all, univalents (Fig. 1D). A single polar body was extruded during meiosis (Fig. 3A,C), and 60% of spo-11 rec-8 dpy-4 embryos inherited both ChrII-RFLPs, consistent with equational sister separation (Fig. 2D). rec-8(RNAi) also increased viability of spo-11 mutants (Supplemental Fig. S4B).
Multiple kleisins mediate meiotic SCC
Animals mutant for rec-8(ok978) display additional phenotypes inconsistent with complete abrogation of SCC. For example, the cohesin subunits SMC-1 and SMC-3 persist on meiotic chromosomes of rec-8(ok978), rec-8(RNAi), and rec-8(ok978, RNAi) animals (Fig. 4A; Supplemental Fig. S3B; Chan et al. 2003; AF Severson unpubl.). Furthermore, far fewer DNA fragments are observed in rec-8 mutants than expected based on the number of DSBs formed. On average, 12 DSBs are made per nucleus in wild-type animals (D Mets and B Meyer, pers. comm.) and likely more in rec-8(ok978) mutants (Hayashi et al. 2007). Eliminating SCC should prevent DSB repair using either a sister chromatid or homolog as a repair template, and rec-8(ok978) diakinesis nuclei should have at least 12 fragments. These were not observed (Figs. 1D, 4A; Supplemental Fig. S3A,B).
The uniform size and shape of univalents in rec-8(ok978) diakinesis nuclei suggests that DSBs are not repaired by nonhomologous end-joining (NHEJ) or single-strand annealing (SSA), error-prone processes that repair DSBs without homologous templates and produce chromosomal rearrangements and abnormalities like those in rad-51 and brca-2 mutants (Martin et al. 2005). Furthermore, DNA fragmentation and embryonic lethality in rec-8(ok978) mutants were not substantially increased by disrupting LIG-4 (Fig. 1F; Supplemental Fig. S3B), the C. elegans ortholog of DNA ligase IV, required for NHEJ (Martin et al. 2005; Clejan et al. 2006). Thus, meiotic DSB repair in rec-8 mutants likely uses the sister chromatid as a template, suggesting that SCC persists in the absence of REC-8.
Because SMC-1 and SMC-3 persist on meiotic chromosomes of rec-8 mutants, we hypothesized that kleisins other than REC-8 might be integral to meiotic cohesin complexes. Four C. elegans kleisins were known: REC-8, SCC-1, and the two predicted kleisins COH-1 and COH-3 (Pasierbek et al. 2001). We identified a fifth predicted kleisin, COH-4, which is 84% identical to COH-3. Our data unambiguously demonstrate the involvement of COH-3 and COH-4 (COH-3/4 henceforth) in meiotic SCC. We found SMC-1 staining to be undetectable in meiotic nuclei of rec-8(ok978); coh-4(tm1857) coh-3(gk112) triple deletion mutants, suggesting that COH-3/4 associate with meiotic chromosomes in the context of cohesin complexes (Fig. 4A). Multiple synergistic defects occurred when COH-3/4 were disrupted in rec-8 mutants (Figs. 1E, 4A–C; Supplemental Fig. S5C); 0.1% of rec-8; coh-4 coh-3 embryos were viable, compared with 89% of rec-8(ok978) and 12% of coh-4 coh-3 embryos (Fig. 1E). AE assembly was severely disrupted. The four known C. elegans AE proteins HTP-3, HTP-1/2, and HIM-3 and the SC central element SYP-1 were present in nucleoplasmic aggregates (polycomplexes) in pachytene nuclei of rec-8; coh-4 coh-3 triple mutants, but associated with meiotic chromosomes of rec-8 single or coh-4 coh-3 double mutants (Fig. 4A–C; Supplemental Figs. S3A,B, S5C). Detached sister chromatids were evident in diakinesis nuclei of rec-8(ok978); coh-4(tm1857) coh-3(gk112) triple mutants (Fig. 4A,B). Defects in AE assembly and meiotic SCC were also observed in rec-8(ok978); coh-4(tm1857) coh-3(ttTi10553) mutants and rec-8(ok978); coh-3/4(RNAi) animals (Fig. 4A,B; Supplemental Fig. S5C). Furthermore, depletion of SMC-1, expected to associate with all C. elegans cohesin complexes, caused defects in AE assembly and SCC. Notably, chromosomal fragmentation was observed in diakinesis nuclei of SMC-1-depleted animals and COH-3/4-depleted rec-8 mutants (Fig. 4A,B). Together, these data indicate that the REC-8 paralogs COH-3/4 associate with meiotic cohesin complexes to mediate AE assembly and SCC in rec-8(ok978) mutants.
REC-8 and COH-3/4 perform specialized functions
COH-3/4 are required for meiosis in animals with wild-type REC-8, and thus do not simply substitute for REC-8 in cohesin complexes of rec-8 mutants. SMC-1 was much reduced on meiotic chromosomes of coh-4 coh-3 deletion mutants (Fig. 4A; Supplemental Fig. S5C). HTP-3, HTP-1/2, HIM-3, and SYP-1 were present in short, fragmented stretches on pachytene chromosomes of coh-4 coh-3 mutants, unlike the long tracks of staining observed in rec-8(ok978) mutants (Fig. 4A,B; Supplemental Fig. S5C). Univalents were present in diakinesis nuclei of coh-4 coh-3 mutants. However, discrete sisters were not visible, and the inheritance of ChrII-RFLPs suggested that random homolog segregation, not equational sister separation, occurred in meiosis I (Figs. 2E, 4A,B; Supplemental Fig. S5C). Thus, COH-3/4 and REC-8 are all important for AE assembly and meiotic SCC, but REC-8 alone can co-orient sister chromatids and mediate SCC until meiosis II. We conclude that multiple kleisins are required during C. elegans meiosis, suggesting the participation of multiple, molecularly distinct cohesin complexes.
The involvement of COH-3/4 in meiosis can explain why sister chromatids remain together in rec-8 mutants, but raises the question of why homologs are apart. Univalents could result from defective SCC at the short arm if, for example, COH-3/4•cohesin associates exclusively with the long arm following recombination. Alternatively, homologs may be apart due to defects in meiotic recombination, as occurs in yeast rec8 mutants (Klein et al. 1999). Indeed, we found that CO recombination fails in C. elegans rec-8 mutants (Supplemental Material). This recombination defect explains the presence of univalents and indicates that COH-3/4•cohesin is insufficient for meiotic recombination.
