Abstract
RNA-binding proteins (RBPs) and microRNAs (miRNAs) are potent post-transcriptional regulators of gene expression. Here, we show that the RBP HuR reduced c-Myc expression by associating with the c-Myc 3′ untranslated region (UTR) next to a miRNA let-7-binding site. Lowering HuR or let-7 levels relieved the translational repression of c-Myc. Unexpectedly, HuR and let-7 repressed c-Myc through an interdependent mechanism, as let-7 required HuR to reduce c-Myc expression and HuR required let-7 to inhibit c-Myc expression. Our findings suggest a regulatory paradigm wherein HuR inhibits c-Myc expression by recruiting let-7-loaded RISC (RNA miRNA-induced silencing complex) to the c-Myc 3′UTR.
Keywords: Post-transcriptional gene regulation, RNA-binding protein, microRNAs, ribonucleoprotein complex
RNA-binding proteins (RBPs) and microRNAs (miRNAs) are potent post-transcriptional regulators of gene expression, acting primarily through 5′ and 3′ untranslated regions (UTRs) (Ambros 2004; Moore 2005; Keene 2007; Filipowicz et al. 2008; Bartel 2009). The RBP HuR (human antigen R, a member of the elav/hu family) stabilizes and modulates the translation of numerous target mRNAs (Hinman and Lou 2008), but the molecular mechanisms of HuR function are not fully understood. HuR is predominantly nuclear but its post-transcriptional actions are linked to its cytoplasmic levels and its association with target mRNAs, two processes that are influenced by HuR-interacting proteins and by post-translational modification of HuR (Hinman and Lou 2008; Kim and Gorospe 2008). The HuR-elicited stabilization of target mRNAs has been extensively documented (Brennan and Steitz 2001; Hinman and Lou 2008); HuR also increases the translation of numerous mRNAs (e.g., p53, TIA-1, cytochrome c, ProTα, CAT-1 [cationic amino acid transporter-1], heme oxygenase-1, GLUT1, MKP-1, HIF-1α) and represses the translation of other mRNAs (e.g., p27, IGF-IR, Wnt-5a) (Kullmann et al. 2002; Meng et al. 2005; Leandersson et al. 2006; Abdelmohsen et al. 2008). Through its actions on target mRNAs, HuR has been implicated in various biological processes, including cell division, immune and stress responses, differentiation, and carcinogenesis (López de Silanes et al. 2005; Hinman and Lou 2008; Kim and Gorospe 2008).
The mRNA encoding the proto-oncogene c-Myc was identified previously as a target of HuR (Lafon et al. 1998), but the influence of HuR on c-Myc expression was not analyzed in detail. Our studies to test the influence of HuR on c-Myc expression revealed that HuR bound the c-Myc 3′UTR at a site proximal to that bound by the miRNA let-7. miRNAs are ∼22-nucleotide (nt) transcripts that associate with the 5′UTR, coding region (CR), and 3′UTR of target mRNAs forming imperfect hybrids, and can reduce target mRNA stability and repress its translation (Ambros 2004; Filipowicz et al. 2008; Bartel 2009). miRNAs are synthesized as longer transcripts [primary (Pri)miRNAs] that are processed by the nuclear RNase III Drosha into 70-nt, hairpin precursor miRNAs [(Pre)miRNAs], and further processed in the cytoplasm by RNase III, giving rise to mature miRNAs that assemble with members of the argonaute (Ago) protein family into the miRNA-induced silencing complex (RISC). The miRNA directs the RISC complex to target mRNAs (Kim et al. 2009).
Analysis of endogenous c-Myc mRNA and c-Myc(3′UTR) reporter constructs showed that reductions in HuR or let-7 levels relieved the repression of c-Myc. However, HuR was required both for let-7 to bind the c-Myc 3′UTR and for let-7 to repress c-Myc expression; conversely, let-7 was required for HuR to repress c-Myc expression, as inhibition by HuR was lost after mutating the c-Myc 3′UTR let-7 site. These findings indicate that RBPs and miRNAs can function jointly in the repression of a shared target mRNA, as exemplified by HuR and let-7 acting on the c-Myc mRNA.
