Abstract
The obligate intracellular human pathogen Chlamydia trachomatis undergoes a complex developmental program involving transition between two forms: the infectious elementary body (EB), and the rapidly dividing reticulate body (RB). However, the regulators controlling this development have not been identified. To uncover potential regulators of transcription in C. trachomatis, we screened a C. trachomatis genomic library for sequences encoding proteins that interact with RNA polymerase (RNAP). We report the identification of one such protein, CT663, which interacts with the β and σ subunits of RNAP. Specifically, we show that CT663 interacts with the flap domain of the β subunit (β-flap) and conserved region 4 of the primary σ subunit (σ66 in C. trachomatis). We find that CT663 inhibits σ66-dependent (but not σ28-dependent) transcription in vitro, and we present evidence that CT663 exerts this effect as a component of the RNAP holoenzyme. The analysis of C. trachomatis-infected cells reveals that CT663 begins to accumulate at the commencement of the RB-to-EB transition. Our findings suggest that CT663 functions as a negative regulator of σ66-dependent transcription, facilitating a global change in gene expression. The strategy used here is generally applicable in cases where genetic tools are unavailable.
Keywords: Chlamydia trachomatis, RNA polymerase, CT663, β subunit, σ factors, anti-σ factors
Chlamydia trachomatis is the most prevalent sexually transmitted bacterial pathogen worldwide. In the United States alone, the annual healthcare cost associated with C. trachomatis infection and its consequences is more than $2 billion (for review, see Brunham and Rey-Ladino 2005). The ability of C. trachomatis to invade and multiply within host cells is essential for pathogenesis, but the precise mechanisms underlying the disease process remain unknown. An obligate intracellular pathogen, C. trachomatis undergoes morphological transition between an elementary body (EB), which is the small, spore-like, metabolically inactive, infectious form, and a reticulate body (RB), which is the large, metabolically active but noninfectious form (for reviews, see Moulder 1991; Hatch 1999). Thus, the growth of C. trachomatis can be divided into stages, which are characterized by specific gene expression profiles: early-stage (EB-to-RB differentiation), mid-stage (RB growth and division), and late-stage (RB-to-EB differentiation and preparation for a subsequent infection cycle) (Shaw et al. 2000; Belland et al. 2003; Nicholson et al. 2003). Elucidating the mechanisms by which stage-specific gene expression is regulated is critical for understanding the intracellular parasitism of C. trachomatis.
Transcription is the first step in gene expression and a key point of regulation. The bacterial RNA polymerase (RNAP) consists of a core enzyme (subunit composition α2ββ′ω) and a σ factor that confers on the enzyme the ability to direct promoter-specific transcription (for reviews, see Gross et al. 1998; Murakami and Darst 2003). RNAP in C. trachomatis resembles other bacterial RNAPs except that it lacks an identifiable ω subunit (Stephens et al. 1998). C. trachomatis genomes encode three σ factors: the primary σ factor (σ66), and two alternative σ factors (σ54 and σ28); both σ66 and σ28 are members of the so-called σ70 (the primary σ factor in Escherichia coli) family, which includes the preponderance of bacterial σ factors. All primary σ factors share four regions of conserved sequence (regions 1–4), whereas only regions 2 and 4 are well conserved among all of the alternative σ factors in the σ70 family (Lonetto et al. 1992; Paget and Helmann 2003). Regions 2 and 4 of σ contain DNA-binding domains that recognize the promoter −10 and −35 elements, respectively. In addition, regions 2 and 4 contain core-binding determinants that mediate interaction with the coiled-coil domain of the β′ subunit (β′cc) and the flexible flap domain of the β subunit (β-flap), respectively (Arthur and Burgess 1998; Young et al. 2001; Kuznedelov et al. 2002). The interaction between σ region 2 and the β′cc is essential for formation of the RNAP holoenzyme, whereas the interaction between σ region 4 and β-flap is required to properly position σ region 4 with respect to σ region 2, permitting σ regions 2 and 4 to make simultaneous contact with the −10 and −35 promoter elements (Kuznedelov et al. 2002).
As well as serving as an anchor point for σ region 4 during transcription initiation, the β-flap helps delineate the RNA exit channel; thus, during transcription elongation the nascent RNA emerges from underneath the β-flap (Vassylyev et al. 2007). Accordingly, the β-flap can influence both transcription pausing and termination through interaction with the nascent RNA (Wang et al. 1997; Toulokhonov et al. 2001; Toulokhonov and Landick 2003). As the focal point of important events during all stages of the transcription cycle, the β-flap is a plausible target for regulatory factors that influence transcription at one or another stage. To date, several bacteriophage encoded transcription regulators have been identified that interact directly with the β-flap, including two T4-encoded regulators, gp33 (Nechaev et al. 2004) and AsiA (Yuan et al. 2009), which each modulate transcription initiation, and the λ Q protein (Deighan et al. 2008), which is a transcription anti-termination factor.
Hypothesizing that global regulators of transcription play important roles in the developmental cycle of C. trachomatis, we sought to identify C. trachomatis proteins that interact with RNAP, using the β-flap as bait. The screen led to the identification of CT663, a hypothetical component of the type III secretion machinery, which interacts with both the β-flap and region 4 of σ66. We provide genetic and biochemical evidence that CT663 serves as a negative regulator of σ66-dependent transcription, likely by disrupting the σ66 region 4/β-flap interaction in the context of the RNAP holoenzyme. CT663 is detected toward the end of the middle stage of C. trachomatis development, and its levels continue to increase throughout the late stage. We suggest that CT663 may play a role in the switch in transcription profile that accompanies the RB-to-EB transition.
Results
A screen for C. trachomatis proteins that interact with the β-flap domain of RNAP
In order to identify proteins that interact with the β-flap domain of C. trachomatis RNAP (β-flapCt, β residues 798–887), we took advantage of a transcription activation-based two-hybrid system (Fig. 1A; Dove et al. 1997; Dove and Hochschild 2001). In this assay, contact between a protein domain fused to a component of RNAP (here the N-terminal domain of the α subunit; αNTD) and a partner protein fused to a DNA-binding protein (here, the CI protein of bacteriophage λ; λCI) activates transcription of a lacZ reporter gene under the control of a test promoter bearing an upstream recognition site for the DNA-binding protein (here, a λ operator). To perform the screen, we transformed reporter strain KS1 (Dove et al. 1997) harboring plasmid pACλCI-β-flapCt (directing the synthesis of λCI fused to the β-flap domain of C. trachomatis RNAP) with a pBRα-based library of C. trachomatis serovar F genomic DNA (see the Materials and Methods). The transformants were plated on indicator medium (see the Materials and Methods) to identify clones expressing the lacZ reporter gene at elevated levels, indicative of an interaction between the β-flap moiety of the λCI-β-flapCt fusion protein and a C. trachomatis-encoded polypeptide fused in-frame to the αNTD (Fig. 1A). This genetic screen led to the identification of four candidate plasmids that encoded in-frame fusion proteins (see the Supplemental Material).
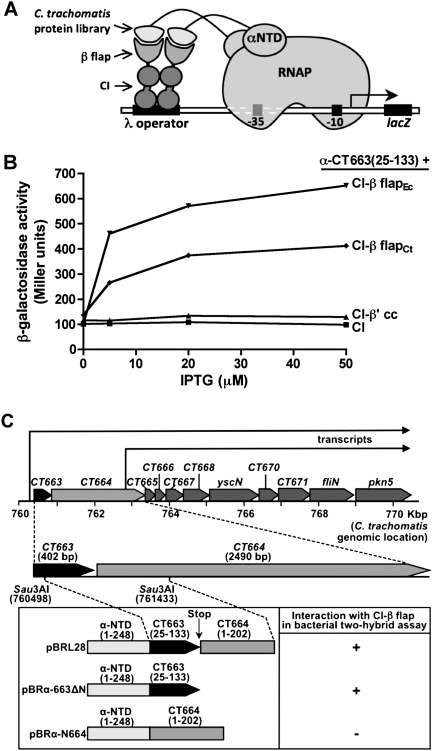
Figure 1.