AE protein HTP-3 promotes the meiotic pattern of chromosome segregation
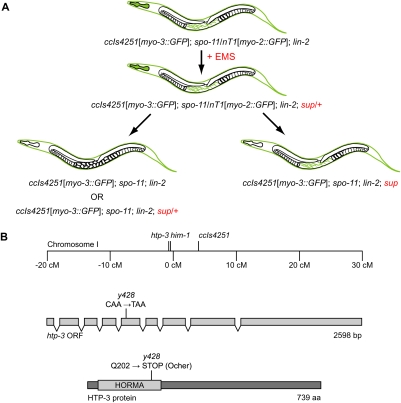
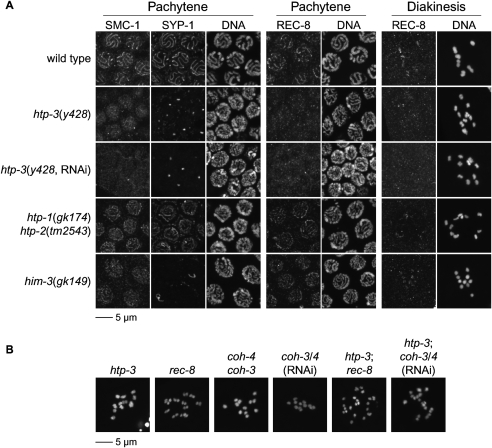
The suppression of spo-11 lethality by rec-8 disruption presented a unique opportunity to identify regulators of cohesin loading and factors that protect SCC at the long arm. We therefore designed and conducted a screen for spo-11 suppressors to identify mutations that allow precocious equational division (Fig. 5A). In other organisms, this screen would identify shugoshin mutations; however, shugoshin is not required for meiosis in C. elegans (Supplemental Fig. S6; de Carvalho et al. 2008), and we therefore expected to identify other factors. From 4000 mutagenized haploid genomes screened, we identified three suppressors. During outcrossing, one suppressor (y428) segregated with ccIs4251, an integrated myo-3∷GFP transgene present in the mutagenized parental strain. Two genes with known meiotic functions reside near ccIs4251: him-1, which encodes the cohesin SMC-1 subunit, and htp-3 (Fig. 5B). Three lines of evidence showed y428 to be an allele of htp-3. First, y428 complemented the conditional lethal allele him-1(e879ts) and the null allele him-1(h55) (data not shown). Second, we identified a single c-to-t transition in the coding region of htp-3 that created Q202Ocher in the 739-amino-acid HTP-3 protein (Fig. 5B). No mutations were found in him-1. The likely null htp-3 mutation disrupts the HORMA domain, conserved among the AE proteins HTP-3, HTP-1/2, and HIM-3 and the yeast ortholog Hop1p. Third, identical phenotypes resulted from y428, the deletion allele htp-3(tm3655), and depletion of HTP-3 by RNAi (Fig. 6B–D; Supplemental Fig. S7). Thus, y428 is a severe loss-of-function or null allele of htp-3.
Figure 5.
A screen for suppressors of spo-11 identified a nonsense allele of htp-3. (A) Hermaphrodites heterozygous for spo-11 were mutagenized with EMS. In the F3 generation, homozygous spo-11 worms were scored for hatching embryos. The chromosomal rearrangement used to balance the spo-11 mutation was marked by an integrated myo-2∷gfp promoter fusion that is expressed in the pharynx. A mutation in lin-2, a gene required for vulval development, was used to confine the progeny of each worm in its uterus. The integrated myo-3∷gfp promoter fusion ccIs4251, expressed in body wall muscles late in embryogenesis, eased the identification of hatching larvae. (B) The spo-11 suppressor y428 segregated with ccIs4251 during outcrossing. Based on their position near ccIs4251 on chromosome I, him-1 and htp-3 were identified as candidates for the mutated gene. A single c-to-t transition was found in the coding region of htp-3. This mutation introduces a premature stop codon that disrupts the conserved HORMA domain.
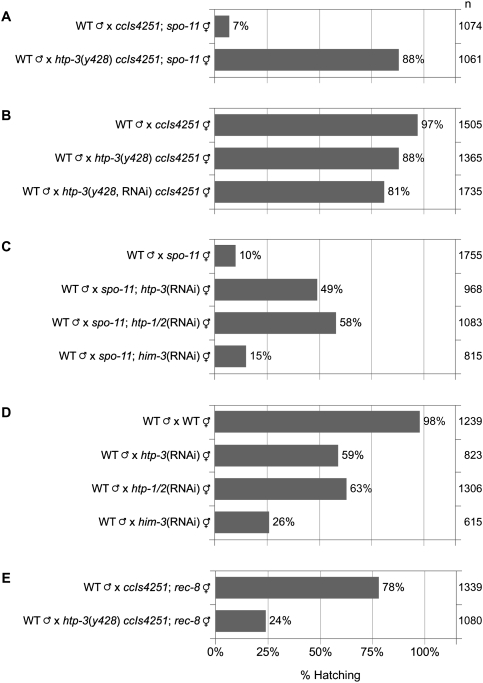
Figure 6.
Disrupting HTP-3 or HTP-1/2 rescues spo-11, but depleting HIM-3 does not. (A) htp-3(y428) reduces the embryonic lethality of spo-11(me44). (A,B) Similar levels of hatching occur in the progeny of htp-3(y428); spo-11(me44) double mutants (A) and htp-3(y428) single mutants (B). Depletion of HTP-3 in htp-3(y428) mutants does not notably increase embryonic lethality. Thus, y428 is a null or strong loss-of-function allele. (C) HTP-3 and HTP-1/2 depletion suppress spo-11(me44), but HIM-3 depletion does not. (D) HIM-3 depletion results in much higher embryonic lethality than depletion of HTP-3 or HTP-1/2. (E) Synergistic embryonic lethality occurs in the broods of htp-3; rec-8 double mutants, implying that HTP-3 is not required for association of all REC-8•cohesin with meiotic chromosomes.
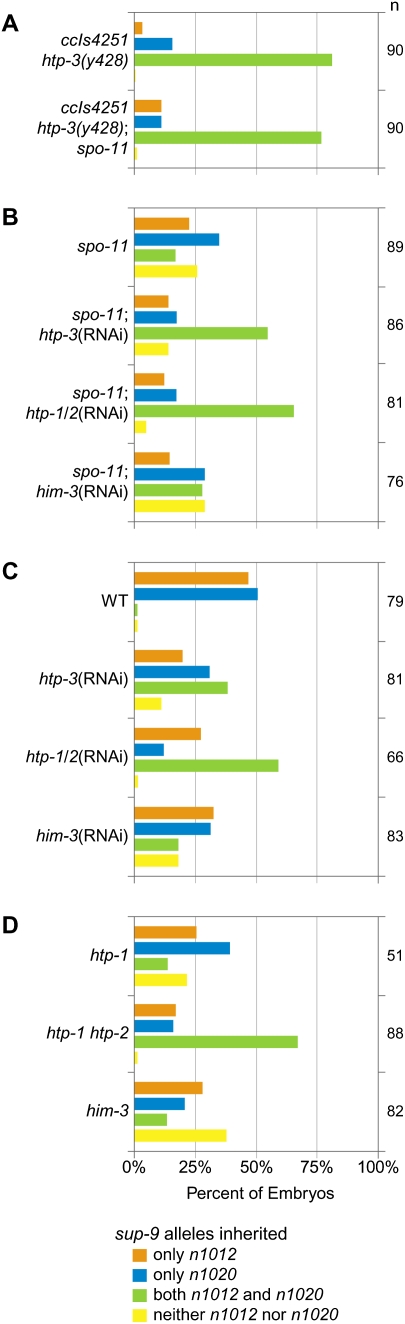
The htp-3(y428) mutation strongly suppressed the lethality of spo-11 worms (Fig. 6A; Supplemental Fig. S4C). While only 7% of spo-11 embryos were viable, 88% of y428; spo-11 embryos were viable. The increased viability of y428; spo-11 embryos resulted from equational sister separation at meiosis I. A single polar body was present in 92% of y428 mutants (Fig. 3D), and both ChrII-RFLPs were detected in 77% of zygotes produced by y428; spo-11 double mutants (Fig. 7A). Strong suppression of spo-11 lethality and equational sister separation also occurred following depletion of htp-3 by RNAi (Figs. 6C, 7B,C). Similar levels of embryonic lethality and equational sister separation occurred in htp-3 single mutants and htp-3; spo-11 double mutants (Figs. 6A–D, 7A–C; Supplemental Fig. S4C,D), as expected because HTP-3 is required for formation of SPO-11-dependent DSBs (Goodyer et al. 2008). We conclude that HTP-3 is required for sister chromatid co-orientation and the persistence of SCC until meiosis II.