Results and Discussion
HuR represses c-Myc expression through the c-Myc 3′UTR
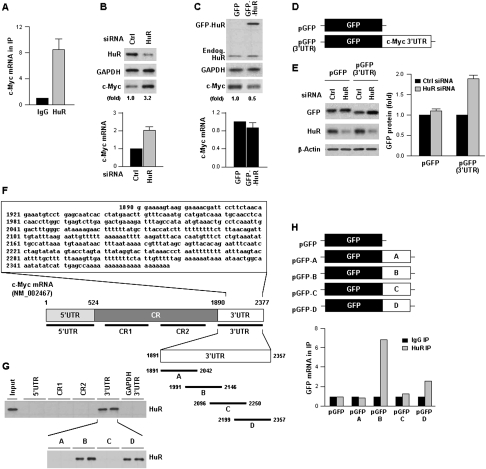
We tested the interaction of HuR with c-Myc mRNA in cervical carcinoma HeLa cells by ribonucleoprotein immunoprecipitation (RNP IP) analysis (Materials and Methods; Kim et al. 2008). As shown in Figure 1A, c-Myc mRNA was enriched more than eightfold in HuR IP samples compared with IgG IP samples. Contrary to our expectation, however, HuR inhibited c-Myc expression. Silencing of HuR by siRNA increased the abundance of c-Myc protein, as measured by Western blot analysis, and c-Myc mRNA, as measured by using RT and real-time quantitative qPCR (qPCR); conversely, increasing HuR levels by transfection of a construct that expressed a GFP-HuR fusion protein lowered c-Myc levels (Fig. 1B,C). To study whether HuR influenced c-Myc expression by acting on the c-Myc 3′UTR, a heterologous reporter construct expressing a chimeric RNA that spanned the GFP CR and the c-Myc 3′UTR (Fig. 1D) was tested. As shown, HuR silencing enhanced the expression of GFP (Fig. 1E) from the reporter chimeric plasmid pGFP(3′UTR), but not from pGFP. Supporting the notion that HuR repressed c-Myc translation was evidence that HuR did not influence c-Myc protein stability and data showing that HuR silencing enhanced c-Myc association with actively translating polysomes and promoted nascent c-Myc biosynthesis (Supplemental Figs. S1, S2).
Figure 1.
HuR represses c-Myc expression. (A) HeLa cell lysates were subjected to RNP IP followed by RT–qPCR analysis to measure the enrichment of c-Myc mRNA in HuR IP compared with control IgG IP (Materials and Methods). (B) Forty-eight hours after transfection of HeLa cells with control (Ctrl) siRNA or HuR-directed siRNA, lysates were prepared to assess the levels of c-Myc, HuR, and loading control GAPDH by Western blot analysis (top) and the levels of c-Myc mRNA by RT–qPCR, using 18S rRNA for normalization (bottom). (C) Forty-eight hours after transfection with a control plasmid expressing GFP or a plasmid overexpressing HuR as fusion protein GFP-HuR, the levels of c-Myc, GFP-HuR, endogenous HuR, and GAPDH (top), and c-Myc mRNA (bottom) were measured as explained in B. (D) Plasmid pGFP-3′UTR was constructed by attaching the entire c-Myc 3′UTR after the GFP CR. (E) By 48 h after transfecting pGFP or pGFP-3′UTR, the levels of reporter GFP protein, HuR, and loading control β-actin (left) and GFP and chimeric GFP-3′UTR mRNAs (right) were measured as explained in B. (F) Sequence of the AU-rich c-Myc 3′UTR and schematic depiction of the 5′UTR, CR (CR1, CR2), and 3′UTR (A–D) biotinylated RNAs (assayed in duplicate) used for biotin pull-down analysis (shown in G) (Materials and Methods). (Input) Positive control; (biotinylated GAPDH 3′UTR) negative control. HuR in biotin pull-down samples was detected by Western blot analysis. (H) Constructs were prepared to express chimeric RNAs spanning the GFP CR and each of the four c-Myc 3′UTR segments shown in F; 48 h after transfection, binding of HuR to each chimeric RNA was tested by RNP IP followed by GFP mRNA detection by RT–qPCR. Values in A–C and E are the means ± SD from three independent experiments. The Western blotting data are representative of three or more experiments. Where indicated, c-Myc Western blotting signals were quantified by densitometry.