CT663 interacts with the β-flap. (A) Diagram shows a transcription activation-based two-hybrid assay that was adopted to screen a C. trachomatis genomic DNA library for proteins that could interact with the β-flap domain of C. trachomatis RNAP. An interaction between a protein from the library, fused in-frame to the αNTD, and the β-flap, fused to the bacteriophage λCI protein, activates transcription from a test promoter that bears a λ operator centered 62 bp upstream of the start site of the lac core promoter. In reporter strain KS1, which was used for the screen, test promoter placOR2-62 is located on the chromosome and drives the expression of a linked lacZ gene. In reporter strain FW102 OL2-62, which was used for the quantitative β-gal assays (shown in B), test promoter placOL2-62 is located on an F′ episome and drives the expression of a linked lacZ gene. (B) Results of β-gal performed with FW102 OL2-62 cells that contained two compatible plasmids: one that encoded the α-CT663(25–133) fusion protein, and another that encoded λCI or one of three fusion proteins: λCI-β-flapCt (C. trachomatis β residues 798–887 fused to λCI), λCI-β-flapEc (E. coli β residues 858–946 fused to λCI) or λCI-β′cc (E. coli β′ residues 262–309 fused to λCI). The plasmids directed the synthesis of the fusion proteins under the control of IPTG-inducible promoters and the cells were grown in the absence of IPTG, or in the presence of 5, 20, or 50 μM IPTG. Shown is a single experiment; the values from three independent experiments differed by <5%. (C) Schematic representation of the CT663 chromosomal region and bacterial two-hybrid fusion proteins. The numbers represent a scale (kilobase pairs) depicting the positions of the various genes (labeled above the box arrows) on the C. trachomatis serovar D genome (Stephens et al. 1998). The CT663 gene is the first gene of a putative type III secretion operon, and the arrows indicate the proposed transcripts of this operon (Hefty and Stephens 2007). The “blow-up” schematic shows the Sau3AI fragment that was cloned in pBRL28. This fragment contains a 330-bp fragment of CT663 (including a stop codon as indicated by the vertical arrow) followed by 3-bp (GGG) of intergenic sequence and a 606-bp 5′-portion of the CT664 gene (including a start codon). As the CT663 fragment was cloned in-frame with the rpoA gene (encodes α subunit of E. coli RNAP) in pBRL28, and contained the native stop codon, the resulting gene fusion directs the synthesis of an α-CT663(25–133) protein chimera, in which residues 25–133 of CT663 are fused to the αNTD (α residues 1–248) as indicated. A derivative of pBRL28 was made that lacks the CT664 gene fragment; thus, the resultant plasmid pBRα-663ΔN directs the synthesis of the same α-CT663(25–133) protein chimera as pBRL28. Plasmid pBRα-N664 directs the synthesis of an α-CT664(1–202) protein chimera, in which residues 1–202 of CT664 are fused in-frame to the αNTD. We detected a stimulatory effect (indicated as plus [+] sign) of the α-CT663(25–133) fusion protein (encoded by both pBRL28 and pBRα-663ΔN) on lacZ transcription in the presence of the λCI-β-flap fusion protein in the bacterial-two hybrid reporter strain, but did not detect a stimulatory effect (indicated as minus [−] sign) of the α-CT664(1–202) fusion protein on lacZ transcription in the presence of the λCI-β-flap fusion protein.
One plasmid (pBRL28) (Fig. 1C) that was isolated twice contained the αNTD coding sequence fused in-frame to a 330-base-pair (bp) gene fragment (including a stop codon) followed by 3 bp of intergenic sequence and a second 606-bp gene fragment (including a start codon). This region corresponds to position 760498–761436 of the C. trachomatis genome sequence of serovar D (GenBank ID: NC_000117) (Stephens et al. 1998) and encodes residues 25–133 of CT663 and residues 1–202 of CT664. Both CT663 and CT664 encode proteins of unknown function. By protein BLAST analysis, CT663 is classified as a member of the CesT (a type III secretion chaperone in enteropathogenic E. coli) (see the Discussion) superfamily, whereas CT664 is classified as a member of the forkhead-associated domain (FHA) family.
We quantified the effect of the α-CT663(25–133) fusion protein encoded by pBRL28 on transcription of the lacZ reporter gene in strain FW102 OL2-62 by performing β-galactosidase (β-gal) assays. Figure 1B indicates that the α-CT663(25–133) fusion protein stimulated transcription up to 3.5-fold in the presence of the λCI-β-flapCt fusion protein and up to 10-fold in the presence of a λCI-β-flap fusion protein bearing residues 858–946 from the E. coli β subunit (λCI-β-flapEc). The C. trachomatis and E. coli β-flap domains used in the two-hybrid analysis share 62.2% amino acid identity and 73.8% similarity (Supplemental Fig. S1). Control assays indicated that there was no stimulatory effect of the α-CT663(25–133) fusion protein on lacZ transcription in the presence of λCI fused to another domain of E. coli RNAP (the β′cc) or λCI lacking any fused moiety (Fig. 1B). Immunoblot analysis with an anti-λCI antibody revealed that λCI and the three λCI fusion proteins were present at similar intracellular levels (data not shown).
In order to confirm that the α-CT663(25–133) fusion protein was solely responsible for the observed stimulatory effect of plasmid pBRL28 on lacZ expression in strain FW102 OL2-62 (Fig. 1), we attempted to construct a plasmid that lacked the CT664 coding sequences and directed the synthesis of the αNTD fused to full-length CT663(1–133). However, we only obtained clones with frameshift mutations within the CT663 gene, suggesting that the full-length α-CT663 fusion protein was toxic to the cell. Nevertheless, we were able to rule out any effect of the downstream CT664 gene on the CT663/β-flap interaction by constructing plasmid pBRα-663ΔN, which encodes the α-CT663(25–133) fusion protein, but lacks the CT664 gene (Fig. 1C). This plasmid mediated a similar stimulatory effect on lacZ transcription in the presence of either the λCI-β-flapCt or the λCI-β-flapEc fusion protein as did pBRL28 (data not shown). We also constructed plasmid pBRα-N664, which encodes the αNTD fused to the N-terminal region of the CT664 protein (residues 1–202) (Fig. 1C). We detected no stimulatory effect of the α-N664 fusion protein on lacZ transcription in the presence of either λCI-β-flap fusion protein, even though stable production of the α-N664 fusion protein was confirmed (data not shown). Collectively, these results indicate that the CT663(25–133) moiety of the α-CT663(25–133) fusion protein can bind to the β-flap domain of RNAP.
The β-flap tip helix (FTH) is the CT663-binding determinant on the β subunit
We next used the two-hybrid assay to identify specific amino acid residues within the β-flap that mediate the CT663/β-flap interaction. First, we tested the effect of removing the FTH (an α helix encompassing β residues 900–909), which is the primary determinant for the binding of σ region 4 to the β-flap (Geszvain et al. 2004). As shown in Figure 2A, removal of the FTH eliminated any detectable interaction between the CT663 moiety of the α-CT663(25–133) fusion protein and the β-flap moiety of the λCI-β-flapEc fusion protein, indicating that CT663 and σ region 4 interact with overlapping determinants of the β-flap.
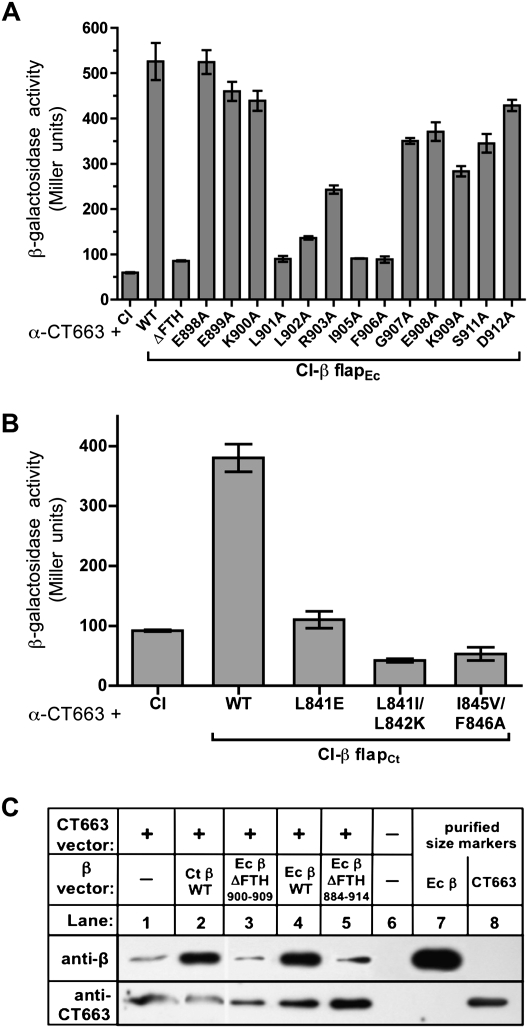
Figure 2.
The β FTH is an essential determinant of the CT663/β-flap interaction. (A) Amino acid substitutions in the E. coli β-flap that weaken the CT663/β-flap interaction. Results of β-gal assays performed with FW102 OL2-62 cells containing one plasmid that encoded the α-CT663(25–133) fusion protein and a compatible plasmid that encoded either λCI or the indicated λCI-β-flap variant. The plasmids directed the synthesis of the fusion proteins under the control of IPTG-inducible promoters and the cells were grown in the presence of 20 μM IPTG. The bar graph shows the averages of three independent measurements and standard deviations. (B) Amino acid substitutions in the C. trachomatis β-flap that weaken the CT663/β-flap interaction. Results of β-gal assays performed with FW102 OL2-62 cells containing one plasmid that encoded the α-CT663(25–133) fusion protein and a compatible plasmid that encoded either λCI or the indicated λCI-β-flap variant. The plasmids directed the synthesis of the fusion proteins under the control of IPTG-inducible promoters and the cells were grown in the presence of 20 μM IPTG. The bar graph shows the averages of three independent measurements and standard deviations. (C). Immunoprecipitation of CT663/β complexes. Immunoblot showing the results of an immunoprecipitation assay from E. coli C2566 cells that contained two compatible vectors: one that encoded CT663, and another that encoded E. coli β (Ecβ), or C. trachomatis β (Ctβ), or one of two deletion derivatives of E. coli β that lacked the β FTH (either β Δ900–909 or β Δ884–914). For each plasmid type, the minus sign (−) refers to an empty vector control. After the induction of protein synthesis, the cells were processed as described in the Supplementary Material, and CT663/β protein complexes were immunoprecipitated with an anti-CT663 antibody. Immunoprecipitated proteins were separated by SDS-PAGE and probed by immunoblotting with an anti-β antibody or an anti-CT663 antibody. Purified CT663 and E. coli β proteins were used as size markers (lanes 7,8).