Figure 7.
HTP-3 and HTP-1/2 promote the persistent association of sister chromatids until anaphase II. (A–D) Assessment of chromosome segregation in mutants using ChrII-RFLP analysis. (A) Sisters separate equationally in meiosis I of htp-3(y428) and htp-3(y428); spo-11(me44) mutants. (B,C) Depletion of HTP-3 or HTP-1/2 by RNAi in spo-11 (B) or wild-type (C) animals causes precocious, equational sister separation. In contrast, sisters remain together while homologs segregate randomly in meiosis I of him-3(RNAi) or spo-11; him-3(RNAi) animals. (D) Similar results were obtained using null or strong loss-of-function alleles. Although precocious sister separation has been observed in htp-1(gk174) single mutants (Martinez-Perez et al. 2008), homologs partition randomly. Thus, HTP-1 may be required for persistence of SCC but not for sister chromatid co-orientation.
Together, our data and the recent report that sisters disjoin before anaphase II of htp-1 htp-2 double mutants (Martinez-Perez et al. 2008) suggested that the AE as a whole might promote the meiotic pattern of chromosome segregation. However, it was unknown whether the detached sisters observed in htp-1 htp-2 mutants were the result of an equational division. Moreover, the involvement of HIM-3 in sister chromatid co-orientation and the protection of SCC had not been examined. We therefore used RNAi to deplete HTP-1/2 and HIM-3 and then quantified embryonic viability and the inheritance of ChrII-RFLPs (Figs. 6C,D, 7B,C). Depletion of HTP-1/2 resulted in equational sister separation in wild-type and spo-11 mutants, caused only moderate embryonic lethality in wild-type worms, and dramatically reduced the lethality of spo-11 mutants. In contrast, transmission of ChrII-RFLPs during meiosis of him-3(RNAi) and spo-11; him-3(RNAi) animals resembled that of spo-11 single mutants, suggesting that intact homologs segregated randomly in meiosis I. Accordingly, HIM-3 depletion caused severe embryonic lethality in wild-type worms and did not markedly alter the lethality of spo-11 worms (Fig. 6C,D). Thus, HTP-3 and HTP-1/2 are required for sister chromatid co-orientation and association of sisters until meiosis II, but HIM-3 is not.
Results similar to these were obtained when HTP-1/2 or HIM-3 were disrupted using deletion alleles instead of RNAi (Fig. 7D; Supplemental Fig. S4E). However, htp-1 single mutants showed random inheritance of ChrII-RFLPs (Fig. 7D), and >90% of htp-1 embryos died (Supplemental Fig. S4E; Martinez-Perez et al. 2008). Thus, both HTP-1 and HTP-2 must be disrupted to allow equational sister separation.
HTP-3 promotes AE assembly and meiotic cohesin loading
Because HTP-3 and the related AE components HTP-1/2 are required to maintain SCC until meiosis II, we examined the association of AE proteins and cohesin subunits with meiotic chromosomes of htp-3 mutants. We found HTP-3 plays a pre-eminent role in AE assembly. HTP-3 associates with meiotic chromosomes in the absence of HIM-3 (Fig. 8A; Supplemental Fig. S8A; Goodyer et al. 2008) and HTP-1/2 (Fig. 8A; Supplemental Fig. S8A). Conversely, HTP-3 is essential for the association of HIM-3 (Fig. 8C; Supplemental Figs. S7B, S8C; Goodyer et al. 2008) and HTP-1/2 (Fig. 8B; Supplemental Figs. S7B, S8B) with meiotic chromosomes. As noted above (Fig. 4A), COH-3/4 and REC-8 are together required for association of HTP-3 with meiotic chromosomes. Unexpectedly, REC-8 and SMC-1 were undetectable on meiotic chromosomes in htp-3(y428), htp-3(tm3655), and htp-3(y428, RNAi) mutants (Fig. 9A; Supplemental Figs. S7C, S8D), suggesting that HTP-3 in turn promotes the association of cohesin with meiotic chromosomes. Nuclei in premeiotic S-phase and all stages of meiotic prophase are visible in all C. elegans germlines. In htp-3 mutants, axial REC-8 and SMC-1 staining were never detected in these nuclei, although nucleoplasmic staining was present in mitotically proliferating germline stem cells. Thus, HTP-3 is likely required for cohesin loading, although we cannot exclude the possibility that cohesin is unstable or loads very transiently at the onset of meiosis.
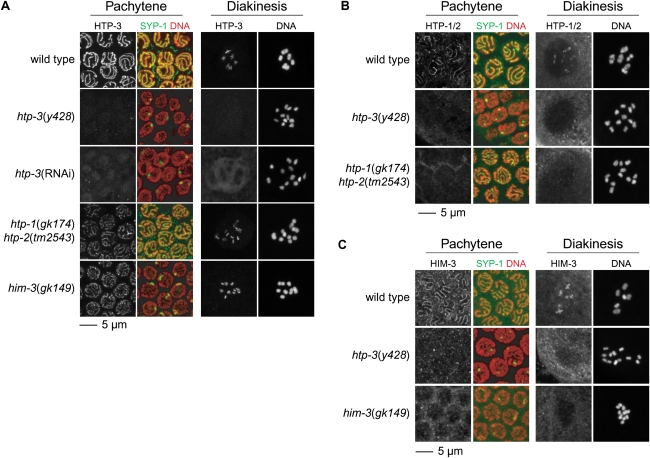
Figure 8.
AE assembly requires HTP-3. (A) HTP-3 and SYP-1 colocalize in pachytene nuclei of wild-type animals. Cruciform HTP-3 staining is observed on diakinesis bivalents. HTP-3 staining persists in htp-1 htp-2 and him-3 mutants. HTP-3 is undetectable in htp-3(y428) animals, and SYP-1 is present in polycomplexes, as previously described in htp-3(RNAi) animals (Goodyer et al. 2008). (A–C) Sister chromatids appear slightly separated in diakinesis univalents of htp-3(y428) and htp-3(RNAi) animals. (B,C) HTP-3 is required for the association of HTP-1/2 and HIM-3 with meiotic chromosomes. HTP-1/2 and HIM-3 colocalize with SYP-1 in pachytene nuclei of wild-type animals. HIM-3 is present along both arms of diakinesis bivalents (C), while HTP-1/2 associate exclusively with the long arm (B). HTP-1/2 and HIM-3 are undetectable on meiotic chromosomes of htp-3(y428) animals. HIM-3 is present in polycomplexes in pachytene nuclei.
Figure 9.
