Abstract
Developmental regulation of the floral meristem ensures that plants of the same species have similarly sized flowers with a fixed number of floral organs. The maintenance of stem cells in the floral meristem is terminated after the production of a fixed number of floral organ primordia. Precise repression of the Arabidopsis thaliana homeobox gene WUSCHEL (WUS) by the floral homeotic protein AGAMOUS (AG) plays a major part in this process. Here we show that KNUCKLES (KNU) mediates the repression of WUS in floral meristem determinacy control. AG directly induces the transcription of KNU, which encodes a C2H2-type zinc finger protein with a conserved transcriptional repression motif. In turn, KNU represses WUS transcription to abolish stem cell activity. We also show that the timing of KNU induction is key in balancing proliferation and differentiation in flower development. Delayed KNU expression results in an indeterminate meristem, whereas ectopic KNU expression prematurely terminates the floral meristem. Furthermore, KNU induction by AG is preceded by changes in repressive histone modification at the KNU locus, which occurs in an AG-dependent manner. This study provides a mechanistic link between transcriptional feedback and epigenetic regulation in plant stem cell proliferation.
Keywords: Arabidopsis thaliana, flower development, homeotic control, MADS protein, AGAMOUS, WUSCHEL, stem cell
How stem cell populations are maintained and how their differentiation potential is regulated are key issues in developmental biology. In plants, organs are created post-embryonically from a self-maintaining stem cell structure termed the meristem (Steeves and Sussex 1989). Meristem cells divide in a stereotypical manner to keep stem cell activity at the central zone and also to give rise to progeny cells that are displaced to the lateral peripheral zone. Stem cell activity is maintained by signals that come from the underlying organizing center. Flowers develop from floral meristems produced at the peripheral zone of the shoot apical meristem (SAM). In Arabidopsis thaliana, floral meristems and SAMs are regulated by overlapping sets of proteins, which include the CLAVATA (CLV) ligand–receptor system and the homeodomain protein WUSCHEL (WUS) (Clark et al. 1997; Mayer et al. 1998; Fletcher et al. 1999; Brand et al. 2000; Schoof et al. 2000; Kondo et al. 2006). Maintenance of stem cells that express CLV3 requires signals from the organizing center, where WUS is expressed. WUS is sufficient to revert differentiating cells back to the stem cells (Lenhard et al. 2001; Lohmann et al. 2001; Gallois et al. 2004; Reddy and Meyerowitz 2005). Expansion of WUS expression is prevented by the CLV signal transduction pathway (Brand et al. 2000; Schoof et al. 2000). Thus, a negative feedback loop between the WUS-expressing organizing center and CLV3-expressing stem cells maintains appropriate meristem size.
In floral meristems, the balance between the rates of differentiation and proliferation dynamically shifts toward organogenesis as flower development proceeds. Proper developmental regulation of the floral meristem leads to a genus- or species-specific size and number of four different types of floral organs: sepals, petals, stamens, and carpels. In contrast to the indeterminate shoot meristem, the floral meristem ceases to maintain a stem cell population after the initiation of carpels (thus, called determinacy) (Steeves and Sussex 1989; Lenhard et al. 2001; Lohmann et al. 2001). The timing of floral meristem termination is precisely controlled by another feedback loop. The Arabidopsis floral homeotic MADS-box gene AGAMOUS (AG) integrates stem cell regulation with floral patterning events (Bowman et al. 1989; Yanofsky et al. 1990) and is a key component of the second feedback loop in stem cell regulation. AG is induced at flower developmental stage 3 by WUS and the floral meristem regulator LEAFY in whorls 3 and 4 of the floral primordia where stamens and carpels will later form (Smyth et al. 1990; Bowman et al. 1991; Busch et al. 1999; Lenhard et al. 2001; Lohmann et al. 2001). About 2 d after the induction of AG (under 24-h light conditions) (Smyth et al. 1990), WUS expression is shut off in an AG-dependent manner at stage 6, when carpel primordia are specified (Lenhard et al. 2001; Lohmann et al. 2001), whereas AG is continuously expressed in developing stamens and carpels to regulate reproductive development (Bowman et al. 1991). In ag-1 mutant flowers, extra whorls of sepals and petals develop from the center of the flower, resulting in a flower-within-flower phenotype (Bowman et al. 1989). The ag mutant flowers show prolonged WUS expression in the center of the floral meristem, and ag wus double-mutant flowers resemble wus flowers, showing that one role of AG is to down-regulate WUS expression, which prevents the floral meristem from growing indefinitely by terminating stem cell activity in the center of the flower (Lenhard et al. 2001; Lohmann et al. 2001).