As other HuR target mRNAs, the c-Myc 3′UTR is highly rich in As and Us (Fig. 1F), and contained two hits of a HuR signature motif (López de Silanes et al. 2004). We further tested the association of HuR with the Myc mRNA by preparing biotinylated segments spanning the c-Myc CR and 5′ and 3′UTRs and by studying their association with HuR by biotin pull-down analysis. As binding was limited to the 3′UTR (Fig. 1G, top), we further subdivided it into four overlapping, ∼150-nt fragments; among them, HuR bound the 3′B and 3′D regions (Fig. 1G, bottom). Validation of these findings using GFP chimeric reporters revealed that fragment B, which contained a 29-nt sequence shown previously to interact with the HuR-related protein HuB/Hel-N1 (Levine et al. 1993; Gao and Keene 1996), preferentially associated with HuR, and it became the focus of subsequent analyses. Together, these findings suggested that HuR interacted with the c-Myc 3′UTR and reduced c-Myc levels by lowering the c-Myc mRNA and by inhibiting its translation. We thus set out to study the mechanism by which HuR represses c-Myc expression.
c-Myc repression by let-7 requires HuR
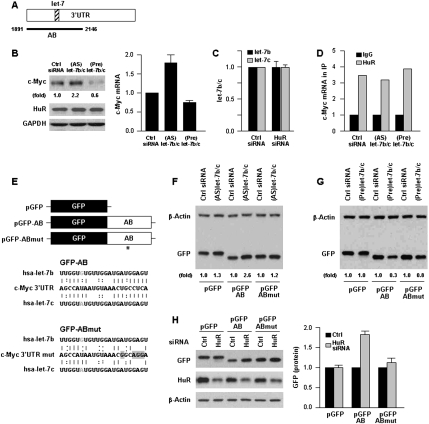
We hypothesized that repression of c-Myc by HuR implicated other factors that associated with the c-Myc mRNA. The miRNA let-7 was predicted to bind the c-Myc 3′UTR at the overlap of segments A and B (Fig. 2A), as reported previously for let-7a (Akao et al. 2006; Sampson et al. 2007); among all computationally identified miRNAs, let-7b and let-7c were predicted to bind with the strongest affinity (Supplemental Fig. S3). Transfection with antagomir (antisense, AS) RNAs to reduce the levels of various let-7 miRNAs revealed that individual reduction of let-7b or let-7c each increased c-Myc expression (Supplemental Fig. S4) and that their joint reduction [(AS)let-7b/c] more potently elevated c-Myc protein and mRNA levels (Fig. 2B). Conversely, transfection of chemically synthesized, precursor duplex RNAs for let-7b and let-7c individually (Supplemental Fig. S4) or jointly [(Pre)let-7b/c] (Fig. 2B) lowered c-Myc levels.
Figure 2.
let-7b/c inhibits c-Myc expression in an HuR-dependent fashion. (A) Schematic of the let-7 interaction site on the c-Myc 3′UTR (hatched). (B) Forty-eight hours after transfection of (AS)let-7b/c or (Pre)let-7b/c, the levels of c-Myc, HuR, and loading control GAPDH (left) were tested by Western blot analysis, and the levels of c-Myc mRNA (right) were measured by RT–qPCR. (C) Forty-eight hours after transfection of Ctrl or HuR siRNAs, the levels of let-7b and let-7c were measured by RT–qPCR analysis. (D) In cells transfected as described in B, the interaction of HuR with c-Myc mRNA was measured by RNP IP followed by RT–qPCR analysis. (E) To test the influence of let-7b/c on c-Myc expression, GFP reporters were prepared in which GFP was linked to the AB segment containing the intact let-7 site (GFP-AB) or a mutant let-7 site (GFP-ABmut) with four point mutations (*) in the seed region that disrupted the interaction of let-7b and let-7c with the c-Myc mRNA (gray highlight). (F,G) Thirty-six hours after transfection of Ctrl siRNA or (AS)let-7b/c (F) or (Pre)let/7b/c (G), plasmid pGFP, pGFP-AB or pGFP-ABmut were transfected and the levels of GFP and loading control β-actin tested 24 h later by Western blot analysis. (H) Following transfection of Ctrl or HuR siRNAs together with plasmids pGFP, pGFP-AB or pGFP-ABmut as explained in F and G, the levels of GFP, HuR, and β-actin were assessed by Western blot analysis (left) and quantified (right). Values in B, C, and H are the means ± SD from three independent experiments. Western blotting data are representative of three or more experiments. Where indicated, “(fold),” and in H, c-Myc or GFP Western blotting signals were quantified by densitometry.