To identify specific amino acid side chains within the β FTH that are important for the binding of CT663, we constructed an alanine scan library of the FTH in the context of the λCI-β-flapEc fusion protein, individually changing E. coli β-residues 898–912 to alanines (except residues 904 and 910, which are already alanines). Plasmids encoding each of these mutant fusion proteins were transformed into reporter strain FW102 OL2-62 harboring the α-CT663(25–133) fusion protein (encoded by pBRL28). As shown in Figure 2A, β-flap substitutions L901A, I905A, and F906A essentially eliminated the CT663(25–133)/β-flap interaction, substitution L902A had a slightly less dramatic effect, and substitution R903A moderately weakened the interaction; several other substitutions, including G907A had more modest effects. Western blot analysis indicated that all of the mutant fusion proteins were stably produced (data not shown). We also tested the effects of amino acid substitutions at positions 841, 842, 845, and 846 of the β-flap moiety of the λCI-β-flapCt fusion protein (corresponding to positions 901, 902, 905, and 906 of the E. coli β-flap) (Supplemental Fig. S1). As expected, these amino acid substitutions strongly disrupted the CT663(25–133)/β-flapCt interaction (Fig. 2B). E. coli β residues L901, L902, I905, and F906, which are predicted to be surface-exposed on the RNAP core enzyme (Vassylyev et al. 2002), make essential contacts with σ region 4 in the context of the RNAP holoenzyme (Kuznedelov et al. 2002; Geszvain et al. 2004), suggesting that CT663 competes with σ region 4 for access to the β-flap.
Next, we sought to confirm the CT663/β-flap interaction detected in the two-hybrid assay by coimmunoprecipitation experiments. Plasmids that directed the inducible synthesis of full-length E. coli β, full-length C. trachomatis β, an E. coli β variant lacking residues 900–909 (encompassing the FTH) or an E. coli β variant lacking residues 884–914 (encompassing the FTH and the flanking β-flap “arms”) were constructed. The wild-type and mutant β proteins (Ec β, Ct β, Ec β Δ900–909, and Ec β Δ884–914) were overproduced in E. coli cells in the presence or absence of overproduced CT663. Any CT663/β complexes that formed were immunoprecipitated from the cell lysates with an anti-CT663 antibody, separated by SDS-PAGE, and probed for the β subunit by immunoblot. As shown in Figure 2C, CT663 was able to coimmunoprecipitate wild-type E. coli β (lane 4) and C. trachomatis β (lane 2). In contrast, the association of CT663 with the β mutants, Ec β Δ900–909 and Ec β Δ884–914, was significantly reduced (Fig. 2C, cf. lanes 3,5 and 4) as only background levels of β (i.e., chromosomally encoded β) were immunoprecipitated (Fig. 2C, cf. lanes 3,5 and 1). Control immunoblots indicated that the Ec β Δ900–909 and Ec β Δ884–914 proteins were present at approximately the same level as wild-type Ec β in the cell lysates (data not shown). These data are thus consistent with the results of the two-hybrid analysis and support the proposal that CT663 interacts with the β-flap in the context of full-length β subunit.
CT663 interacts with regions 4 of C. trachomatis σ66 and E. coli σ70
In the context of the RNAP holoenzyme, the σ region 4/β-flap interaction positions region 4 for contact with the −35 element (Kuznedelov et al. 2002). The bacteriophage T4-encoded AsiA protein, an anti-σ factor that inhibits σ70-dependent transcription from −10/−35 promoters as a component of the σ70-containing RNAP holoenzyme, interacts with both σ70 region 4 (Severinova et al. 1996; Gregory et al. 2004; Lambert et al. 2004) and the β-flap (Yuan et al. 2009). We considered the possibility that CT663 might also interact with both σ region 4 and the β-flap in the context of a particular holoenzyme species. To investigate this possibility, we used the two-hybrid assay to test whether CT663 could interact with σ region 4. We found that the α-CT663(25–133) fusion protein stimulated lacZ reporter gene slightly, but reproducibly, in the presence of a λCI fusion protein bearing region 4 of C. trachomatis σ66 (data not shown). Although we were unable to fuse full-length CT663 to the αNTD, we were able to construct a full-length λCI-CT663 fusion protein. We therefore tested this λCI-CT663 fusion protein for its ability to stimulate lacZ reporter gene transcription in the presence of an α-Ct σ66 region 4 fusion protein, an α-Ec σ70 region 4 fusion protein, or an α-Ct σ28 region 4 fusion protein (Fig. 3A). With the fused protein moieties in this orientation, we were able to demonstrate that CT663 interacted significantly with regions 4 from C. trachomatis σ66 and E. coli σ70 (which exhibit 60% amino acid identity) (Supplemental Fig. S2), but not with region 4 from C. trachomatis σ28 (Fig. 3B). All the fusion proteins were stably produced as revealed by immunoblot (data not shown). These results imply that full-length CT663 is able to interact specifically with region 4 of the primary σ factors (σ70 and σ66), and that the N-terminal region of CT663 is likely involved in this interaction.
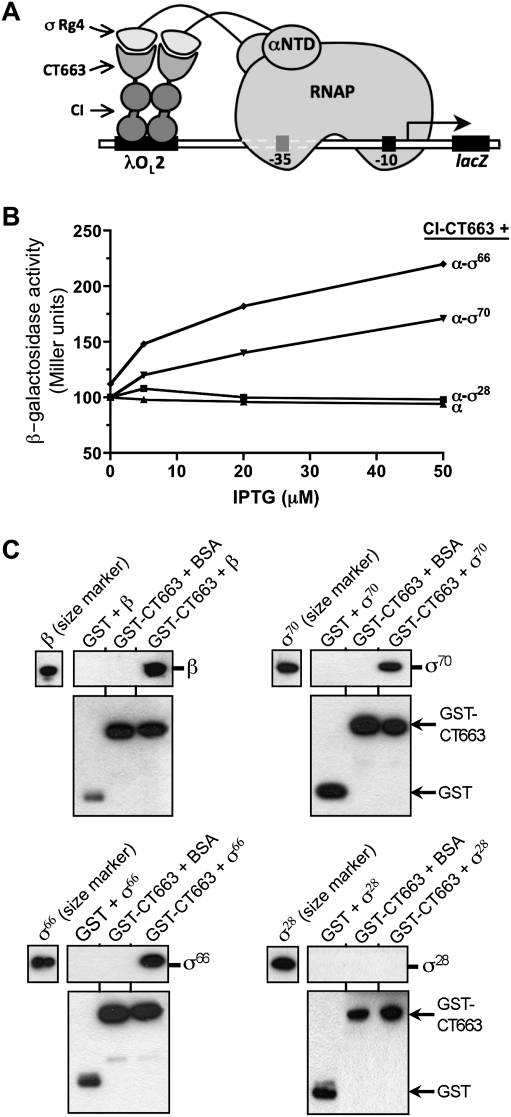
Figure 3.
CT663 interacts with region 4 of the primary σ factors σ66 and σ70, but not with region 4 of the alternative σ factor σ28. (A) Bacterial two-hybrid assay used to test whether region 4 of various σ factors interacts with CT663. Cartoon depicts how the interaction between CT663, fused to the bacteriophage λCI protein, and region 4 (Rg4) from various σ factors, fused to the αNTD, activates transcription from test promoter placOL2-62 in reporter strain FW102 OL2-62 (see Fig. 1 legend). (B) Results of β-gal assays performed with FW102 OL2-62 cells containing one plasmid that encoded the λCI-CT663 fusion protein and a compatible plasmid that encoded either α or the indicated σ region 4 (from E. coli σ70, C. trachomatis σ66, or C. trachomatis σ28) fused to the αNTD. The plasmids directed the synthesis of the fusion proteins under the control of IPTG-inducible promoters and the cells were grown in the absence of IPTG, or in the presence of 5, 20, or 50 μM IPTG. Shown is a single experiment; the values from three independent experiments differed by <5%. (C) GST-mediated pull-down of CT663/σ region 4 complexes. GST-tagged CT663 or GST was immobilized onto glutathione beads and incubated with BSA or with purified E. coli σ70, C. trachomatis σ66, or σ28, or with E. coli β as a positive control (see Fig 1). GST-CT663-interacting proteins were detected by immunoblotting with antibodies to β, σ70, σ66, or σ28, as indicated (top panels), and GST-CT663 (or GST) was detected by immunoblotting with antibody to GST (bottom panels). The control reactions performed with GST lacking the fused CT663 moiety confirmed that the RNAP subunits were interacting specifically with the CT663 component of the GST-CT663 fusion protein. Purified β, σ70, σ66, and σ28 were loaded on the appropriate gels to serve as molecular size markers.