Meiotic cohesin loading requires HTP-3. (A) REC-8 and SMC-1 are present along the axis of pachytene chromosomes and between sister chromatids of diakinesis nuclei. REC-8 and SMC-1 associate with meiotic chromosomes independently of HTP-1/2 and HIM-3, but are undetectable on meiotic chromosomes of htp-3(y428) and htp-3(y428; RNAi) animals. Stretches of SYP-1 are present along chromosomes in htp-1 htp-2 and him-3 mutants, but not htp-3 mutants, in which SYP-1 is only detected in polycomplexes. (B) Residual meiotic cohesin complexes tether sisters together in htp-3 mutants. Discrete sisters are visible in diakinesis univalents of htp-3(y428) mutants but are more tightly associated than sister chromatids in rec-8(ok978) mutants. Sister chromatids cannot be resolved in diakinesis univalents of coh-4 coh-3 double mutants or coh-3/4(RNAi) animals. Disrupting either kleisin in htp-3 mutants further weakens SCC. The gap separating sister chromatids is increased in htp-3; rec-8 and htp-3; coh-3/4(RNAi) double mutants compared with single mutants, and complete detachment of sisters may occur in some univalents of htp-3; rec-8 double mutants. HTP-3 is required for DSB formation, and DNA fragments were not observed in diakinesis nuclei of htp-3; rec-8 or htp-3; coh-3/4(RNAi) mutants, as expected.
HTP-3 is the only known C. elegans AE protein that regulates cohesin early in meiosis. REC-8 and SMC-1 associated with meiotic chromosomes throughout prophase I in him-3 and htp-1 htp-2 mutants (Fig. 9A, Supplemental Fig. S8D). Our data show that HTP-3 and cohesin are interdependent for their association with meiotic chromosomes, and together may form the fundamental core of the meiotic axis, the scaffold with which HTP-1/2 and HIM-3 associate.
Although REC-8 and SMC-1 were undetectable on meiotic chromosomes of htp-3(y428) animals, sister chromatids remained together until anaphase I, indicating that SCC had been established (Figs. 8A–C, 9A,B). COH-3/4•cohesin alone could mediate the remaining SCC. Alternatively, some REC-8•cohesin and COH-3/4•cohesin may both load independently of HTP-3. Our data support the second possibility. Disrupting REC-8 in htp-3(y428) animals caused synergistic lethality, suggesting that REC-8 contributes to SCC in htp-3 mutants (Fig. 6E; Supplemental Fig. S4F,G). While 88% and 78% of embryos from htp-3 and rec-8, mutants, respectively, were viable, only 24% of htp-3; rec-8 mutant embryos were viable. Immunofluorescence observations indicated that REC-8 and COH-3/4 both mediate HTP-3-independent SCC. Sister chromatids appeared more loosely associated in htp-3; coh-3/4(RNAi) diakinesis nuclei than in htp-3 nuclei or coh-3/4(RNAi) nuclei, and some sisters appeared completely detached in htp-3; rec-8 diakinesis nuclei (Fig. 9B). Thus, HTP-3 is required for the chromosomal association of most meiotic cohesin, but some cohesin complexes containing COH-3/4 or REC-8 likely associate with chromosomes independently of HTP-3. Antibodies that recognize COH-3/4 are not currently available, leaving open the possibility that some COH-3/4 loading also requires HTP-3. Nevertheless, the finding that REC-8•cohesin loading occurs by HTP-3-dependent and HTP-3-independent mechanisms raises the intriguing possibility that cohesin complexes with identical subunit composition may perform distinct meiotic roles dictated by the mechanism of their loading onto chromosomes.
Discussion
Our work reveals unexpected roles for cohesin and the AE in meiosis. We show that the paralogous cohesin kleisins REC-8, COH-3, and COH-4 are all required for SCC during worm meiosis, challenging the view of Rec8 as the key meiotic kleisin. Of these kleisins, only REC-8 can maintain SCC after meiosis I. Thus, the C. elegans kleisins are not redundant but rather perform specialized functions. Moreover, molecularly distinct cohesin complexes underlie AE assembly. AE assembly fails in C. elegans only when REC-8, COH-3, and COH-4 are simultaneously disrupted. The involvement of multiple kleisins is likely a common theme in meiosis and may have obscured a widely conserved requirement for cohesin in AE assembly (see below). Finally, we showed that beyond the known roles of AE protein HTP-3 in DSB formation, homolog pairing, and synapsis (Goodyer et al. 2008), HTP-3 is critical for the chromosomal association of all other AE components and REC-8•cohesin, providing the first demonstration of a requirement for AE proteins in cohesin loading. HTP-3 is therefore a molecular link between multiple events that distinguish meiosis from mitosis and ensure the reduction of ploidy during gametogenesis.
AE proteins regulate cohesin to co-orient sister chromatids and maintain SCC until meiosis II
In Saccharomyces cerevisiae, the monopolin complex co-orients sister chromatids independently of cohesin (Toth et al. 2000; Monje-Casas et al. 2007). However, monopolin components have not been identified in other organisms, and cohesin regulation alone may underlie sister chromatid unification. Our data enhance this possibility, since REC-8 and the cohesin regulators HTP-3 and HTP-1/2 are all required for sister chromatid co-orientation in C. elegans.
In fission yeast, tension created when sisters form bipolar attachments on the meiosis I spindle can trigger precocious SCC release (Vaur et al. 2005). The premature loss of SCC in rec-8, htp-3, and htp-1 htp-2 mutants could therefore be an indirect result of defective sister chromatid co-orientation. However, active AIR-2 kinase accumulates between homologs of wild-type animals and between sisters of rec-8 mutants before NEBD, spindle assembly, and tension generation occur. Phosphorylation by AIR-2 is thought to tag REC-8 for destruction. Thus, the pattern of meiotic chromosome segregation is established independently of tension in C. elegans, indicating that REC-8, HTP-3, and HTP-1/2 are required directly to keep sisters together after anaphase I, in addition to their role in sister chromatid co-orientation.
Around the time of fertilization and NEBD, sister chromatids become more tightly associated in bivalents of wild-type worms (Supplemental Fig. S9). This dramatic change in chromosome structure may underlie sister chromatid co-orientation. A comparable change occurs in the univalents of spo-11 mutants, but not those of rec-8 or htp-3 animals. The loose association of sister chromatids following NEBD in rec-8 and htp-3 mutants (Supplemental Fig. S9) correlates with equational sister separation in meiosis I. In contrast, intact homologs of spo-11 and him-3 mutants segregate randomly in meiosis I. Thus, defective sister chromatid co-orientation and precocious loss of SCC are not general properties of univalents but genuinely reflect the need for REC-8, HTP-3 and HTP-1/2 in establishing the meiotic pattern of chromosome segregation.
rec-8 mutants make diploid oocytes but aneuploid sperm
Embryos produced by rec-8, htp-3, and htp-1 htp-2 mutant hermaphrodites inherit a chromatid from each homolog as a result of equational sister separation in meiosis I. Random segregation of these chromatids in meiosis II would cause aneuploidy, but polar body extrusion fails and all chromatids remain within the embryo. Fertilization by a wild-type haploid sperm yields a viable triploid. In contrast, markedly higher levels of embryonic lethality occur when oocytes are fertilized by sperm from rec-8, htp-3, or htp-1 htp-2 mutants. In fact, sperm produced by rec-8 males are highly aneuploid (Supplemental Fig. S10).
In contrast to htp-1 htp-2 (our data and Martinez-Perez et al. 2008), htp-3, or rec-8 mutants, in which premature SCC loss disrupts extrusion of the second polar body, two polar bodies were extruded during meiosis of htp-1 single mutants (Fig. 3D). This was surprising, since detached sister chromatids have been observed prior to anaphase of meiosis II in htp-1 zygotes, suggesting that SCC is precociously lost (Martinez-Perez et al. 2008). Our experiments show that any detached sisters in htp-1 mutants do not result from equational sister separation in meiosis I, which occurs only when both HTP-1 and HTP-2 are disrupted (Fig. 7D). The extent to which sister chromatids come apart in anaphase I of htp-1 single mutants is not known. Perhaps weak SCC persists between some or all sisters until meiosis II, when they separate equationally. This model is consistent with our ChrII-RFLP analysis and would explain the formation of two polar bodies in htp-1 zygotes.