First, we hypothesized that HuR might repress c-Myc translation by elevating let-7 levels; however, the levels of let-7b/c, as measured by using RT–qPCR (Materials and Methods; Lal et al. 2008), were unaffected by HuR silencing (Fig. 2C). Next, we tested the possibility that the association of HuR with c-Myc mRNA may vary depending on let-7b/c levels, but this also was not the case, as binding of HuR to c-Myc mRNA remained essentially unchanged in the (AS)let-7b/c and (Pre)let-7b/c groups (Fig. 2D). To test directly whether HuR and let-7 regulated c-Myc expression, we prepared two GFP reporter constructs: one containing the wild-type c-Myc 3′UTR AB segment, the other containing four point mutations in the let-7 “seed region” to disrupt the let-7 interaction site (pGFP-AB and pGFP-ABmut, respectively) (Fig. 2E). As shown, GFP-AB expression remained responsive to the influence of (Pre)let-7b/c or (AS)let-7b/c, while GFP-ABmut did not (Fig. 2F,G). Importantly, while HuR silencing enhanced GFP-AB levels, in keeping with the data described thus far, it did not enhance GFP-ABmut levels (Fig. 2H). These results indicated that HuR promoted the association of let-7 with the c-Myc mRNA, while let-7 did not affect HuR binding to the c-Myc mRNA. The data further suggested that let-7 binding to c-Myc mRNA is required for HuR-mediated inhibition of c-Myc expression.
HuR is necessary for Ago2/let7 interaction with c-Myc mRNA
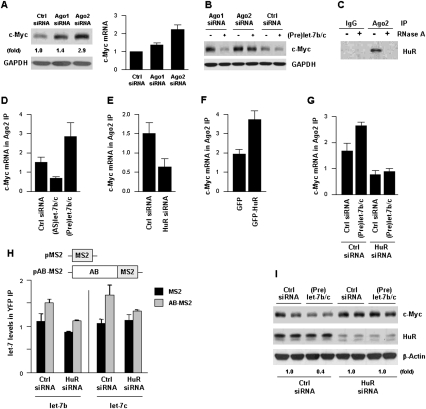
To further test the working model that HuR repressed c-Myc levels by recruiting let-7b/c to the c-Myc 3′UTR, we examined the involvement of the RISC complex in this process. Argonaute (Ago) proteins are integral components of the RISC complex; four Ago proteins are reported, Ago1 through 4, but only Ago2 appears to be implicated in cleaving target mRNAs (Liu et al. 2004; Hutvagner and Simard 2008). Individually silencing Ago1 or Ago2 (Supplemental Fig. S5) revealed that c-Myc protein and, to a lesser extent, the c-Myc mRNA, were preferentially elevated in the Ago2 silencing group (Fig. 3A). In addition, (Pre)let-7 repressed c-Myc levels principally in cells with reduced Ago1, not Ago2 (Fig. 3B), and Ago1 showed relatively lower association with c-Myc mRNA (Supplemental Fig. S6). Collectively, these findings suggested that Ago1 had a lesser influence on c-Myc expression. In co-IP analyses using an anti-Ago2 antibody, we observed that HuR associated with Ago2 in an RNA-dependent manner, as digestion with RNase A disrupted this interaction (Fig. 3C). Further analysis revealed that the interaction of Ago2 with c-Myc mRNA was reduced in cells with silenced HuR or with low let-7b/c levels, while Ago2 showed increased association with c-Myc mRNA in cells overexpressing let-7b/c (Fig. 3D,E); conversely, HuR overexpression increased the interaction of c-Myc mRNA with Ago2 (Fig. 3F). Supporting the notion that the suppressive effect of let-7b/c was dependent on HuR binding to c-Myc mRNA, the enhanced interaction of c-Myc mRNA with Ago2 in the (Pre)let-7b/c group was abrogated if HuR was silenced (Fig. 3G).
Figure 3.