CT663 interacts with full-length primary σ factors in vitro
We used a pull-down assay to test the ability of CT663 to associate with full-length σ factor. Purified GST-tagged CT663 (GST-CT663) was immobilized on glutathione beads and was coincubated with each of the following purified RNAP subunits: E. coli σ70, C. trachomatis σ66, C. trachomatis σ28, and E. coli β. Bound proteins were fractionated by SDS-PAGE and detected by immunoblot with specific antisera. As expected, we found that the GST-CT663 protein was able to pull-down the E. coli β subunit (Fig. 3C, top left panel), consistent with the results of the coimmunoprecipitation experiments (Fig. 2C). GST-CT663 also associated specifically with the primary σ factors from both E. coli (σ70) and C. trachomatis (σ66) (Fig. 3C, top right and bottom left panels); however, as expected, GST-CT663 did not interact with C. trachomatis σ28 (Fig. 3C, bottom right panel).
CT663 inhibits σ66-dependent transcription but not σ28-dependent transcription in vitro
The direct interaction of CT663 with components of the transcription machinery (E. coli β and σ70, and C. trachomatis β and σ66) suggested that CT663 might affect RNAP function. We tested this hypothesis by performing the in vitro transcription analysis. Attempts to form a functional hybrid holoenzyme using recombinant C. trachomatis σ66 and E. coli core were not successful, in agreement with a previous report (Douglas et al. 1994). Therefore, we used RNAP holoenzyme reconstituted with E. coli σ70 or hybrid σ66, which contains regions 1–3 from E. coli σ70 and region 4 from C. trachomatis σ66 (Supplemental Table S1). The RNAP holoenzyme reconstituted with C. trachomatis σ28 was functional (Shen et al. 2004) and was used as a control. We tested the effect of purified CT663 (Supplemental Fig. S3) on transcription by these holoenzymes using four well-defined promoters (E. coli PlacUV5, C. trachomatis P1rRNA, P1tuf, and P3tuf) recognized by both C. trachomatis σ66 and E. coli σ70 (Tan and Engel 1996; Shen et al. 2000). For the control assays, we used σ28-dependent promoters from C. trachomatis (PhctB) and E. coli (PfliC) (Shen et al. 2006). We found that both σ70- and σ66-dependent transcription from PlacUV5, P1rRNA, P1tuf, and P3tuf decreased in the presence of CT663 (Fig. 4A, panels 1–6; Supplemental Fig. S4A). With σ70RNAP the inhibition of transcription was greatest at P1rRNA (93%), and was at least 76% on the other templates at the highest concentration of CT663 used; with σ66RNAP the inhibition of transcription was greatest at PlacUV5 (93%), and reached at least 61% on the other templates. In contrast, we observed very little inhibition of transcription from the σ28-dependent promoters PhctB (15%) and PfliC (3%), even at the highest concentration of CT663 tested (Fig. 4A, panel 7; Supplemental Fig. S4A). To confirm that the effect of CT663 on σ66-dependent transcription was mediated solely by the CT663 protein, we showed that incubating CT663 with an anti-CT663 antibody prior to adding it to the in vitro transcription reactions blocked its ability to inhibit σ66-dependent transcription (data not shown).
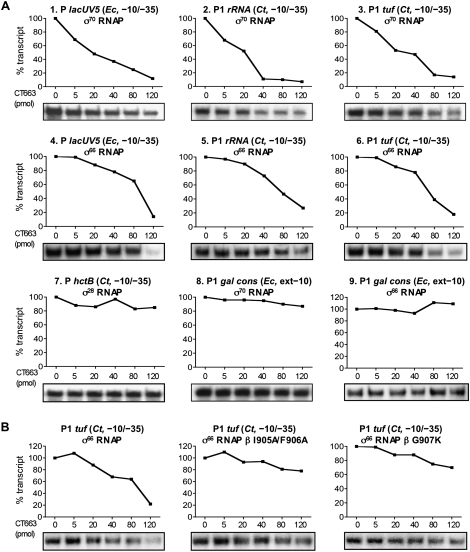
Figure 4.
CT663-mediated inhibition of transcription in vitro. (A) CT663 inhibits σ70- and σ66-dependent, but not σ28-dependent, transcription specifically from −10/−35 promoters. Holoenzymes containing E. coli core RNAP reconstituted with E. coli σ70 (panels 1–3,8), hybrid C. trachomatis σ66 (panels 4–6,9) or C. trachomatis σ28 (panel 7) were incubated with supercoiled plasmid templates bearing the indicated promoter. Single-round in vitro transcription assays were done as described in the Materials and Methods in the absence of CT663 or in the presence of increasing concentrations of CT663 (5, 20, 40, 80, and 120 pmol) as indicated. Transcripts were labeled with [α-32P]CTP and quantified following urea-PAGE and autoradiography. For each promoter the percentage of transcripts in the presence of CT663 is plotted compared with that observed in the absence of CT663, which was set to 100%. Assays were repeated three times with similar results. (B) Mutations in the β-flap that disrupt the CT663/β-flap interaction weaken the inhibitory effect of CT663 on σ66-dependent transcription from the P1 tuf promoter from C. trachomatis. Single-round in vitro transcription assays were done as described above using either wild-type RNAP or mutant RNAPs carrying substitutions in the β-flap (I905A/F906A or G907K) that weaken the CT663/β-flap interaction (see Supplemental Fig. S5B). For each holoenzyme the percentage of transcripts in the presence of CT663 (5, 20, 40, 80, and 120 pmol) is plotted compared with that observed in the absence of CT663, which was set to 100%. Assays were repeated three times with similar results.
We next examined whether CT663 exerts its inhibitory effect on σ66-dependent transcription from an extended −10 promoter (galP1/cons). Extended −10 promoters are defined by the presence of a TG dinucleotide 1 bp upstream of the −10 hexamer and typically lack a recognizable −35 element; additional contacts between σ and the TG dinucleotide compensate for the lack of a −35 element and, consequently, σ region 4 is dispensable for the recognition of extended −10 promoters (Kumar et al. 1993; Kuznedelov et al. 2002). We found that the addition of CT663 did not decrease transcription from the galP1/cons promoter even at the highest concentration of CT663 tested (Fig. 4A, panels 8,9) in contrast to the results with the −10/−35 promoters (Fig. 4A, panels 1–6). These data suggest that CT663 carries out its inhibitory effect on σ66-dependent transcription in the context of the σ66-containing RNAP holoenzyme, presumably by disrupting the σ66 region 4/β-flap interaction.
To test whether the ability of CT663 to inhibit σ66-dependent transcription from −10/−35 promoters depends on its interaction with the β-flap, we performed in vitro transcription experiments using the hybrid σ66 together with E. coli RNAP core enzymes reconstituted with mutant β subunits bearing amino acid substitutions that weaken the CT663/β-flap interaction. Specifically, we tested the effect of β substitution G907K, which moderately weakened the CT663/β-flap interaction (without affecting the σ region 4/β-flap interaction), and β substitutions I905A and F906A (in combination), which strongly weakened the CT663/β-flap interaction in the two-hybrid assay (Supplemental Fig. S5B–D). As shown in Figure 4B, weakening the CT663/β-flap interaction with the β-flap substitutions (either G907K or I905A and F906A) significantly compromised the ability of CT663 to inhibit σ66-dependent transcription. Similarly, we observed that weakening the CT663/β-flap interaction compromised the ability of CT663 to inhibit σ70-dependent transcription from the lacUV5 promoter (Supplemental Fig. S4B).
Developmental expression of CT663 in C. trachomatis
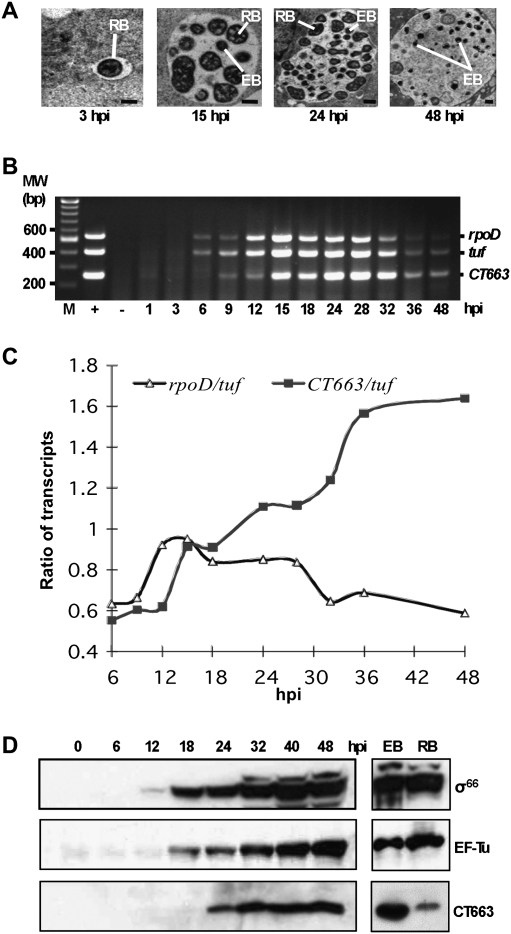
As a first step in investigating a role for CT663 in inhibiting σ66-dependent transcription in the physiologically relevant context of C. trachomatis-infected cells, we monitored CT663 mRNA levels by RT–PCR and CT663 protein levels by immunoblot at various time points across the developmental cycle from 1 to 48 h post-infection (hpi). We also monitored the levels of rpoD mRNA and its product (σ66) as well as the levels of tuf mRNA and its product (EF-Tu), the latter serving as a reference gene that is constitutively expressed in growing C. trachomatis (Shen et al. 2000). Micrographs of C. trachomatis-infected cells were taken to monitor morphology change across the C. trachomatis developmental cycle (Fig. 5A). By 3 hpi, the EBs were already beginning to differentiate into the RBs. By 15 hpi, many growing RBs were visible; by 24 hpi, the RBs were seen to differentiate back into EBs and by 48 hpi, predominantly EBs were observable.