An AE component promotes meiotic cohesin loading
HTP-3 and HTP-1/2 control the association of cohesin with chromosomes at different stages of meiosis. HTP-1/2 act at or near the time of chromosome segregation in meiosis I to prevent kleisin removal (Martinez-Perez et al. 2008). Cohesin appears normal earlier in meiosis of htp-1 htp-2 mutants (Fig. 9A; Supplemental Fig. S8D). In contrast, cohesin was undetectable in all stages of meiosis in htp-3 mutants, indicating that HTP-3 promotes association of cohesin with meiotic chromosomes.
HTP-3 is expressed at the right time for a factor involved in meiotic cohesin loading. Rec8•cohesin loads prior to premeiotic S-phase (Watanabe and Nurse 1999; Watanabe et al. 2001; Eijpe et al. 2003). HTP-3 and REC-8 are present in germline stem cells and nuclei in premeiotic S-phase, but both proteins first appear in axial structures in the transition zone, the C. elegans germline region containing nuclei in leptotene and zygotene of prophase I (Supplemental Fig. S8A,D; Pasierbek et al. 2001; Chan et al. 2003; Hansen et al. 2004; MacQueen et al. 2005; Hayashi et al. 2007; Goodyer et al. 2008). Thus, HTP-3 and cohesin may load onto chromosomes concurrently. Consistent with this model, REC-8 and SMC-1 appeared normal in premeiotic nuclei of htp-3(y428) animals, but were undetectable on meiotic chromosomes (Fig. 9A; Supplemental Fig. S8D; data not shown).
We showed that REC-8•cohesin is required for the persistent association of sister chromatids after anaphase I. The synergistic lethality and increased sister separation in diakinesis nuclei of htp-3; rec-8 double mutants suggests that some REC-8•cohesin associates with meiotic chromosomes independently of HTP-3, yet sister chromatids still separate equationally at anaphase I of htp-3 mutants. HTP-3-independent REC-8•cohesin may be destroyed at anaphase I of htp-3 mutants because HTP-1/2, which normally protect SCC until anaphase II (Martinez-Perez et al. 2008), require HTP-3 for their association with the meiotic axis.
Multiple kleisins function during C. elegans meiosis and perform distinct functions in SCC
Sister chromatids remain together until anaphase I of rec-8 mutants, indicating the persistence of SCC. The visible gap between sisters in rec-8 mutants has been interpreted as evidence of complete loss of SCC and separation of the 12 homologs into 24 individual sister chromatids. However, the published images of diakinesis chromosomes in rec-8(ok978) and rec-8(RNAi) animals are consistent with our interpretation that SCC persists, since pairs of DAPI-staining structures, likely pairs of sister chromatids, are evident (Pasierbek et al. 2001; Kaitna et al. 2002; Alpi et al. 2003; Chan et al. 2003, 2004; Colaiácovo et al. 2003; Reddy and Villeneuve 2004; Hayashi et al. 2007; Penkner et al. 2007; Smolikov et al. 2007). Also, phosH3(S10) resides between sister chromatids in univalents of rec-8(ok978) and rec-8(RNAi) animals (Fig. 1D; Kaitna et al. 2002; AF Severson, unpubl.), and sisters separate equationally in meiosis I, not randomly as expected if sisters were detached. Few chromosome fragments are present relative to the number of meiotic DSBs formed, indicating that some manner of DSB repair occurs, and our data are consistent with the model that meiotic DSBs in rec-8 mutants are repaired using a homologous sequence (e.g., a tethered sister) as a template.
Our data show that the REC-8 paralogs COH-3/4 are also required for meiotic SCC, indicating the role of multiple kleisins in SCC and suggesting the involvement of multiple, distinct cohesin complexes in C. elegans meiosis. All known AE proteins and the SC central element component SYP-1 were undetectable on meiotic chromosomes of rec-8; coh-4 coh-3 triple mutants, demonstrating that worms, like budding yeast have a requirement for cohesin in AE assembly (see below). HTP-3 is present in a similar aggregate in animals deficient for the cohesin subunit SCC-3 (Goodyer et al. 2008) or SMC-1 (Fig. 4A). Thus, SCC is not eliminated in meiosis of rec-8 mutants because COH-3/4 also mediate meiotic SCC.
The existence of meiotic cohesin complexes with different subunit composition, shown by multiple lines of evidence, raised the possibility that meiotic cohesin complexes might contain distinct kleisins to mediate specific meiotic functions. Indeed, COH-3/4 and REC-8 appear to perform different roles in AE assembly, since short, fragmented stretches of AE proteins associate with pachytene chromosomes of coh-4 coh-3 animals but long tracks of AE components are visible in rec-8(ok978) mutants. Moreover, REC-8 alone can co-orient sister chromatids and establish SCC that persists after anaphase I. Thus, the meiotic kleisins of C. elegans are not functionally redundant but perform specialized functions.
Involvement of multiple kleisins is likely to be a common theme in meiosis. Although budding yeast Rec8 is essential for meiotic SCC, the mitotic kleisin Scc1 associates with meiotic chromosomes and potentially has meiotic functions (Klein et al. 1999; Kateneva et al. 2005). The mouse Scc1 ortholog Rad21 may also act during meiosis, since it associates with AE structures of wild-type animals (Xu et al. 2004). Equational sister separation occurs in rec8 mutants of fission yeast, Arabidopsis, and maize, indicating that SCC persists in the absence of Rec8 (Watanabe and Nurse 1999; Chelysheva et al. 2005; Golubovskaya et al. 2006). In fission yeast, this persistent SCC depends on Rad21 (Yokobayashi et al. 2003). The factors that mediate Rec8-independent SCC in Arabidopsis and maize are unknown; however, four kleisin genes have been identified in both organisms (for review, see Hamant et al. 2006). While one kleisin appears to be the true Rec8 ortholog, it is likely that other kleisins also function in meiosis, as we showed for C. elegans COH-3/4.
Interdependent loading of an AE protein and cohesin is fundamental for meiotic axis formation
We have provided the first evidence that an axis protein, HTP-3, promotes meiotic cohesin loading. HTP-3 is also required for assembly of all known AE components. HTP-3 is therefore an important determinant of the molecular composition of the meiotic axis. Mitotic chromosomes are organized around a proteinaceous core that contains SMC proteins, topoisomerase II, and high-mobility group (HMG) proteins (Zickler and Kleckner 1999). The meiotic axis includes Rec8•cohesin and the AE and may be an elaboration of the mitotic chromosomal core.
The relationship between the AE and cohesin has remained controversial. AEs do not form in yeast rec8 mutants, suggesting that AE assembly requires cohesin (Klein et al. 1999). Similarly, SCC-3 depletion disrupts the chromosomal association of HTP-3 and HIM-3 (Pasierbek et al. 2003; Goodyer et al. 2008). However, AE components associate with meiotic axes of rec8 mutants in mice, worms, Arabidopsis, and maize (Bhatt et al. 1999; Colaiácovo et al. 2003; Bannister et al. 2004; Xu et al. 2005; Golubovskaya et al. 2006; Martinez-Perez et al. 2008). AEs form in Coprinus cinereus spo22 mutants and S. cerevisiae cdc6-mn mutants with impaired premeiotic DNA replication, indicating that SCC is not required for AE assembly (Pukkila et al. 1995; Merino et al. 2000; Brar et al. 2009). It has therefore been suggested that AE assembly may require not cohesin but rather other, shared components of mitotic and meiotic chromosomal cores (Kleckner 2006). However, Rec8 associates with the unreplicated chromosomes of cdc6-mn mutants (Brar et al. 2009) and may also associate with meiotic chromosomes in spo22 mutants (H Palmerini and M Zolan, pers. comm.). Thus, cohesin may be essential for AE assembly even if SCC is not.