The AG and CLV pathways appear to function at least partially independently to repress WUS, because the effects of ag and clv1 mutations on floral meristem determinacy are additive and WUS is expressed in larger domain in ag clv1 flowers than in ag flowers (Clark et al. 1993; Lohmann et al. 2001). The WUS/CLV regulatory loop takes place between adjacent cells inside the meristem, with activation and repression occurring simultaneously (Brand et al. 2000; Schoof et al. 2000). In contrast, in the WUS/AG pathway, activation and repression are temporally separated in the same domain (Lenhard et al. 2001; Lohmann et al. 2001). Thus, AG is necessary for the temporal balance between differentiation and proliferation of stem cells. However, almost nothing is known about mechanisms by which AG represses WUS at the proper developmental time in the feedback loop. Moreover, whether AG directly controls WUS or induces a mediator to repress WUS and how developmental timing is measured to ensure complete flower development are questions that remain unanswered.
Mutations in several other genes besides AG cause floral meristem determinacy defects (Fletcher 2001; Payne et al. 2004; Zhao et al. 2004; Carles et al. 2005; Prunet et al. 2008), but none of these genes have been shown to function downstream from AG. The flowers of knuckles (knu) produce ectopic stamens and carpels in a reiterating pattern inside fourth-whorl carpels (Payne et al. 2004). KNU encodes a C2H2-type zinc finger protein with a C-terminal EAR-like active repression motif. Expression of KNU starts at stage 6 in the center of the floral meristem, when and where WUS expression is repressed, indicating that KNU may be an upstream repressor of WUS. KNU expression continues in developing stamens and carpels to promote maturation of reproductive organs, as is suggested by the male sterile phenotypes of knu (Payne et al. 2004).
To understand molecular mechanisms by which the floral homeotic protein AG controls stem cell maintenance and differentiation, we performed genetic and biochemical analyses based on a working hypothesis that KNU might be a link between AG and WUS in the transcriptional cascade. First, we performed detailed timed analysis of AG function in floral meristem determinacy control, and then we examined the genetic and molecular interactions among AG, KNU, and WUS. We show by expression analyses, chromatin immunoprecipitation (ChIP), and promoter-mutagenesis assays that AG directly regulates KNU, a potent repressor of WUS transcription. We further show that the timing of KNU induction is key in balancing proliferation and differentiation in flower development. We furthermore report that proper KNU regulation requires AG-dependent changes of a repressive histone modification. Based on these data, we propose a molecular mechanism controlling the developmental timing of stem cell maintenance and differentiation in Arabidopsis floral meristems.
Results
Timing of AG-dependent meristem determinacy
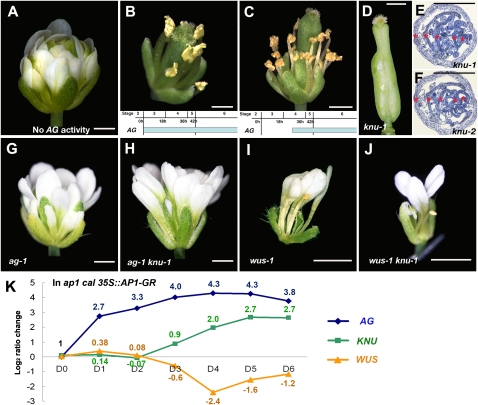
To examine the action of AG in floral meristem determinacy control, we performed a series of timed activation experiments using an established ag-1 35S∷AG-GR line (Ito et al. 2004). This line contains a chemically inducible gene activity by a translational fusion of the AG protein and the steroid-binding domain of the rat glucocorticoid receptor (GR) (Lloyd et al. 1994). The fusion gene is expressed ubiquitously under the Cauliflower Mosaic Virus 35S promoter independently of floral stage and rescues the ag-1 mutant phenotype when dexamethasone (DEX) is continuously provided (Fig. 1A,B; Supplemental Fig. 1A–D; Ito et al. 2004, 2007). When initiated at floral stage 3, repeated DEX treatments led to normal-looking stamens and carpels in whorls 3 and 4, respectively (Fig. 1B; Ito et al. 2007). In contrast, DEX treatments of flowers at stage 4 with a 1-d delay from stage 3 gave a partially indeterminate phenotype, with (from whorl 1 inward) a sepal–stamen–stamen–sepal–stamen–stamen–carpel structure (n > 20) (Fig. 1C; Supplemental Fig. 1B,D). These phenotypes indicate that flowers that had been at stage 3 at the time of the initial DEX treatment produced carpels in whorl 4 (Fig. 1B). In contrast, floral buds at stage 4 at the time of DEX treatment had already passed the stage when it is possible to establish meristem determinacy in a normal timing (Fig. 1C). This result shows that the initial AG expression starting from stage 3 is necessary for floral meristem determinacy.