Figure 5.
C. trachomatis developmental gene expression. (A) Transmission electron micrographs of C. trachomatis serovar F within an inclusion. C. trachomatis-infected cells were sampled at different time points post-infection as indicated. Representatives of the two major C. trachomatis forms within the inclusion, the EB and the RB, are labeled (bar, 0.5 μm). (B) Results of multiplex RT–PCR analysis. Ethidium bromide-stained agarose gel showing the developmental expression pattern of the CT663, tuf, and rpoD transcripts harvested from C. trachomatis-infected L929 cells. Genomic DNA extracted from purified C. trachomatis EBs was used as a positive control to show that the three primer pairs amplified their targets with similar efficiencies (lane +) when used in this multiplex format. RNA isolated from L929 cells uninfected with C. trachomatis was used as negative control (lane −). A fixed quantity of total RNA (host cell RNA + C. trachomatis RNA) isolated at the indicated times in hours post-infection (hpi) was treated with DNase I and was subjected to RT–PCR. The products of the RT–PCRs migrated at the expected sizes. (C) Graph showing the ratio of rpoD and CT663 transcripts to the tuf transcript at different time points during the C. trachomatis developmental cycle as revealed by the RT–PCR assay in B. Note that there is a general decrease in transcript levels that occurs late in the developmental cycle due to the conversion of the metabolically active RB form to the relatively inactive EB form; nevertheless, when compared with the tuf transcript there is relatively more CT663 transcript and relatively less rpoD transcript than in the earlier stages of infection. (D) Developmental protein synthesis pattern for CT663, EF-Tu (tuf gene product), and σ66 (rpoD gene product) harvested from C. trachomatis-infected L929 cells. A normalized amount of total protein (host cell protein + C. trachomatis protein) isolated from C. trachomatis-infected L929 cells at the indicated time points in hours post-infection (hpi) was separated by 12% SDS-PAGE and probed with antibodies specific to CT663, σ66, or EF-Tu. A normalized amount of protein isolated from purified EBs (lane EB) and purified RBs (lane RB) was also immunoblotted as indicated. All protein bands migrated at the expected sizes.
For the RT–PCR analysis we took a multiplex approach, using three primer pairs designed to amplify partial coding regions of the CT663, tuf, and rpoD genes in the same reaction tube. With C. trachomatis genomic DNA as a template, we observed that each of these primer pairs worked with comparable efficiency and independently of the other primer pairs, returning similar amounts of amplification product (Fig. 5B, lane +). With C. trachomatis RNA as a template, we observed no amplification products in the absence of a cDNA synthesis step, indicating no detectable DNA contamination in the samples (data not shown). In this RT–PCR analysis the rpoD and tuf transcripts were detected at 6 hpi, reached their highest levels at 15 hpi, and remained at high levels up to 28 hpi, and decreased thereafter. The CT663 transcript was first detected at a later time point, 9 hpi, but sharply increased by 15 hpi, peaking at 24–28 hpi, and gradually decreasing at later times (Fig. 5B). When normalized against the tuf transcript, the relative levels of the rpoD transcript peaked at ∼15 hpi, correlating with the exponential growth phase of the RBs, but declined later, as the metabolically inactive EBs became more predominant (Fig. 5A,C). However, the relative levels of the CT663 transcript increased during the latter part of the developmental cycle (from ∼18–36 hpi), coincident with the gradual shift from the RB back to the EB phase (Fig. 5A,C).
Next, we monitored the protein levels of CT663, σ66, and EF-Tu in lysates of C. trachomatis-infected cells. At the early time points we detected little (EF-Tu) or no (CT663 and σ66) protein by immunoblot analysis, consistent with the presence of only very few metabolically active C. trachomatis at the start of the infection. ET-Tu levels remained low until 18 hpi, and accumulated steadily thereafter. The σ66 protein was first detected 12 hpi, and then accumulated for the duration of the experiment. The CT663 protein became detectable at 24 hpi and then remained at a constant level from 32 hpi to the end of the experiment at 48 hpi (Fig. 5D). Moreover, both the EF-Tu and σ66 proteins were easily detected in samples containing either purified EBs or purified RBs, whereas, CT663 was significantly more abundant in the EB sample. The accumulation of CT663 at the late stage of the C. trachomatis developmental cycle raises the possibility that CT663 may inhibit σ66-dependent transcription during the RB-to-EB transition, perhaps facilitating a transition to σ28-dependent gene expression.
Discussion
The understanding of gene regulation in C. trachomatis is limited due to the absence of a tractable genetic system for this organisn. To uncover novel regulators of transcription in C. trachomatis, we performed a two-hybrid screen to identify C. trachomatis proteins that interact with the β-flap, a potential regulatory target on the RNAP core enzyme. We report the isolation and characterization of CT663, a hypothetical type III secretion chaperone, which inhibits transcription in vitro by RNAP reconstituted with either σ70 or a σ70/66 hybrid, and presumably σ66-dependent transcription in C. trachomatis.
CT663 interacts with both the β and σ66 subunits of RNAP
We found CT663 associates with both the β and σ66 subunits of C. trachomatis RNAP, and inhibits transcription from promoters that are recognized by both C. trachomatis σ66 and E. coli σ70 (Fig. 4; Supplemental Fig. S4). As would be expected for an anti-σ factor that inhibits σ66-dependent transcription (Hughes and Mathee 1998; Helmann 1999; Campbell et al. 2008), CT663 interacts specifically with σ66 (and σ70), targeting conserved region 4. However, unlike most previously described anti-σ factors (except the bacteriophage T4 AsiA protein), CT663 also interacts directly with a component of the RNAP core enzyme, the β-flap. We obtained several lines of evidence in support of this interaction. First, CT663 interacted with β-flap from both C. trachomatis and E. coli in the two-hybrid assay and specific conserved amino acid residues were identified in the FTH that are critical determinants of the interaction (Figs. 1, 2). Second, CT663 formed a complex with the full-length β subunit from either E. coli or C. trachomatis in vivo, as detected by coimmunoprecipitation, and complex formation was dependent on the β FTH (Fig. 2C). Third, CT663 (fused to GST) associated with full-length E. coli β in vitro, as detected using a pull-down assay (Fig. 3C). Finally, weakening the interaction of CT663 with the β-flap compromised the ability of CT663 to inhibit σ66-dependent transcription in vitro (Fig. 4B).
Comparison between CT663 and the bacteriophage T4 AsiA protein
The T4 AsiA protein inhibits σ70-dependent transcription from −10/−35 promoters as a stable component of the σ70-containing RNAP holoenzyme in E. coli (Severinova et al. 1998). Its association with the holoenzyme is stabilized by interactions with both σ70 region 4 (Severinova et al. 1996; Gregory et al. 2004; Lambert et al. 2004) and the β-flap (Yuan et al. 2009). We suggest that CT663 may similarly exert its effects on transcription in the context of the RNAP holoenzyme. Consistent with this view and like AsiA, CT663 was unable to inhibit transcription (<13% inhibition) from an extended −10 promoter (gal P1/cons) lacking a −35 element. If CT663 functioned to prevent formation of the σ66-containing RNAP holoenzyme, then its inhibitory effect on σ66-dependent transcription should have been observed with both −10/−35 and extended −10 promoters. As a component of the σ70-containing RNAP holoenzyme, AsiA functions not only as an inhibitor of transcription, but also as a coactivator of T4 middle promoters, which bear a near-consensus −10 element and a binding site for the T4-encoded activator MotA in place of a −35 element (Stitt and Hinton 1994). Whether or not CT663 might also have additional function(s) remains to be investigated.
AsiA inhibits transcription from −10/−35 promoters both by disrupting the σ70 region 4/β-flap interaction (Gregory et al. 2004) and by stabilizing a conformation of σ70 region 4 that is incompatible with sequence-specific binding to the −35 element (Lambert et al. 2004). Our genetic evidence suggests that CT663 also disrupts the σ66 region 4/β-flap interaction. Specifically, we identified four hydrophobic amino acid residues in the β FTH that are critical determinants of both the CT663/β-flap (Fig. 2; Supplemental Fig. S5) and the σ70 region 4/β-flap interactions (Geszvain et al. 2004; Deighan et al. 2008). We also demonstrated that these residues are important for the σ66 region 4/β-flap interaction (Supplemental Fig. S5C; data not shown). Moreover, each of these residues maps to the σA region 4/β-flap interface, based on the crystal structure of the T. thermophilus RNAP holoenzyme (PDB ID 1IW7). We note that despite its apparent functional resemblance to AsiA, CT663 does not bear any significant amino acid similarity to AsiA.