Our data unambiguously demonstrate that AE assembly in worms requires an intact cohesin complex, even though AE components associate with meiotic chromosomes of rec-8 mutants. Normal AE assembly requires COH-3/4 in addition to REC-8. Thus, cohesin is essential for AE assembly in worms as it is in yeast. The requirement for cohesin in AE assembly is likely widely conserved but masked by the involvement of multiple kleisins. In worms, cohesin and the AE protein HTP-3 are interdependent for their association with meiotic chromosomes, suggesting that cohesin may be an integral part of the AE and that interdependent loading of cohesin and HTP-3 is a principal step in assembly of the meiotic chromosomal axis.
Materials and methods
Strains
Worms strains were grown at 20°C as described in Brenner (1974). N2 Bristol was used as wild type. Strains used in this study are listed in Supplemental Table S3. Because many strains used produce viable polyploids, all experiments were done using known homozygous progeny of heterozygous mothers.
RNAi
Templates for dsRNA production were amplified from genomic DNA by PCR using primers listed in Supplemental Table S4 and then gel purified and reamplified using T7 primers. L4 hermaphrodites were injected with dsRNA at concentrations of 2.5–5 mg/mL and then mated with him-8; mIs10 males. Worms were fixed and stained 72 h post-injection. Embryos were collected 72 h post-injection for ChrII-RFLP assays or 60–84 h post-injection for viability counts.
Microscopy
Immunofluorescence analysis and live imaging were performed as described previously (Chan et al. 2004). The following antibodies were used: rabbit and guinea pig anti-SYP-1 (MacQueen et al. 2002), rabbit anti-HTP-1/2 (Martinez-Perez et al. 2008), guinea pig anti-HTP-3 (MacQueen et al. 2005), rabbit anti-HIM-3 (Zetka et al. 1999), mouse anti-REC-8 (CIM, Arizona State University), rat anti-SMC-1 and rat anti-SMC-3 (Chan et al. 2003), rabbit anti-AIR-2 (Schumacher et al. 1998), and rabbit anti-phosphorylated histone H3 (S10) (Millipore). Polar bodies were counted in one- and two-cell stage embryos fixed as described in Moore et al. (1999), and then mounted in Vectashield (Vector Laboratories) with 2 μg/mL DAPI (Sigma).
Screen for spo-11 suppressors
ccIs4251[myo-3∷GFP] I; spo-11(me44)/nT1[unc-?(n754) let-?(m435)] IV; +/nT1[qIs51] V; lin-2(e1309) X L4 hermaphrodites were mutagenized at room temperature for 4 h in 20 mM EMS (Sigma), washed in M9, and then plated. Two days later, F1 embryos were synchronized by hatchoff. One-thousand heterozygous L4 F1 hermaphrodites were singled and grown two generations at 25°C. Plates were scored for the presence of spo-11 homozygotes with many hatching larvae in their uterus.
ChrII-RFLP assay
To quantify defects in meiotic chromosome segregation, we tracked ChrII-RFLPs produced by the sup-9 alleles n1012 and n1020, which cause no obvious phenotypes other than suppression of certain unc-93 alleles (Greenwald and Horvitz 1986). Hermaphrodites homozygous for the allele being assayed and trans-heterozygous for n1012 and n1020 were mated with him-8; mIs10 males. The next day, embryos were collected for 2–4 h from working crosses. Twenty-four hours later, dead embryos and hatched L1s were transferred to a 96-well plate and lysed in PCR buffer containing 150 μg/mL proteinase K (Roche). DNA spanning the n1012 and n1020 lesions was PCR-amplified with the oligos GACGGAGAATGAGATTCTGCAGG and CGGCTCGTCTTATGAAACGGA. PCR product was digested in three reactions that contained SpeI (diagnostic for n1012), XbaI (diagnostic for n1020), or SpeI and XbaI together (an uncut band is diagnostic for the paternally derived wild-type allele). Data was eliminated from analysis if it did not pass certain criteria (Supplemental Material).
Acknowledgments
We thank A. Dernburg and J. Schumacher for antibodies; A. Villeneuve, the Caenorhabditis Genetics Center, NemaGENETAG, the Gene Knockout Consortium, and the National Bioresource Project for strains; and D. Mets for reading the manuscript. A.F.S was supported by a Helen Hay Whitney Foundation post-doctoral fellowship. B.J.M. is an Investigator of the Howard Hughes Medical Institute.
Footnotes
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.1808809.
Supplemental material is available at http://www.genesdev.org.
References
- Albertson D, Rose A, Villeneuve A. Chromosome organization, mitosis, and meiosis. C. elegans II. In: Riddle DL, et al., editors. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 1997. pp. 47–78. [PubMed] [Google Scholar]
- Alpi A, Pasierbek P, Gartner A, Loidl J. Genetic and cytological characterization of the recombination protein RAD-51 in Caenorhabditis elegans. Chromosoma. 2003;112:6–16. doi: 10.1007/s00412-003-0237-5. [DOI] [PubMed] [Google Scholar]
- Bannister LA, Reinholdt LG, Munroe RJ, Schimenti JC. Positional cloning and characterization of mouse mei8, a disrupted allele of the meiotic cohesin Rec8. Genesis. 2004;40:184–194. doi: 10.1002/gene.20085. [DOI] [PubMed] [Google Scholar]
- Bergerat A, de Massy B, Gadelle D, Varoutas PC, Nicolas A, Forterre P. An atypical topoisomerase II from Archaea with implications for meiotic recombination. Nature. 1997;386:414–417. doi: 10.1038/386414a0. [DOI] [PubMed] [Google Scholar]
- Bhatt AM, Lister C, Page T, Fransz P, Findlay K, Jones GH, Dickinson HG, Dean C. The DIF1 gene of Arabidopsis is required for meiotic chromosome segregation and belongs to the REC8/RAD21 cohesin gene family. Plant J. 1999;19:463–472. doi: 10.1046/j.1365-313x.1999.00548.x. [DOI] [PubMed] [Google Scholar]
- Brar GA, Hochwagen A, Ee LS, Amon A. The multiple roles of cohesin in meiotic chromosome morphogenesis and pairing. Mol Biol Cell. 2009;20:1030–1047. doi: 10.1091/mbc.E08-06-0637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buonomo SB, Clyne RK, Fuchs J, Loidl J, Uhlmann F, Nasmyth K. Disjunction of homologous chromosomes in meiosis I depends on proteolytic cleavage of the meiotic cohesin Rec8 by separin. Cell. 2000;103:387–398. doi: 10.1016/s0092-8674(00)00131-8. [DOI] [PubMed] [Google Scholar]
- Chan RC, Chan A, Jeon M, Wu TF, Pasqualone D, Rougvie AE, Meyer BJ. Chromosome cohesion is regulated by a clock gene paralogue TIM-1. Nature. 2003;423:1002–1009. doi: 10.1038/nature01697. [DOI] [PubMed] [Google Scholar]