Figure 1.
Genetic interactions of AG, KNU, and WUS. Floral phenotypes of ag-1 35S∷AG-GR mock-treated (A) or treated four times with DEX (B,C). AG induction on a stage-3 flower generates a fully rescued flower after 12 d (B), whereas induction on a stage-4 flower after 11 d generates a flower with a partial indeterminate phenotype with extra whorls of stamens (C). Temporal induction of AG is shown at the bottom of B and C. (D) knu-1 silique. (E,F) Cross-sections of knu-1 (E) and knu-2 (F) siliques. Each silique contains five reiterations, as shown by red asterisks. (G–J) Mutant flowers of ag-1 (G), ag-1 knu-1 (H), wus-1 (I), and wus-1 knu-1 (J). (K) Comparison of AG, KNU, and WUS transcripts in ap1 cal 35S∷AP1-GR 0 d (D0), 1 d (D1), 2 d (D2), 3 d (D3), 4 d (D4), 5 d (D5), and 6 d (D6) after one time of 1 μM DEX treatment. The value of the fold change is shown by log2. Fold change values relative to that in D0 with standard deviations are shown in Supplemental Table 1. Bars, 1 mm.
Genetic relationships of AG, KNU, and WUS
WUS is necessary for stem cell maintenance, and its expression is repressed at stage 6 in an AG-dependent manner (Lenhard et al. 2001; Lohmann et al. 2001). We investigated a series of AG targets (Ito et al. 2004, 2007), and some lines of evidence, including our AG-binding assay, indicated that AG might not directly repress WUS (Mizukami and Ma 1997; Sieburth et al. 1998; data not shown). We hypothesized that KNUCKLES (KNU) might be an intermediate factor in the AG–WUS pathway because of our preliminary overexpression assay suggesting a link with WUS (data not shown; see below).
There was only one mutant allele for knu. The temperature-sensitive knu-1 grown at 16°C were indistinguishable from wild type, but when grown at 25°C, the flowers showed indeterminate phenotypes with four to five reiterations of ectopic stamens and carpels in the primary carpels (Fig. 1D,E; Supplemental Fig. 2; Payne et al. 2004). To test if knu-1 shows null mutant phenotypes, we obtained the second allele from the Arabidopsis Biological Resource Center with a T-DNA insertion in the coding region, and designated it as knu-2 (Fig. 1F; Supplemental Fig. 2). The knu-2 flowers grown under different temperatures (16°C, 22°C, and 25°C) showed sterile and indeterminate mutant phenotypes similar to knu-1 flowers grown at 25°C (n > 20 for each) (Fig. 1F; Supplemental Fig. 2), suggesting that the phenotype of knu-1 grown at 25°C appears to be a consequence of the loss of KNU function. Thus, we used fertile knu-1 grown at 16°C for genetic crosses and observed mutant phenotypes by growing them at 25°C.
The ag-1 knu-1 double-mutant flowers (n > 20) showed the same phenotypes as ag-1 (Fig. 1G,H). The combination of knu-1 and a partial AG loss-of-function allele ag-4 (knu-1 ag-4; n > 20) that has almost normal stamens and indeterminate floral meristems (Sieburth et al. 1995) showed similar indeterminate flowers to ag-4, but with sterile stamens (data not shown). These results indicate that the ag mutation is epistatic to knu. Next, we created wus-1 knu-1 and found that wus knu flowers (n > 5) were indistinguishable from wus, showing that wus is epistatic to knu in the floral meristem and that the indeterminate phenotype of knu is dependent on the activity of WUS (Fig. 1I,J). These genetic interactions suggest that AG, KNU, and WUS function in the same pathway in floral meristem control and support our hypothesis that KNU is an intermediate factor between AG and WUS in the transcriptional regulation of meristem determinacy.