Possible effects of CT663 on post-initiation stages of the transcription cycle
We cannot rule out the possibility that CT663 may also exert effects on transcription downstream from initiation. Because our data indicate that CT663 interacts with the β-flap, it is possible that CT663 could associate with (or remain associated with) the RNAP core enzyme during the elongation phase of transcription. A variety of regulators have been described that associate with the transcription elongation complex and affect transcription pausing and/or termination (Roberts et al. 2008), including the bacteriophage λ Q protein, which has been shown to interact with the β-flap (Deighan et al. 2008). Current efforts are underway to determine whether or not CT663 affects the elongation properties of RNAP.
The potential physiological roles of CT663 in the developmental cycle of C. trachomatis
We demonstrated that purified CT663 inhibits the activity of hybrid σ66RNAP in vitro, and we hypothesize that CT663 may function to inhibit the activity of σ66RNAP in C. trachomatis. We detected active synthesis of CT663 mRNA and high levels of CT663 protein at the middle–late stage (Fig. 5), suggesting that CT663 functions relatively late in the developmental cycle of C. trachomatis. We found that RBs contain a low abundance of CT663, whereas EBs contain much higher levels of CT663, although both EBs and RBs contain high levels of σ66 protein. Thus, the levels of σ66 protein revealed by immunoblot may not necessarily reflect the levels of functional σ66 RNAP. Moreover, this difference in CT663 abundance provides an explanation for the previous observation that RB extracts, but not EB extracts, support transcription from C. trachomatis promoters in vitro (Douglas et al. 1994). Presumably, however, an additional regulatory mechanism must exist to reverse the negative effect of CT663 on transcription so that active σ66RNAP can be liberated to transcribe the growth-related genes needed for initiating a new round of infection.
It is known that σ66RNAP is required for transcription of most housekeeping genes during the developmental cycle of C. trachomatis (in both the RB and EB phases) (Koehler et al. 1990; Fahr et al. 1995; Hatch 1999). We note that the strong inhibitory effect of CT663 on σ66-dependent transcription directed from −10/−35 promoters, but not extended −10 promoters, raises the possibility of another level of regulation in C. trachomatis. The selective inhibition of some σ66-dependent promoters (i.e., those with strong −35 elements), but not others (i.e., those that are relatively independent of a −35 element) (Fig. 4A), might allow C. trachomatis to maintain expression of certain genes that are required for viability and/or for completion of the developmental cycle. We find that the C. trachomatis genome includes candidate extended −10 σ66-dependent promoters (X Hu, X Feng, and L Shen, unpubl.). Experiments are underway to test the functional relevance of their putative extended −10 motifs.
We demonstrated previously that σ28 accumulates in C. trachomatis when RBs convert to EBs (Shen et al. 2004). However, the presence of high concentrations of σ28 was insufficient to displace σ66 from the RNAP holoenzyme in RB extracts (Yu and Tan 2003). Possibly the accumulation of CT663 during the RB-to-EB transition facilitates formation of the σ28-containing holoenzyme. This could occur if CT663 destabilized the σ66-containing holoenzyme, allowing σ28 to compete more effectively for access to the core enzyme. Thus, CT663 may indirectly facilitate σ28-dependent transcription, which is required for late-stage morphological differentiation in C. trachomatis (Shen et al. 2004). A similar function has been proposed for the E. coli stationary-phase-specific anti-σ70 factor Rsd, overproduction of which increased the activity of some σ38-dependent promoters (Jishage and Ishihama 1998; Mitchell et al. 2007).
Based on its position at the start of an operon encoding putative structural components of a type III secretion apparatus in C. trachomatis (Hefty and Stephens 2007; Betts et al. 2008) and its weak homology with the type III secretion chaperone CesT, CT663 has been designated as a hypothetical type III secretion chaperone. All 10 of the C. trachomatis type III secretion operons are preceded by σ70-like promoters that are presumably recognized by σ66RNAP (Hefty and Stephens 2007). Our finding that CT663 acts as an inhibitor of σ66RNAP therefore suggests a role for CT663 in the regulation of virulence gene expression in C. trachomatis. As a putative regulator of virulence gene expression, CT663 is reminiscent of the anti-σ70 factor AlgQ, which regulates the synthesis of several virulence factors in Pseudomonas aeruginosa, including alginate (Deretic and Konyecsni 1989; Kato et al. 1989), pyoverdine (Ambrosi et al. 2005), and pyocyanin (Yuan et al. 2008). In enteropathogenic E. coli, CesT supports the type III secretion of the translocated intimin receptor (Tir) protein (and other effectors) into host cells (Abe et al. 1999; Elliott et al.1999; Thomas et al. 2005), which confers upon bacteria the ability to alter host cell morphology following intimate bacterial attachment. CT663 has a theoretical molecular weight of 14.7 kDa and an isoelectric point of 4.06, compatible with most known type III secretion-chaperones that are small in size and have a low isoelectric point (∼4–5.6) (Parsot et al. 2003). An intriguing possibility is that CT663 functions both as a type III secretion chaperone for one or more effector proteins and as a transcription regulator.
Materials and methods
Strains, plasmids, and primers
A complete list of strains and plasmids is provided in Supplemental Table S1, and primers are listed in Supplemental Table S2.
Screen for C. trachomatis proteins that interact with the β-flap domain of RNAP
To identify proteins from C. trachomatis that could interact with the β-flap domain of RNAP, we used a bacterial two-hybrid assay (Fig. 1A; Dove et al. 1997). A C. trachomatis serovar F genomic DNA library was prepared by cloning Sau3AI-digested chromosomal DNA (≈500–1500 bp) into BamHI-digested pRBR (Supplemental Table S1). This plasmid library, which directed the synthesis of C. trachomatis protein fragments fused to the αNTD, was transformed into E. coli strain KS1 containing plasmid pACλCI-β-flapCt (Hua et al. 2009). KS1 contains test promoter placOR2-62 fused to a lacZ reporter gene and integrated into the chromosome on a prophage. Plating the transformation mix onto indicator medium containing isopropyl-β-D-thiogalactoside (IPTG; 100 μM) and X-gal (100 μg/mL), permitted the identification of colonies derived from cells containing α fusion proteins that increased the λCI-β-flapCt-dependent activation of the two-hybrid test promoter (see the Supplemental Material).
Coimmunoprecipitation assay
E. coli C2566 cells were cotransformed with plasmid pXDCM (directs the synthesis of CT663) and a plasmid directing the synthesis of a wild-type or mutant β subunit (pIA545, pIA545Δ900–909, pIA545Δ884–914, or pBAD-Ctβ) (see Supplemental Table S1). Proteins were induced and CT663/β complexes were immunoprecipitated from cell lysates as described in the Supplemental Material.
GST pull-down assay
Purified GST-tagged CT663 protein (or GST) was prebound to glutathione beads, then incubated with equimolar amounts of E. coli β or σ70, or C. trachomatis σ66 or σ28, and the CT663-bound proteins detected as described in the Supplemental Material.
In vitro transcription
The supercoiled plasmids used as templates for in vitro transcriptions reactions were pMT504, pP1tufG, pP3tufG, pGHR, pPlacUV5G, pGLC, and pGPgalP1/cons (see Supplemental Table S1). RNAP holoenzymes were reconstituted by mixing E. coli core enzymes (for Fig 4A, wild-type core was purchased from Epicentre; for Fig 4B, wild-type and mutant core enzymes [β I905A/F906A and β G907K] were purified as described) (Deighan et al. 2008), with a threefold molar excess of σ70 or σ70/66 and incubated for 10 min at 37°C. The C. trachomatis σ28-containing RNAP holoenzyme was prepared as described (Shen et al. 2004). When required CT663 (5, 20, 40, 80, and 120 pmol) was preincubated with σ for 20 min at 37°C, before adding the RNAP core enzymes. Single-round in vitro transcription reactions were carried out with 0.5 pmol RNAP holoenzyme, supercoiled plasmid templates (1 μg), in reaction buffer containing 10 mM Tris-HCl (pH 8), 200 mM NaCl, 10 mM MgCl2 and 5 mM dithiothreitol. Transcription was initiated by adding 400 μM ATP, 400 μM UTP, 1.2 μM CTP, 0.20 μM [α-32P]CTP (3000 μCi/mmol), and 100 μM 3-O-methylguanosine 5′-triphosphate (GE Healthcare) and continued for 20 min at 37°C. Transcripts were analyzed by electrophoresis on 6% polyacrylamide gels containing 8 M urea. Gels were exposed to film and signal intensities from autoradiographs were quantified using Quantity One software (Bio-Rad).
β-Gal assay
β-Gal assays were performed on mid-log-phase cultures using a plate assay (Thibodeau et al. 2004) as detailed in the Supplemental Material.
RT–PCR and Western blot analysis of C. trachomatis gene expression in C. trachomatis-infected L929 cells
Total RNA from C. trachomatis-infected L929 cells was isolated using the RNeasy Kit (Qiagen), treated with DNase I (USB) for 1 h at 37°C to remove contaminating DNA, followed by heat inactivation of DNase I for 5 min at 75°C. Primer pairs are listed in Supplemental Table S2 and were designed to amplify products of 550 bp for rpoD, 410 bp for tuf, and 250 bp for CT663. Five micrograms of total RNA were reverse-transcribed in a reaction containing appropriate reverse primers and 5 U AMV reverse transcriptase (Upstate Biotechnologies) for 1 h at 42°C. Then PCRs (0.5 min at 94°C, 0.5 min at 54°C, and 1 min at 72°C for 30 cycles) were performed using the cDNA products, primer pairs as above and Taq DNA polymerase (New England Biolabs). PCR products were separated on a 1.5% agarose gel, stained with ethidium bromide, and photographed with Gel Doc 3000 (Bio-Rad).