- Chan RC, Severson AF, Meyer BJ. Condensin restructures chromosomes in preparation for meiotic divisions. J Cell Biol. 2004;167:613–625. doi: 10.1083/jcb.200408061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chelysheva L, Diallo S, Vezon D, Gendrot G, Vrielynck N, Belcram K, Rocques N, Marquez-Lema A, Bhatt AM, Horlow C, et al. AtREC8 and AtSCC3 are essential to the monopolar orientation of the kinetochores during meiosis. J Cell Sci. 2005;118:4621–4632. doi: 10.1242/jcs.02583. [DOI] [PubMed] [Google Scholar]
- Clejan I, Boerckel J, Ahmed S. Developmental modulation of nonhomologous end joining in Caenorhabditis elegans. Genetics. 2006;173:1301–1317. doi: 10.1534/genetics.106.058628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colaiácovo MP, MacQueen AJ, Martinez-Perez E, McDonald K, Adamo A, La Volpe A, Villeneuve AM. Synaptonemal complex assembly in C. elegans is dispensable for loading strand-exchange proteins but critical for proper completion of recombination. Dev Cell. 2003;5:463–474. doi: 10.1016/s1534-5807(03)00232-6. [DOI] [PubMed] [Google Scholar]
- de Carvalho CE, Zaaijer S, Smolikov S, Gu Y, Schumacher JM, Colaiácovo MP. LAB-1 antagonizes the Aurora B kinase in C. elegans. Genes & Dev. 2008;22:2869–2885. doi: 10.1101/gad.1691208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Cruz IP, Levin JZ, Cummins C, Anderson P, Horvitz HR. sup-9, sup-10, and unc-93 may encode components of a two-pore K+ channel that coordinates muscle contraction in Caenorhabditis elegans. J Neurosci. 2003;23:9133–9145. doi: 10.1523/JNEUROSCI.23-27-09133.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dernburg AF, McDonald K, Moulder G, Barstead R, Dresser M, Villeneuve AM. Meiotic recombination in C. elegans initiates by a conserved mechanism and is dispensable for homologous chromosome synapsis. Cell. 1998;94:387–398. doi: 10.1016/s0092-8674(00)81481-6. [DOI] [PubMed] [Google Scholar]
- Dunleavy E, Pidoux A, Allshire R. Centromeric chromatin makes its mark. Trends Biochem Sci. 2005;30:172–175. doi: 10.1016/j.tibs.2005.02.007. [DOI] [PubMed] [Google Scholar]
- Eijpe M, Offenberg H, Jessberger R, Revenkova E, Heyting C. Meiotic cohesin REC8 marks the axial elements of rat synaptonemal complexes before cohesins SMC1β and SMC3. J Cell Biol. 2003;160:657–670. doi: 10.1083/jcb.200212080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golubovskaya IN, Hamant O, Timofejeva L, Wang CJ, Braun D, Meeley R, Cande WZ. Alleles of afd1 dissect REC8 functions during meiotic prophase I. J Cell Sci. 2006;119:3306–3315. doi: 10.1242/jcs.03054. [DOI] [PubMed] [Google Scholar]
- Goodyer W, Kaitna S, Couteau F, Ward JD, Boulton SJ, Zetka M. HTP-3 links DSB formation with homolog pairing and crossing over during C. elegans meiosis. Dev Cell. 2008;14:263–274. doi: 10.1016/j.devcel.2007.11.016. [DOI] [PubMed] [Google Scholar]
- Greenwald I, Horvitz HR. A visible allele of the muscle gene sup-10 of C. elegans. Genetics. 1986;113:63–72. doi: 10.1093/genetics/113.1.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamant O, Ma H, Cande WZ. Genetics of meiotic prophase I in plants. Annu Rev Plant Biol. 2006;57:267–302. doi: 10.1146/annurev.arplant.57.032905.105255. [DOI] [PubMed] [Google Scholar]
- Hansen D, Hubbard E, Schedl T. Multi-pathway control of the proliferation versus meiotic development decision in the Caenorhabditis elegans. Dev Biol. 2004;268:342–357. doi: 10.1016/j.ydbio.2003.12.023. [DOI] [PubMed] [Google Scholar]
- Hayashi M, Chin GM, Villeneuve AM. C. elegans germ cells switch between distinct modes of double-strand break repair during meiotic prophase progression. PLoS Genet. 2007;3:e191. doi: 10.1371/journal.pgen.0030191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollingsworth NM, Byers B. HOP1: A yeast meiotic pairing gene. Genetics. 1989;121:445–462. doi: 10.1093/genetics/121.3.445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu JY, Sun ZW, Li X, Reuben M, Tatchell K, Bishop DK, Grushcow JM, Brame CJ, Caldwell JA, Hunt DF, et al. Mitotic phosphorylation of histone H3 is governed by Ipl1/aurora kinase and Glc7/PP1 phosphatase in budding yeast and nematodes. Cell. 2000;102:279–291. doi: 10.1016/s0092-8674(00)00034-9. [DOI] [PubMed] [Google Scholar]
- Kaitna S, Pasierbek P, Jantsch M, Loidl J, Glotzer M. The aurora B kinase AIR-2 regulates kinetochores during mitosis and is required for separation of homologous chromosomes during meiosis. Curr Biol. 2002;12:798–812. doi: 10.1016/s0960-9822(02)00820-5. [DOI] [PubMed] [Google Scholar]
- Kateneva AV, Konovchenko AA, Guacci V, Dresser ME. Recombination protein Tid1p controls resolution of cohesin-dependent linkages in meiosis in Saccharomyces cerevisiae. J Cell Biol. 2005;171:241–253. doi: 10.1083/jcb.200505020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeney S, Giroux CN, Kleckner N. Meiosis-specific DNA double-strand breaks are catalyzed by Spo11, a member of a widely conserved protein family. Cell. 1997;88:375–384. doi: 10.1016/s0092-8674(00)81876-0. [DOI] [PubMed] [Google Scholar]
- Kleckner N. Chiasma formation: Chromatin/axis interplay and the role(s) of the synaptonemal complex. Chromosoma. 2006;115:175–194. doi: 10.1007/s00412-006-0055-7. [DOI] [PubMed] [Google Scholar]
- Klein F, Mahr P, Galova M, Buonomo SB, Michaelis C, Nairz K, Nasmyth K. A central role for cohesins in sister chromatid cohesion, formation of axial elements, and recombination during yeast meiosis. Cell. 1999;98:91–103. doi: 10.1016/S0092-8674(00)80609-1. [DOI] [PubMed] [Google Scholar]
- MacQueen AJ, Colaiácovo MP, McDonald K, Villeneuve AM. Synapsis-dependent and -independent mechanisms stabilize homolog pairing during meiotic prophase in C. elegans. Genes & Dev. 2002;16:2428–2442. doi: 10.1101/gad.1011602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacQueen AJ, Phillips CM, Bhalla N, Weiser P, Villeneuve AM, Dernburg AF. Chromosome sites play dual roles to establish homologous synapsis during meiosis in C. elegans. Cell. 2005;123:1037–1050. doi: 10.1016/j.cell.2005.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marston AL, Amon A. Meiosis: Cell-cycle controls shuffle and deal. Nat Rev Mol Cell Biol. 2004;5:983–997. doi: 10.1038/nrm1526. [DOI] [PubMed] [Google Scholar]