Interaction of c-Myc mRNA with Ago2 requires HuR and is enhanced by let-7b/c. (A) Forty-eight hours after silencing Ago1 or Ago2 by using siRNAs (Supplemental Fig. S5), the levels of c-Myc protein was evaluated by Western blot analysis (left) and the levels of c-Myc mRNA by RT–qPCR analysis (right). (B) Twenty-four hours after transfecting HeLa cells with Ago1, Ago2, and Ctrl siRNAs, cells were further transfected with Ctrl siRNA or (Pre)let-7b/c for an additional 24 h, whereupon the levels of c-Myc were determined by Western blot analysis. (C) The interaction of Ago2 with HuR was tested by IP with IgG or anti-Ago2 antibodies, followed by HuR detection by Western blot analysis in lysates that were either left untreated or digested with RNase A. (D–F) Forty-eight hours after transfection with the indicated RNAs (D,E) and plasmids (F), the levels of c-Myc mRNA present in Ago2 IP were measured by RT–qPCR analysis. (G) Cells were transfected as described in B, except that the influence of (Pre)let-7b/c on c-Myc mRNA interaction with Ago2 was tested in cells with normal (Ctrl siRNA) and reduced (HuR siRNA) HuR levels. (H) Plasmids pAB-MS2 [pcDNA-MS2(12X)-AB] and pMS2 [pcDNA-MS2(12X)] were derived from plasmid pSL-MS2(12X) (Fusco et al. 2003) and the levels of let-7b and let-7c were measured by RT–qPCR in cells expressing either normal (Ctrl siRNA) or reduced (HuR siRNA) HuR by 48 h after transfection (graph). (I) Forty-eight hours after transfection of cells with either Ctrl or HuR siRNAs, in the absence or presence of (Pre)let-7b/c, the levels of c-Myc, HuR, and β-actin were tested by Western blot analysis; c-Myc levels were quantified by densitometry. Values in A and D–H are the means ± SD from three independent experiments. The Western blotting data are representative of three or more experiments.
The dependence of let-7b/c binding to c-Myc mRNA was tested by constructing an artificial reporter RNA bearing the AB fragment linked to multiple MS2 RNA hairpins (Fig. 3H; described by Fusco et al. 2003). By 24 h after transfection of Ctrl or HuR siRNAs, cells were transfected with pMS2 or pAB-MS2 along with a plasmid that expressed an MS2-YFP fusion protein; 24 h later, let-7b and let-7c miRNAs were measured in YFP IP samples. As shown, both let-7b and let-7c were significantly enriched in YFP IP samples in the presence of normal HuR levels, but not if HuR was silenced (Fig. 3H, graph); although we could not totally rule out the possibility of post-lysis rearrangement of these RNP complexes, the data supported the view that HuR promoted the association of let-7b/c to the c-Myc 3′UTR. Finally, c-Myc protein levels were significantly lower in the (Pre)let-7b/c group only in the presence of normal HuR levels; HuR silencing rendered cells refractory to the inhibition of c-Myc expression by (Pre)let-7b/c (Fig. 3I).