To monitor σ66, EF-Tu and CT663 protein levels in C. trachomatis infected cells, total protein lysates were prepared by boiling cell pellets in SDS loading buffer for 10 min. Protein lysates were also prepared from purified RBs and EBs (Shen et al. 2004), and were analyzed in parallel; protein lysates from uninfected L929 cells served as a negative control for antibody specificity. After centrifugation of the denatured lysates, a normalized amount of protein was separated on 12% SDS-PAGE and transferred to Immobilon-P membrane (Millipore) and immunoblotted with antibodies against σ66, C. trachomatis EF-Tu (mAb GP-45) and CT663. Blots were developed using a horseradish peroxidase-conjugated secondary antibody (Sigma) and the SuperSignal Chemiluminescent Detection kit (Pierce).
Additional methods
Antibodies used in this study are described in the Supplemental Material. Details on purification of recombinant proteins (E. coli β and derivatives, C. trachomatis β, CT663, σ66 and σ28) and on the experimental procedure for the transmission electron microscopy analysis can also be found in the Supplemental Material.
Acknowledgments
We thank Dr. David Wothylake and Louis LeCour for help in protein purification, Drs. You-xun Zhang and Tom P. Hatch for antibodies and helpful discussions, Dr. Irina Artsimovitch for plasmids, Dr. Ronald Luftig for comments, and Donald Gantz and Jonathan A. Brex for technical assistance. This work was supported by National Institutes of Health Grants AI055869 (to L.S.) and GM44025 (to A.H.).
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1784009.
Supplemental material is available at http://www.genesdev.org.
References
- Abe A, de Grado M, Pfuetzner RA, Sánchez-Sanmartín C, Devinney R, Puente JL, Strynadka NC, Finlay BB. Enteropathogenic Escherichia coli translocated intimin receptor, Tir, requires a specific chaperone for stable secretion. Mol Microbiol. 1999;33:1162–1175. doi: 10.1046/j.1365-2958.1999.01558.x. [DOI] [PubMed] [Google Scholar]
- Ambrosi C, Tiburzi F, Imperi F, Putignani L, Visca P. Involvement of AlgQ in transcriptional regulation of pyoverdine genes in Pseudomonas aeruginosa PAO1. J Bacteriol. 2005;187:5097–5107. doi: 10.1128/JB.187.15.5097-5107.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arthur TM, Burgess RR. Localization of a σ70 binding site on the N terminus of the Escherichia coli RNA polymerase β′ subunit. J Biol Chem. 1998;273:31381–31387. doi: 10.1074/jbc.273.47.31381. [DOI] [PubMed] [Google Scholar]
- Belland RJ, Zhong G, Crane DD, Hogan D, Sturdevant D, Sharma J, Beatty WL, Caldwell HD. Genomic transcriptional profiling of the developmental cycle of Chlamydia trachomatis. Proc Natl Acad Sci. 2003;100:8478–8483. doi: 10.1073/pnas.1331135100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betts HJ, Twiggs LE, Sal MS, Wyrick PB, Fields KA. Bioinformatic and biochemical evidence for the identification of the type III secretion system needle protein of Chlamydia trachomatis. J Bacteriol. 2008;190:1680–1690. doi: 10.1128/JB.01671-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunham RC, Rey-Ladino J. Immunology of Chlamydia infection: Implications for a Chlamydia trachomatis vaccine. Nat Rev Immunol. 2005;5:149–161. doi: 10.1038/nri1551. [DOI] [PubMed] [Google Scholar]
- Campbell EA, Westblade LF, Darst SA. Regulation of bacterial RNA polymerase σ factor activity: A structural perspective. Curr Opin Microbiol. 2008;11:121–127. doi: 10.1016/j.mib.2008.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deighan P, Diez CM, Leibman M, Hochschild A, Nickels BE. The bacteriophage λQ antiterminator protein contacts the β-flap domain of RNA polymerase. Proc Natl Acad Sci. 2008;105:15305–15310. doi: 10.1073/pnas.0805757105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deretic V, Konyecsni WM. Control of mucoidy in Pseudomonas aeruginosa: Transcriptional regulation of algR and identification of the second regulatory gene, algQ. J Bacteriol. 1989;171:3680–3688. doi: 10.1128/jb.171.7.3680-3688.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douglas AL, Saxena NK, Hatch TP. Enhancement of in vitro transcription by addition of cloned, overexpressed major σ factor of Chlamydia psittaci 6BC. J Bacteriol. 1994;176:3033–3039. doi: 10.1128/jb.176.10.3033-3039.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dove SL, Hochschild A. Bacterial two-hybrid analysis of interactions between region 4 of the σ70 subunit of RNA polymerase and the transcriptional regulators Rsd from Escherichia coli and AlgQ from Pseudomonas aeruginosa. J Bacteriol. 2001;183:6413–6421. doi: 10.1128/JB.183.21.6413-6421.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dove SL, Joung JK, Hochschild A. Activation of prokaryotic transcription through arbitrary protein–protein contacts. Nature. 1997;386:627–630. doi: 10.1038/386627a0. [DOI] [PubMed] [Google Scholar]
- Elliott SJ, Hutcheson SW, Dubois MS, Mellies JL, Wainwright LA, Batchelor M, Frankel G, Knutton S, Kaper JB. Identification of CesT, a chaperone for the type III secretion of Tir in enteropathogenic Escherichia coli. Mol Microbiol. 1999;33:1176–1189. doi: 10.1046/j.1365-2958.1999.01559.x. [DOI] [PubMed] [Google Scholar]
- Fahr MJ, Douglas AL, Xia W, Hatch TP. Characterization of late gene promoters of Chlamydia trachomatis. J Bacteriol. 1995;177:4252–4260. doi: 10.1128/jb.177.15.4252-4260.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geszvain K, Gruber TM, Mooney RA, Gross CA, Landick R. A hydrophobic patch on the flap-tip helix of E. coli RNA polymerase mediates σ70 region 4 function. J Mol Biol. 2004;343:569–587. doi: 10.1016/j.jmb.2004.08.063. [DOI] [PubMed] [Google Scholar]
- Gregory BD, Nickels BE, Garrity SJ, Severinova E, Minakhin L, Urbauer RJ, Urbauer JL, Heyduk T, Severinov K, Hochschild A. A regulator that inhibits transcription by targeting an intersubunit interaction of the RNA polymerase holoenzyme. Proc Natl Acad Sci. 2004;101:4554–4559. doi: 10.1073/pnas.0400923101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross CA, Chan C, Dombroski A, Gruber T, Sharp M, Tupy J, Young B. The functional and regulatory roles of σ factors in transcription. Cold Spring Harb Symp Quant Biol. 1998;63:141–155. doi: 10.1101/sqb.1998.63.141. [DOI] [PubMed] [Google Scholar]
- Hatch TP. Development biology. Chlamydia: Intracellular biology, pathogenesis and immunology. In: Stephens RS, editor. ASM Press; Washington, DC: 1999. pp. 29–67. [Google Scholar]
- Hefty PS, Stephens RS. Chlamydial type III secretion system is encoded on ten operons preceded by σ70-like promoter elements. J Bacteriol. 2007;189:198–206. doi: 10.1128/JB.01034-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helmann JD. Anti-σ factors. Curr Opin Microbiol. 1999;2:135–141. doi: 10.1016/S1369-5274(99)80024-1. [DOI] [PubMed] [Google Scholar]
- Hua Z, Rao X, Feng X, Luo X, Liang Y, Shen L. Mutagenesis of region 4 of σ28 from Chlamydia trachomatis defines determinants for protein–protein and protein–DNA interactions. J Bacteriol. 2009;191:651–660. doi: 10.1128/JB.01083-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes KT, Mathee K. The anti-σ factors. Annu Rev Microbiol. 1998;52:231–286. doi: 10.1146/annurev.micro.52.1.231. [DOI] [PubMed] [Google Scholar]
- Jishage M, Ishihama A. A stationary phase protein in Escherichia coli with binding activity to the major σ subunit of RNA polymerase. Proc Natl Acad Sci. 1998;95:4953–4958. doi: 10.1073/pnas.95.9.4953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato J, Chu L, Kitano K, DeVault JD, Kimbara K, Chakrabarty AM, Misra TK. Nucleotide sequence of a regulatory region controlling alginate synthesis in Pseudomonas aeruginosa: Characterization of the algR2 gene. Gene. 1989;84:31–38. doi: 10.1016/0378-1119(89)90136-4. [DOI] [PubMed] [Google Scholar]
- Koehler JE, Burgess RR, Thompson NE, Stephens RS. Chlamydia trachomatis RNA polymerase major σ subunit. Sequence and structural comparison of conserved and unique regions with Escherichia coli σ70 and Bacillus subtilis σ43. J Biol Chem. 1990;265:13206–13214. [PubMed] [Google Scholar]