- Martin JS, Winkelmann N, Petalcorin MI, McIlwraith MJ, Boulton SJ. RAD-51-dependent and -independent roles of a Caenorhabditis elegans BRCA2-related protein during DNA double-strand break repair. Mol Cell Biol. 2005;25:3127–3139. doi: 10.1128/MCB.25.8.3127-3139.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Perez E, Schvarzstein M, Barroso C, Lightfoot J, Dernburg AF, Villeneuve AM. Crossovers trigger a remodeling of meiotic chromosome axis composition that is linked to two-step loss of sister chromatid cohesion. Genes & Dev. 2008;22:2886–2901. doi: 10.1101/gad.1694108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merino ST, Cummings WJ, Acharya SN, Zolan ME. Replication-dependent early meiotic requirement for Spo11 and Rad50. Proc Natl Acad Sci. 2000;97:10477–10482. doi: 10.1073/pnas.190346097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monje-Casas F, Prabhu VR, Lee BH, Boselli M, Amon A. Kinetochore orientation during meiosis is controlled by Aurora B and the monopolin complex. Cell. 2007;128:477–490. doi: 10.1016/j.cell.2006.12.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore LL, Morrison M, Roth MB. HCP-1, a protein involved in chromosome segregation, is localized to the centromere of mitotic chromosomes in Caenorhabditis elegans. J Cell Biol. 1999;147:471–480. doi: 10.1083/jcb.147.3.471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nabeshima K, Villeneuve AM, Colaiácovo MP. Crossing over is coupled to late meiotic prophase bivalent differentiation through asymmetric disassembly of the SC. J Cell Biol. 2005;168:683–689. doi: 10.1083/jcb.200410144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onn I, Heidinger-Pauli JM, Guacci V, Unal E, Koshland DE. Sister chromatid cohesion: A simple concept with a complex reality. Annu Rev Cell Dev Biol. 2008;24:105–129. doi: 10.1146/annurev.cellbio.24.110707.175350. [DOI] [PubMed] [Google Scholar]
- Padmore R, Cao L, Kleckner N. Temporal comparison of recombination and synaptonemal complex formation during meiosis in S. cerevisiae. Cell. 1991;66:1239–1256. doi: 10.1016/0092-8674(91)90046-2. [DOI] [PubMed] [Google Scholar]
- Pasierbek P, Jantsch M, Melcher M, Schleiffer A, Schweizer D, Loidl J. A Caenorhabditis elegans cohesion protein with functions in meiotic chromosome pairing and disjunction. Genes & Dev. 2001;15:1349–1360. doi: 10.1101/gad.192701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasierbek P, Fodermayr M, Jantsch V, Jantsch M, Schweizer D, Loidl J. The Caenorhabditis elegans SCC-3 homologue is required for meiotic synapsis and for proper chromosome disjunction in mitosis and meiosis. Exp Cell Res. 2003;289:245–255. doi: 10.1016/s0014-4827(03)00266-0. [DOI] [PubMed] [Google Scholar]
- Penkner A, Portik-Dobos Z, Tang L, Schnabel R, Novatchkova M, Jantsch V, Loidl J. A conserved function for a Caenorhabditis elegans Com1/Sae2/CtIP protein homolog in meiotic recombination. EMBO J. 2007;26:5071–5082. doi: 10.1038/sj.emboj.7601916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pukkila PJ, Shannon KB, Skrzynia C. Independent synaptic behavior of sister chromatids in Coprinus cinereus. Can J Bot. 1995;73:S215–S220. [Google Scholar]
- Reddy KC, Villeneuve AM. C. elegans HIM-17 links chromatin modification and competence for initiation of meiotic recombination. Cell. 2004;118:439–452. doi: 10.1016/j.cell.2004.07.026. [DOI] [PubMed] [Google Scholar]
- Rogers E, Bishop JD, Waddle JA, Schumacher JM, Lin R. The aurora kinase AIR-2 functions in the release of chromosome cohesion in Caenorhabditis elegans meiosis. J Cell Biol. 2002;157:219–229. doi: 10.1083/jcb.200110045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumacher JM, Golden A, Donovan PJ. AIR-2: An Aurora/Ipl1-related protein kinase associated with chromosomes and midbody microtubules is required for polar body extrusion and cytokinesis in Caenorhabditis elegans embryos. J Cell Biol. 1998;143:1635–1646. doi: 10.1083/jcb.143.6.1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smolikov S, Eizinger A, Schild-Prufert K, Hurlburt A, McDonald K, Engebrecht J, Villeneuve AM, Colaiácovo MP. SYP-3 restricts synaptonemal complex assembly to bridge paired chromosome axes during meiosis in Caenorhabditis elegans. Genetics. 2007;176:2015–2025. doi: 10.1534/genetics.107.072413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toth A, Rabitsch KP, Galova M, Schleiffer A, Buonomo SB, Nasmyth K. Functional genomics identifies monopolin: A kinetochore protein required for segregation of homologs during meiosis i. Cell. 2000;103:1155–1168. doi: 10.1016/s0092-8674(00)00217-8. [DOI] [PubMed] [Google Scholar]
- Vagnarelli P, Ribeiro SA, Earnshaw WC. Centromeres: Old tales and new tools. FEBS Lett. 2008;582:1950–1959. doi: 10.1016/j.febslet.2008.04.014. [DOI] [PubMed] [Google Scholar]
- Vaur S, Cubizolles F, Plane G, Genier S, Rabitsch PK, Gregan J, Nasmyth K, Vanoosthuyse V, Hardwick KG, Javerzat JP. Control of Shugoshin function during fission-yeast meiosis. Curr Biol. 2005;15:2263–2270. doi: 10.1016/j.cub.2005.11.034. [DOI] [PubMed] [Google Scholar]
- Watanabe Y, Nurse P. Cohesin Rec8 is required for reductional chromosome segregation at meiosis. Nature. 1999;400:461–464. doi: 10.1038/22774. [DOI] [PubMed] [Google Scholar]
- Watanabe Y, Yokobayashi S, Yamamoto M, Nurse P. Pre-meiotic S phase is linked to reductional chromosome segregation and recombination. Nature. 2001;409:359–363. doi: 10.1038/35053103. [DOI] [PubMed] [Google Scholar]
- Xu H, Beasley M, Verschoor S, Inselman A, Handel MA, McKay MJ. A new role for the mitotic RAD21/SCC1 cohesin in meiotic chromosome cohesion and segregation in the mouse. EMBO Rep. 2004;5:378–384. doi: 10.1038/sj.embor.7400121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H, Beasley MD, Warren WD, van der Horst GT, McKay MJ. Absence of mouse REC8 cohesin promotes synapsis of sister chromatids in meiosis. Dev Cell. 2005;8:949–961. doi: 10.1016/j.devcel.2005.03.018. [DOI] [PubMed] [Google Scholar]
- Yokobayashi S, Yamamoto M, Watanabe Y. Cohesins determine the attachment manner of kinetochores to spindle microtubules at meiosis I in fission yeast. Mol Cell Biol. 2003;23:3965–3973. doi: 10.1128/MCB.23.11.3965-3973.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zetka MC, Kawasaki I, Strome S, Muller F. Synapsis and chiasma formation in Caenorhabditis elegans require HIM-3, a meiotic chromosome core component that functions in chromosome segregation. Genes & Dev. 1999;13:2258–2270. doi: 10.1101/gad.13.17.2258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zickler D, Kleckner N. Meiotic chromosomes: Integrating structure and function. Annu Rev Genet. 1999;33:603–754. doi: 10.1146/annurev.genet.33.1.603. [DOI] [PubMed] [Google Scholar]