Conclusions and perspective
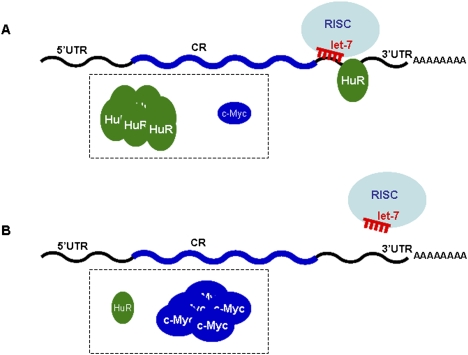
Taken together, our findings indicate that HuR represses c-Myc expression by recruiting let-7 miRNA to an adjacent site on the c-Myc 3′UTR (Fig. 4). Jing et al. (2005) previously reported that tristetraprolin (TTP), another RBP with affinity for AU-rich mRNAs, promoted the decay of the TNF-α mRNA by targeting miR-16-loaded Ago/RISC onto the TNF-α mRNA. Our findings differ from those in the Jing et al. (2005) report in two significant respects. First, we did not find that HuR interacted directly with the Ago/RISC complex, and instead HuR appeared to facilitate the targeting of let-7-loaded RISC to an adjacent region of the c-Myc 3′UTR; the underlying mechanism is unclear, but perhaps HuR binding changes the local RNA conformation, unmasking the let-7 recognition site. Second, the HuR-elicited recruitment of c-Myc mRNA to let-7/RISC reduced both c-Myc mRNA levels and c-Myc translation (Fig. 1; Supplemental Fig. S2). The specific contribution of mRNA decay could not be measured accurately in untreated HeLa cells; perhaps a stressor that enhances or reduces c-Myc mRNA turnover needs to be tested in order to establish whether let-7/RISC affects c-Myc mRNA stability. In contrast to our results that HuR cooperates with a miRNA to repress a shared target mRNA, an elegant study by Bhattacharyya et al. (2006) illustrated an opposite interaction mode for HuR, as it prevented the inhibitory influence of another miRNA. Bhattacharyya et al. (2006) showed that the miR-122-mediated repression of CAT-1 translation was reversed by binding of HuR to a shared site on the AU-rich CAT-1 3′UTR; they further showed that the stress-triggered elevation in cellular HuR facilitated the release of CAT-1 mRNA from processing (P) bodies, suggesting a model whereby HuR and miR-122 associated in a mutually exclusive fashion with the CAT-1 mRNA.
Figure 4.
Proposed model of HuR repression of c-Myc by recruiting let-7/RISC to c-Myc 3′UTR. (A) The presence of elevated HuR promotes the association of let-7-loaded RISC on the c-Myc 3′UTR, in turn lowering c-Myc mRNA levels and translation. (B) In the presence of low HuR levels, let-7-loaded RISC has lower affinity for the c-Myc 3′UTR, and c-Myc production is enhanced. See the text for details.
AU-rich elements such as those present in the TNFα, CAT-1, and c-Myc 3′UTRs and the RBPs that bind to them are widely believed to modulate mRNA stability and translation. Further studies are needed to establish whether the regulation of RNPs by AU-binding RBPs (such as HuR and TTP) broadly implicates miRNA/RISC action. As proposed by George and Tenenbaum (2006), additional interaction schemes involving miRNAs and RBPs are likely to emerge, as our studies deepen into the functions of these potent and versatile regulatory factors. A thorough knowledge of the interplay among RBPs, miRNAs, and other mRNA-interacting molecules is needed, as we strive to understand the complexity of post-transcriptional gene control; only then will we be able to intervene in diseases increasingly recognized to involve aberrant mRNA metabolism.
Materials and methods
Cell culture and transfection
HeLa cells were transfected with Ctrl or HuR siRNAs, with 2-O-methyl (AS)let-7b and (AS)let-7c, or with chemically synthesized duplex (Pre)RNAs (Pre)let-7b and (Pre)let-7c (Ambion); RNAs (at 10 nM final) were transfected using Oligofectamine (Kim et al. 2008; Lal et al. 2008). By 24 h after transfection with Ctrl or HuR siRNAs, or (Pre)let-7b/c was transfected and c-Myc levels assessed by Western blot analysis 24 h later. Plasmid uptake, monitored by examining the numbers of green fluorescent cells and by Western blot analysis, was unchanged among the various transfection groups.
Western blot
Western blot analysis of whole-cell lysates was performed using standard procedures and the following antibodies: monoclonal anti-HuR and anti-GFP from Santa Cruz Biotechnologies; monoclonal anti-c-Myc from BD Pharmingen; and monoclonal anti-Ago2, anti-GAPDH, and anti-β-actin antibodies from Abcam.
RT–qPCR
RNA was isolated from using Trizol (Invitrogen), reverse-transcribed using random hexamers and SSII reverse transcriptase; qPCR was performed using gene-specific primer pairs and SYBR Green (Lal et al. 2008). Primer pairs are listed as Supplemental Material. let-7b/c levels were determined using a miRNA detection kit (mirVana, Ambion) following the manufacturer's protocol.