- Kumar A, Malloch RA, Fujita N, Smillie DA, Ishihama A, Hayward RS. The minus 35-recognition region of Escherichia coli σ70 is inessential for initiation of transcription at an ‘extended minus 10’ promoter. J Mol Biol. 1993;232:406–418. doi: 10.1006/jmbi.1993.1400. [DOI] [PubMed] [Google Scholar]
- Kuznedelov K, Minakhin L, Niedziela-Majka A, Dove SL, Rogulja D, Nickels BE, Hochschild A, Heyduk T, Severinov K. A role for interaction of the RNA polymerase flap domain with the σ subunit in promoter recognition. Science. 2002;295:855–857. doi: 10.1126/science.1066303. [DOI] [PubMed] [Google Scholar]
- Lambert LJ, Wei Y, Schirf V, Demeler B, Werner MH. T4 AsiA blocks DNA recognition by remodeling σ70 region 4. EMBO J. 2004;23:2952–2962. doi: 10.1038/sj.emboj.7600312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lonetto M, Gribskov M, Gross CA. The σ70 family: Sequence conservation and evolutionary relationships. J Bacteriol. 1992;174:3843–3849. doi: 10.1128/jb.174.12.3843-3849.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell JE, Oshima T, Piper SE, Webster CL, Westblade LF, Karimova G, Ladant D, Kolb A, Hobman JL, Busby SJ, et al. The Escherichia coli regulator of σ70 protein, Rsd, can up-regulate some stress-dependent promoters by sequestering σ70. J Bacteriol. 2007;189:3489–3495. doi: 10.1128/JB.00019-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moulder JW. Interaction of chlamydiae and host cells in vitro. Microbiol Rev. 1991;55:143–190. doi: 10.1128/mr.55.1.143-190.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami KS, Darst SA. Bacterial RNA polymerases: The wholo story. Curr Opin Struct Biol. 2003;13:31–39. doi: 10.1016/s0959-440x(02)00005-2. [DOI] [PubMed] [Google Scholar]
- Nechaev S, Kamali-Moghaddam M, André E, Léonetti JP, Geiduschek EP. The bacteriophage T4 late-transcription coactivator gp33 binds the flap domain of Escherichia coli RNA polymerase. Proc Natl Acad Sci. 2004;101:17365–17370. doi: 10.1073/pnas.0408028101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholson TL, Olinger L, Chong K, Schoolnik G, Stephens RS. Global stage-specific gene regulation during the developmental cycle of Chlamydia trachomatis. J Bacteriol. 2003;185:3179–3189. doi: 10.1128/JB.185.10.3179-3189.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paget MSB, Helmann JD. The σ70 family of σ factors. Genome Biol. 2003;4:203. doi: 10.1186/gb-2003-4-1-203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsot C, Hamiaux C, Page AL. The various and varying roles of specific chaperones in type III secretion systems. Curr Opin Microbiol. 2003;6:7–14. doi: 10.1016/s1369-5274(02)00002-4. [DOI] [PubMed] [Google Scholar]
- Roberts JW, Shankar S, Filter JJ. RNA polymerase elongation factors. Annu Rev Microbiol. 2008;62:211–233. doi: 10.1146/annurev.micro.61.080706.093422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Severinova E, Severinov K, Fenyö D, Marr M, Brody EN, Roberts JW, Chait BT, Darst SA. Domain organization of the Escherichia coli RNA polymerase σ70 subunit. J Mol Biol. 1996;263:637–647. doi: 10.1006/jmbi.1996.0604. [DOI] [PubMed] [Google Scholar]
- Severinova E, Severinov K, Darst SA. Inhibition of Escherichia coli RNA polymerase by bacteriophage T4 AsiA. J Mol Biol. 1998;279:9–18. doi: 10.1006/jmbi.1998.1742. [DOI] [PubMed] [Google Scholar]
- Shaw EI, Dooley CA, Fischer ER, Scidmore MA, Fields KA, Hackstadt T. Three temporal classes of gene expression during the Chlamydia trachomatis developmental cycle. Mol Microbiol. 2000;37:913–925. doi: 10.1046/j.1365-2958.2000.02057.x. [DOI] [PubMed] [Google Scholar]
- Shen L, Shi Y, Douglas AL, Hatch TP, O'Connell CM, Chen JM, Zhang YX. Identification and characterization of promoters regulating tuf expression in Chlamydia trachomatis serovar F. Arch Biochem Biophys. 2000;379:46–56. doi: 10.1006/abbi.2000.1854. [DOI] [PubMed] [Google Scholar]
- Shen L, Li M, Zhang YX. Chlamydia trachomatis σ28 recognizes the fliC promoter of Escherichia coli and responds to heat shock in Chlamydiae. Microbiology. 2004;150:205–215. doi: 10.1099/mic.0.26734-0. [DOI] [PubMed] [Google Scholar]
- Shen L, Feng X, Yuan Y, Luo X, Hatch TP, Hughes KT, Liu JS, Zhang YX. Selective promoter recognition by chlamydial σ28 holoenzyme. J Bacteriol. 2006;188:7364–7377. doi: 10.1128/JB.01014-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens RS, Kalman S, Lammel C, Fan J, Marathe R, Aravind L, Mitchell W, Olinger L, Tatusov RL, Zhao Q, et al. Genome sequence of an obligate intracellular pathogen of humans: Chlamydia trachomatis. Science. 1998;282:754–759. doi: 10.1126/science.282.5389.754. [DOI] [PubMed] [Google Scholar]
- Stitt B, Hinton D. Regulation of middle-mode transcription. In: Karam JD, et al., editors. Molecular biology of bacteriophage T4. American Society for Microbiology; Washington, DC: 1994. pp. 142–160. [Google Scholar]
- Tan M, Engel JN. Identification of sequences necessary for transcription in vitro from the Chlamydia trachomatis rRNA P1 promoter. J Bacteriol. 1996;178:6975–6982. doi: 10.1128/jb.178.23.6975-6982.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thibodeau SA, Fang R, Joung JK. High-throughput β-galactosidase assay for bacterial cell-based reporter systems. Biotechniques. 2004;36:410–415. doi: 10.2144/04363BM07. [DOI] [PubMed] [Google Scholar]
- Thomas NA, Deng W, Puente JL, Frey EA, Yip CK, Strynadka NC, Finlay BB. CesT is a multi-effector chaperone and recruitment factor required for the efficient type III secretion of both LEE- and non-LEE-encoded effectors of enteropathogenic Escherichia coli. Mol Microbiol. 2005;57:1762–1779. doi: 10.1111/j.1365-2958.2005.04802.x. [DOI] [PubMed] [Google Scholar]
- Toulokhonov I, Landick R. The flap domain is required for pause RNA hairpin inhibition of catalysis by RNA polymerase and can modulate intrinsic termination. Mol Cell. 2003;12:1125–1136. doi: 10.1016/s1097-2765(03)00439-8. [DOI] [PubMed] [Google Scholar]
- Toulokhonov I, Artsimovitch I, Landick R. Allosteric control of RNA polymerase by a site that contacts nascent RNA hairpins. Science. 2001;292:730–733. doi: 10.1126/science.1057738. [DOI] [PubMed] [Google Scholar]
- Vassylyev DG, Sekine S, Laptenko O, Lee J, Vassylyeva MN, Borukhov S, Yokoyama S. Crystal structure of a bacterial RNA polymerase holoenzyme at 2.6 A resolution. Nature. 2002;417:712–719. doi: 10.1038/nature752. [DOI] [PubMed] [Google Scholar]
- Vassylyev DG, Vassylyeva MN, Perederina A, Tahirov TH, Artsimovitch I. Structural basis for transcription elongation by bacterial RNA polymerase. Nature. 2007;448:157–162. doi: 10.1038/nature05932. [DOI] [PubMed] [Google Scholar]
- Wang D, Severinov K, Landick R. Preferential interaction of the his pause RNA hairpin with RNA polymerase β-subunit residues 904–950 correlates with strong transcriptional pausing. Proc Natl Acad Sci. 1997;95:8433–8438. doi: 10.1073/pnas.94.16.8433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young BA, Anthony LC, Gruber TM, Arthur TM, Heyduk E, Lu CZ, Sharp MM, Heyduk T, Burgess RR, Gross CA. A coiled-coil from the RNA polymerase β′ subunit allosterically induces selective nontemplate strand binding by σ70. Cell. 2001;105:935–944. doi: 10.1016/s0092-8674(01)00398-1. [DOI] [PubMed] [Google Scholar]
- Yu HH, Tan M. σ28 RNA polymerase regulates hctB, a late developmental gene in Chlamydia. Mol Microbiol. 2003;50:577–584. doi: 10.1046/j.1365-2958.2003.03708.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan AH, Gregory BD, Sharp JS, McCleary KD, Dove SL, Hochschild A. Rsd family proteins make simultaneous interactions with regions 2 and 4 of the primary σ factor. Mol Microbiol. 2008;70:1136–1151. doi: 10.1111/j.1365-2958.2008.06462.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan AH, Nickels BE, Hochschild A. The bacteriophage T4 AsiA protein contacts the β-flap domain of RNA polymerase. Proc Natl Acad Sci. 2009;106:6597–6602. doi: 10.1073/pnas.0812832106. [DOI] [PMC free article] [PubMed] [Google Scholar]