Expression correlation between AG, KNU, and WUS
Next, we compared the expression levels of AG, KNU, and WUS by quantitative PCR using plants with the synchronized floral induction system ap1 cal 35S∷AP1-GR (Wellmer et al. 2006), which allows the collection of a large number of synchronized floral buds at distinct developmental stages (Fig. 1K; Supplemental Table 1). AG was rapidly induced after 1 μM DEX treatment and continuously increased until day 4. In contrast, KNU and WUS transcript levels did not change very much from day 0 until day 2 and showed opposite trends thereafter. KNU induction started to be observed on day 3, and at the same time, reduction of WUS transcription started. Later in flower development, WUS RNA level started to increase, which may be due to WUS expression in specific parts of developing anthers and ovules (Gross-Hardt et al. 2002; Deyhle et al. 2007). This correlation between AG, KNU, and WUS expression in early flowers further indicates that AG expression starting at stage 3 may induce KNU ∼2 d later when floral buds reach stage 6, and KNU may then immediately repress WUS in the floral meristem.
KNU is necessary for the repression of WUS
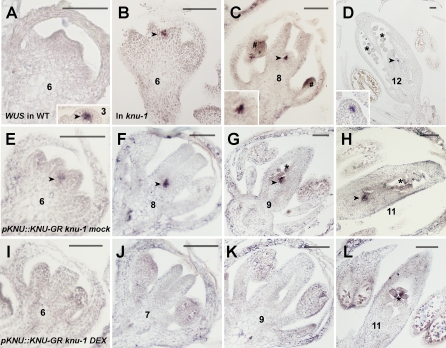
To examine whether KNU functions upstream of WUS and represses its transcription, we examined WUS expression in knu-1 flowers by in situ hybridization. In wild-type flowers, WUS is expressed in the organizing center underlying the stem cells in floral meristems, and the expression is abolished at stage 6 (Fig. 2A; Mayer et al. 1998). However, in knu-1, the dome of cells is present between two developing carpels of flowers later than stage 6, and the prolonged WUS expression was observed in a small group of cells of the dome-shaped meristem (Fig. 2B–D, insets of C,D). This result shows that KNU is necessary to repress WUS transcription.
Figure 2.
KNU represses WUS in the floral meristem. (A) WUS expression in a wild-type flower at stage 3 (arrowhead) and stage 6. (B–D) Prolonged WUS expression in knu-1 flowers at stage 6 (B), stage 8 (C) and stage 12 (D). Arrowheads and insets in C and D show WUS expression in meristematic cells. The pound sign (#) and asterisks (*) show WUS expression in developing anthers and ovules, respectively. (E–H) Ectopic WUS expression in mock-treated knu-1 pKNU∷KNU-GR flowers at stage 6 (E), stage 8 (F), stage 9 (G), and stage 11 (H). Arrowheads show ectopic WUS expression in meristematic cells. Asterisks indicate normal WUS expression in ovules. (I–L) Ectopic WUS expression disappears 6 d after four DEX treatments in knu-1 pKNU∷KNU-GR flowers at stage 6 (I), stage 7 (J), stage 9 (K), and stage 11 (L). The asterisk in L indicates WUS expression in ovules. Bars, 50 μm.
To further test the requirement of KNU for the repression of WUS, we created steroid-inducible lines expressing a translational fusion of KNU with GR under the control of the KNU promoter in knu-1 plants, rather than using functional KNU in knu-1 at slow-growing low-temperature conditions. We selected 10 independent T1 lines for pKNU∷KNU-GR in the knu-1 background that showed knu mutant phenotypes in the uninduced condition. In these lines, continuous WUS expression was detected inside the developing carpels at later stages (Fig. 2E–H; Supplemental Fig. 3). Four DEX treatments at 1-d intervals completely rescued the mutant phenotypes ∼10–12 d later in six lines out of 10 (Supplemental Fig. 3), suggesting that the KNU fusion protein can retain its endogenous function. Six days after the initial DEX treatments, WUS expression in the meristem was fully terminated in the corresponding regions of developing carpels before the morphological rescue was observed (Fig. 2I–L). WUS expression in developing ovules was not suppressed by DEX treatments (Fig. 2L), as expected from the nonoverlapping expression patterns of KNU and WUS in wild-type ovules (Gross-Hardt et al. 2002; Payne et al. 2004). These results indicate that KNU represses WUS expression in the floral meristem from stage 6.