RNP analysis: RNP IP and biotin pull-down
Analysis of mRNA in RNP IP samples by RT–qPCR was performed as described (López de Silanes et al. 2004), using the housekeeping GAPDH mRNA for normalization. The risk of post-lysis RNP rearrangement was minimized by performing the IP reactions on ice and for shorter time periods. Mouse IgG or monoclonal Ago2 (Abcam) antibodies were used to study the enrichment in c-Myc mRNA in Argonaute2 (Ago2) IP. Biotinylated transcripts that spanned the 5′UTR, CR, and 3′UTR of c-Myc mRNA (NM_002467) were prepared using the primers listed in the Supplemental Material; biotin pull-down assays were performed as described (López de Silanes et al. 2004).
GFP and MS2 constructs
pGFP plasmids were derived from pEGFP-C1 (Clontech) (Supplemental Material). Plasmid pGFP-ABmut was made from pGFP-AB; four point mutations in let-7 seed region were introduced by site-directed mutagenesis. pSL-MS2(12X) vector and pMS2-YFP vector were kindly provided by Dr. R.H. Singer (Fusco et al. 2003). MS2(12X) was cloned into pcDNA3 (Invitrogen) to generate pcDNA-MS2(12X) (pMS2) (Fig. 3H). PCR-amplified c-MYC AB was cloned into pcDNA3-MS2(12X) to generate pcDNA-MS2(12X)-AB (pAB-MS2) (Fig. 3H). By 24 h after siRNA transfections, plasmids pMS2 or pAB-MS2 were transfected and cell lysates prepared for RNP IP 24 h after that.
Acknowledgments
We thank R.H. Singer (Albert Einstein College of Medicine) for providing reagents. This research was supported by the Intramural Research Program of the NIA, NIH.
Footnotes
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.1812509.
Supplemental material is available at http://www.genesdev.org.
References
- Abdelmohsen K, Kuwano Y, Kim HH, Gorospe M. Posttranscriptional gene regulation by RNA-binding proteins during oxidative stress: Implications for cellular senescence. Biol Chem. 2008;389:243–255. doi: 10.1515/BC.2008.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akao Y, Nakagawa Y, Naoe T. let-7 microRNA functions as a potential growth suppressor in human colon cancer cells. Biol Pharm Bull. 2006;29:903–906. doi: 10.1248/bpb.29.903. [DOI] [PubMed] [Google Scholar]
- Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: Target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharyya SN, Habermacher R, Martine U, Closs EI, Filipowicz W. Relief of microRNA-mediated translational repression in human cells subjected to stress. Cell. 2006;125:1036–1038. doi: 10.1016/j.cell.2006.04.031. [DOI] [PubMed] [Google Scholar]
- Brennan CM, Steitz JA. HuR and mRNA stability. Cell Mol Life Sci. 2001;58:266–277. doi: 10.1007/PL00000854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: Are the answers in sight? Nat Rev Genet. 2008;9:102–114. doi: 10.1038/nrg2290. [DOI] [PubMed] [Google Scholar]
- Fusco D, Accornero N, Lavoie B, Shenoy SM, Blanchard JM, Singer RH, Bertrand E. Single mRNA molecules demonstrate probabilistic movement in living mammalian cells. Curr Biol. 2003;13:161–167. doi: 10.1016/s0960-9822(02)01436-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao FB, Keene JD. Hel-N1/Hel-N2 proteins are bound to poly(A)+ mRNA in granular RNP structures and are implicated in neuronal differentiation. J Cell Sci. 1996;109:579–589. doi: 10.1242/jcs.109.3.579. [DOI] [PubMed] [Google Scholar]
- George AD, Tenenbaum SA. MicroRNA modulation of RNA-binding protein regulatory elements. RNA Biol. 2006;3:57–59. doi: 10.4161/rna.3.2.3250. [DOI] [PubMed] [Google Scholar]
- Hinman MN, Lou H. Diverse molecular functions of Hu proteins. Cell Mol Life Sci. 2008;65:3168–3181. doi: 10.1007/s00018-008-8252-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutvagner G, Simard MJ. Argonaute proteins: Key players in RNA silencing. Nat Rev Mol Cell Biol. 2008;9:22–32. doi: 10.1038/nrm2321. [DOI] [PubMed] [Google Scholar]
- Jing Q, Huang S, Guth S, Zarubin T, Motoyama A, Chen J, Di Padova F, Lin SC, Gram H, Han J. Involvement of microRNA in AU-rich element-mediated mRNA instability. Cell. 2005;120:623–634. doi: 10.1016/j.cell.2004.12.038. [DOI] [PubMed] [Google Scholar]
- Keene JD. RNA regulons: Coordination of post-transcriptional events. Nat Rev Genet. 2007;8:533–543. doi: 10.1038/nrg2111. [DOI] [PubMed] [Google Scholar]
- Kim HH, Gorospe M. Phosphorylated HuR shuttles in cycles. Cell Cycle. 2008;7:3124–3126. doi: 10.4161/cc.7.20.6884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HH, Abdelmohsen K, Lal A, Pullmann R, Jr, Yang X, Galban S, Srikantan S, Martindale JL, Blethrow J, Shokat KM, et al. Nuclear HuR accumulation through phosphorylation by Cdk1. Genes & Dev. 2008;22:1804–1815. doi: 10.1101/gad.1645808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim VN, Han J, Siomi MC. Biogenesis of small RNAs in animals. Nat Rev Mol Cell Biol. 2009;10:126–139. doi: 10.1038/nrm2632. [DOI] [PubMed] [Google Scholar]
- Kullmann M, Göpfert U, Siewe B, Hengst L. ELAV/Hu proteins inhibit p27 translation via an IRES element in the p27 5′UTR. Genes & Dev. 2002;16:3087–3099. doi: 10.1101/gad.248902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lafon I, Carballès F, Brewer G, Poiret M, Morello D. Developmental expression of AUF1 and HuR, two c-myc mRNA binding proteins. Oncogene. 1998;16:3413–3421. doi: 10.1038/sj.onc.1201895. [DOI] [PubMed] [Google Scholar]
- Lal A, Kim HH, Abdelmohsen K, Kuwano Y, Pullmann R, Jr, Lal A, Srikantan S, Subrahmanyam R, Martindale JL, Yang X, et al. p16(INK4a) translation suppressed by miR-24. PLoS One. 2008;3:e1864. doi: 10.1371/journal.pone.0001864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leandersson K, Riesbeck K, Andersson T. Wnt-5a mRNA translation is suppressed by the Elav-like protein HuR in human breast epithelial cells. Nucleic Acids Res. 2006;34:3988–3999. doi: 10.1093/nar/gkl571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine TD, Gao F, King PH, Andrews LG, Keene JD. Hel-N1: An autoimmune RNA-binding protein with specificity for 3′ uridylate-rich untranslated regions of growth factor mRNAs. Mol Cell Biol. 1993;13:3494–3504. doi: 10.1128/mcb.13.6.3494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Carmell MA, Rivas FV, Marsden CG, Thomson JM, Song JJ, Hammond SM, Joshua-Tor L, Hannon GJ. Argonaute2 is the catalytic engine of mammalian RNAi. Science. 2004;305:1437–1441. doi: 10.1126/science.1102513. [DOI] [PubMed] [Google Scholar]
- López de Silanes I, Zhan M, Lal A, Yang X, Gorospe M. Identification of a target RNA motif for RNA-binding protein HuR. Proc Natl Acad Sci. 2004;101:2987–2992. doi: 10.1073/pnas.0306453101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- López de Silanes I, Lal A, Gorospe M. HuR: Post-transcriptional paths to malignancy. RNA Biol. 2005;2:11–13. doi: 10.4161/rna.2.1.1552. [DOI] [PubMed] [Google Scholar]
- Meng Z, King PH, Nabors LB, Jackson NL, Chen CY, Emanuel PD, Blume SW. The ELAV RNA-stability factor HuR binds the 5′-untranslated region of the human IGF-IR transcript and differentially represses cap-dependent and IRES-mediated translation. Nucleic Acids Res. 2005;33:2962–2979. doi: 10.1093/nar/gki603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore MJ. From birth to death: The complex lives of eukaryotic mRNAs. Science. 2005;309:1514–1518. doi: 10.1126/science.1111443. [DOI] [PubMed] [Google Scholar]
- Sampson VB, Rong NH, Han J, Yang Q, Aris V, Soteropoulos P, Petrelli NJ, Dunn SP, Krueger LJ. MicroRNA let-7a down-regulates MYC and reverts MYC-induced growth in Burkitt lymphoma cells. Cancer Res. 2007;67:9762–9770. doi: 10.1158/0008-5472.CAN-07-2462. [DOI] [PubMed] [Google Scholar]