Ectopic KNU expression leads to wus flowers by repressing WUS transcription
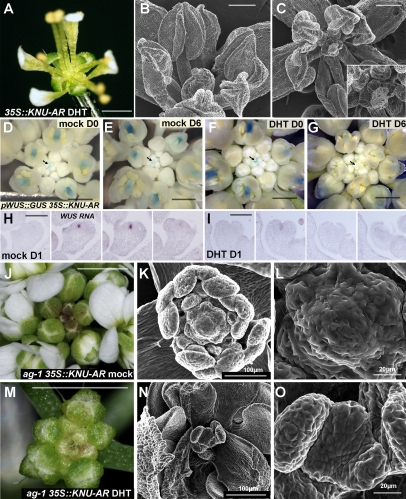
To examine whether KNU is sufficient to repress WUS expression, we ectopically expressed the fusion protein between KNU and the steroid hormone ligand-binding domain (GR or androgen receptor, AR) (Chang et al. 1988) under the control of the constitutive 35S promoter in wild-type plants. The 35S∷KNU-GR/AR plants looked normal without induction, but after continuous hormone treatments (three to four daily treatments with DEX for the GR fusion or with 5α-androstan-17β-ol-3-one [DHT] in the case of the AR fusion), the plants showed wus mutant–like flowers that lacked carpels in the center (Fig. 3A). We examined 15 T1 lines for each construct, and at least eight lines with each construct showed various degrees of wus-like flowers after the repeated hormone treatments (Fig. 3A–C). In early-stage floral buds of 35S∷KNU-AR lines, tiny carpel primordia (Fig. 3B) and a degenerated bulged mass of cells (Fig. 3C) were observed. To determine if the repression motif located at the C-terminal end of KNU is necessary for WUS repression, we produced a fusion protein with truncated KNU (15-amino-acid deletion as shown in Supplemental Fig. 2) and the steroid hormone ligand-binding domain. All of the >20 T1 transgenic plants appeared normal even after the induction (data not shown), suggesting that the repression motif may be necessary for termination of the floral meristem or that truncation of the C-terminal end may affect the conformation of KNU. These results indicate that overexpression of KNU is sufficient to precociously terminate floral meristems, possibly functioning as a repressor.
Figure 3.
KNU overexpression confers a determinate meristem by repressing WUS. (A–C) 35S∷KNU-AR flowers after repeated DHT treatments. (B,C) Degenerated tissue was observed in the center of the flower. The inset in C is the close-up of the central organ. (D–G) GUS staining patterns in pWUS∷GUS 35S∷KNU-AR inflorescences 0 d (D,F) or 6 d (E,G) after three mock (D,E) or DHT (F,G) treatments. (G) WUS expression in young floral buds (arrows) and in developing pistils (arrowheads) almost completely disappeared 6 d after ectopic KNU induction. (H,I) Serial sections of 35S∷KNU-AR flowers at stage 4 hybridized with a WUS antisense probe 1 d after mock (H) or DHT (I) treatment. (J–O) Inflorescences (J,M) and floral meristems (K,L,N,O) of ag-1 35S∷KNU-AR plants 12 d after four mock (J–L) or DHT (M–O) treatments. In the mock-treated ag-1 35S∷KNU-AR plants, the inflorescence remained unchanged (J) and the floral meristem was dome shaped (K,L). (M) In contrast, DHT-treated plants showed retarded inflorescence growth and displayed a discontinuous size increment between young flower buds and old ones grown after the treatments. (N,O) After ectopic expression of KNU, the floral meristem became flat, and a reduced number of developing organs were observed. Bars: A,J,M, 1 mm; B,C, 200 μm; D–G, 500 μm; H,I,K,N, 100 μm; L,O, 20 μm.
To examine if KNU transcriptionally represses WUS, the 35S∷KNU-AR line was crossed with the pWUS∷GUS reporter line, which reflects the endogenous expression pattern of WUS (Gross-Hardt et al. 2002). Mock treatment did not affect WUS promoter activity (Fig. 3D,E). In contrast, KNU induction by three DHT treatments led to full suppression of the WUS reporter within 6 d in inflorescence meristem (data not shown) and in early-stage flowers (n > 20) (arrows in Fig. 3F,G). The repression was also observed in the developing pistils by ectopic KNU activity (arrowheads in Fig. 3F,G). We did not detect rapid repression of GUS staining, possibly due to the stability of GUS activity (data not shown). By in situ hybridization with a WUS probe using the 35S∷KNU-AR inflorescences, the almost complete repression of WUS transcript was observed at stage 4 floral buds 1 d after the KNU induction (Fig. 3H,I). This shows that ectopic KNU expression precociously represses WUS transcription, leading to the termination of stem cell activity in the floral meristem.
Ectopic KNU is sufficient to rescue indeterminate phenotypes of ag-1 flowers
To examine whether KNU plays a major role downstream from AG for the control of floral meristem determinacy, we introduced the 35S∷KNU-AR construct into the ag-1 loss-of-function mutant (Fig. 3J–O). Mock-treated plants (n = 10) showed ag-1 mutant flowers with a dome-shaped meristem in the center of the flower (Fig. 3J–L). In contrast, KNU ectopic expression resulted in smaller flowers with arrested reiteration (Fig. 3M,N) (n = 10). In the center of the flower, the meristematic region became flat, and stem cell maintenance appeared to be terminated (Fig. 3N,O). This shows that KNU is sufficient to rescue indeterminate phenotypes of ag-1.
Taken together, these data suggest that KNU is a key mediator downstream from AG to control floral meristem determinacy.
AG induces KNU after ∼2 d of delay
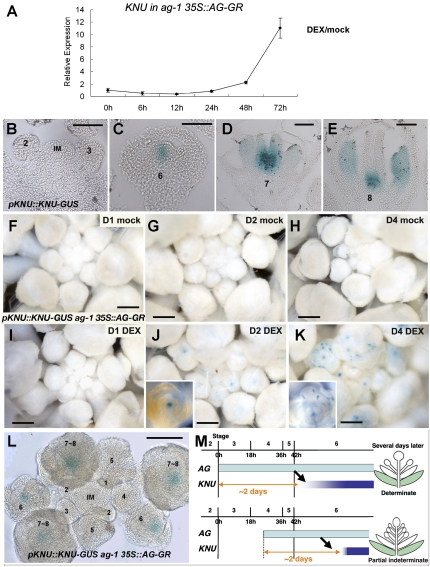
To examine the link between AG and KNU, we used the inducible line of AG activity, ag-1 35S∷AG-GR, for quantitative PCR expression analysis (Fig. 4A). In contrast to the quick induction of another AG target SPOROCYTELESS (SPL) by single DEX treatment (Supplemental Fig. 4; Ito et al. 2004), KNU level remains unchanged for 24 h. KNU was only noticeably induced to 2.3-fold in 48 h. In 72 h, the KNU transcript was up-regulated >10-fold. This KNU expression timing is consistent with the endogenous KNU expression in wild-type flowers shown by KNU reporter lines (Fig. 4B–E; Payne et al. 2004). pKNU∷KNU-GUS plants (n > 20) exhibited strong staining in the organizing center and weak staining in the outermost stem cell layers of the floral buds at stage 6 after an ∼2-d delay from the induction of AG at stage 3 (Fig. 4B,C). KNU induction in 35S∷AG-GR (Fig. 4A) was slightly earlier than that in ap1 cal 35S∷AP1-GR (Fig. 1K), possibly due to the fact that the post-translational induction of AG activity in the 35S∷AG-GR plants happens quicker than transcriptional induction.
Figure 4.
KNU is up-regulated 2 d after AG induction. (A) KNU expression in ag-1 35S∷AG-GR. The inflorescences were treated with mock or 10 μM DEX once and harvested 0, 6, 12, 24, 48, and 72 h after the treatment for expression analysis. The relative expression value was calculated by comparing the amount of KNU transcripts between mock- and DEX-treated samples. (B–E) pKNU∷KNU-GUS staining in wild-type inflorescence (IM) and flowers at stages 2 and 3 (B), stage 6 (C), stage7 (D), and stage 8 (E). (F–K) GUS staining in ag-1 35S∷AG-GR pKNU∷KNU-GUS plants 1 d (F,I), 2 d (G,J), and 4 d (H,K) with one mock (F–H) or one DEX (I–K) treatments at day 0. (J and inset) KNU-GUS was induced in the floral meristem 2 d after the DEX treatments. (L) Cleared image of GUS staining in ag-1 35S∷AG-GR pKNU∷KNU-GUS inflorescence 2 d after the initial DEX treatments. IM and numbers represent the inflorescence meristem and floral stages, respectively. (M) Diagrams showing up-regulation of KNU 2 d after AG induction. AG initiated at stage 3 led to KNU induction 2 d later at stage 6, as occurs in normal flower development, which resulted in a normal-patterned flower. If AG was induced with a 1-d delay, it still took 2 d for AG to induce KNU. This delayed induction of KNU resulted in partial indeterminate flowers (Fig. 1C). Bars: B–E,L, 50 μm; F–K, 500 μm.
The KNU promoter continued to be active in the basal central part of developing pistils and in developing stamen anthers (Fig. 4D,E), which overlapped with the expression pattern of AG. The KNU expression pattern in developing pistils of wild-type flowers corresponded with the pattern of ectopic WUS expression in the knu flowers (cf. Figs. 4C–E and 2B,C), which further suggests that KNU is necessary to prevent prolonged WUS expression.
To address how KNU is induced by AG, we generated ag-1 35S∷AG-GR pKNU∷KNU-GUS transgenic plants (n > 40), conducted with one mock or DEX treatment and stained 1, 2, or 4 d later (Fig. 4F–K). The mock-treated plants did not express the reporter, indicating that AG activity is indispensable for KNU expression (Fig. 4F–H). On day 1 after DEX treatment, GUS staining was not detectable (Fig. 4I), indicating that ectopic AG expression does not immediately induce KNU even in the floral buds older than stage 6, as was also shown by quantitative PCR analysis (Fig. 4A,I). On day 2, strong GUS staining was observed in the center of the floral buds older than stage 6 but not in floral buds at earlier stages (Fig. 4J,L). On day 4, when anther-like characteristics start to develop (Ito et al. 2007), GUS activity was observed not only in the floral meristem but also at the lateral edges of the developing organs (Fig. 4K). The staining in developing organs corresponded with wild-type KNU expression in developing anthers (Fig. 4D,E,K). In summary, the transcriptional induction of KNU leading to WUS repression is dependent on ∼2 d of AG activity. AG expression that starts at stage 3 is necessary for floral meristem determinacy control (Fig. 1B,C). Therefore, the partial indeterminate phenotype resulting from the AG induction at stage 4 with a 1-d delay could be explained by the 1-d delay of KNU induction (Figs. 1C, 4M).
AG directly binds to the KNU promoter region
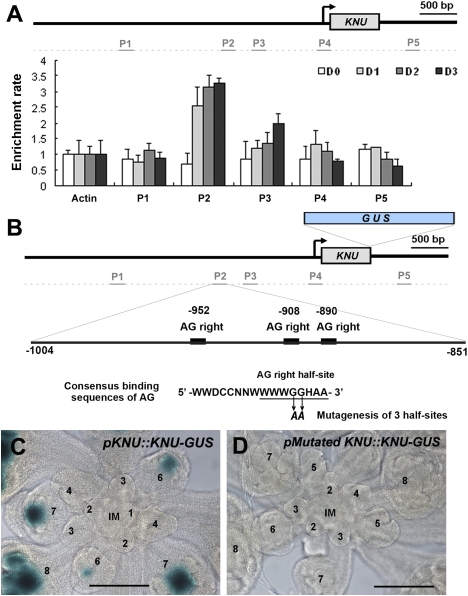
To examine whether AG directly regulates KNU, we performed ChIP analysis using ap1 cal 35S∷AP1-GR inflorescences and an antibody against AG (Fig. 5A; Ito et al. 1997). We treated the plants with DEX once and harvested the inflorescence materials with synchronized floral buds at days 0, 1, 2, and 3 after the treatment. Using the same samples, we showed that induction of KNU starts after day 2 (Fig. 1K). To compare the enrichment ratio in the time course assay, all samples were processed simultaneously in the ChIP assay and quantitative PCR assay using KNU genomic primer sets. On day 0 after DEX treatment, no enrichment was observed by any of the primer sets tested (Fig. 5A). The same was observed in the negative control ChIP assay using ag-1 inflorescences (Supplemental Fig. 5). Interestingly, on day 1, we detected enrichment in the region ∼900 base pairs (bp) upstream of the transcriptional start (primer set P2 spanning from −851 to −1004 from the transcription start site) (Fig. 5A). On days 2 and 3, a slight increase in the enrichment rate was observed using the same primer set. Modest enrichment was also detected by the primer set for the neighboring region (P3 set, −673 to −529). Notably there are no perfect consensus binding sequences of AG (Huang et al. 1993; Shiraishi et al. 1993) around the enrichment site of P2, but there are three half-sites for the AG consensus (Fig. 5B), which can be weakly bound by AG in vitro (Ito et al. 1997).
Figure 5.
KNU is a direct target of AG. (A) ChIP assay using ap1 cal 35S∷AP1-GR inflorescences harvested 0, 1, 2, and 3 d after a single DEX treatment. Nuclear protein complexes were immunoprecipitated with anti-AG (Ito et al. 1997), and the enriched DNA was used for quantitative PCR analysis. Positions of the primer sets in the KNU locus used for the analysis are shown at the top. The Y-axis shows relative enrichment using IgG as a control. Error bars represent SD of three PCR replicates. Actin was used as a control gene for calibration. (B) Diagram of KNU promoter and the positions of conserved half-sites of AG binding consensus. Around the AG-binding site (primer set P2) determined by the ChIP assay, there are three half-sites of the palindromic consensus sequences for AG binding (Huang et al. 1993; Shiraishi et al. 1993). The conserved GG nucleotides were mutagenized to AA. (C,D) GUS staining of inflorescences with the wild-type construct pKNU∷KNU-GUS (C) and the mutated AG-binding sites pMutated KNU∷ KNU-GUS (D). IM and numbers represent the inflorescence meristem and floral stages, respectively. Bars, 100 μm.
To examine whether these AG-binding sites have cis-regulatory activities, we mutagenized the half-sites (GG to AA) (Fig. 5B). Most of the T1 transgenic lines (80%, 12 out of 15) with the wild-type pKNU∷KNU-GUS construct showed the normal GUS expression (Fig. 5C; Table 1). In contrast, the mutation of three half-sites (pMutatedKNU∷KNU-GUS) resulted in no GUS expression at all in the majority of T1 lines (88%, 22 out of 25) (Fig. 5D; Table 1). Taken together, these results show that AG binds the KNU promoter region located ∼900 bp upstream of the transcription start site and directly activates the expression of KNU. Our results also show that the binding of AG to the KNU promoter is evident on day 1 when KNU transcription is not induced, suggesting that AG might require a cofactor or that KNU induction is inhibited by some unknown mechanism.
Table 1.
Distribution of primary transgenic plants for KNU reporter constructs
T1 transgenic plants for each KNU-reporter construct were categorized into three groups by their expression patterns—Normal, Ectopic, or No expression. N shows the total number of plants examined.
aThese three lines showed faint expression in the floral buds at stage 6 and later.
bOut of 11 lines, eight showed ectopic and precocious expression in inflorescence meristems and young floral buds at stages 1–5. The remaining three lines showed precocious expression in the floral buds younger than stage 6, but not in the inflorescence meristem.
The repressive histone modification H3K27me3 prevents precocious KNU expression
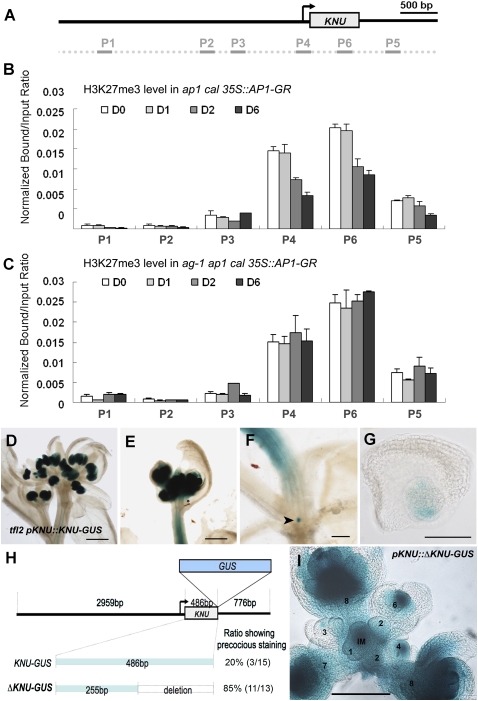
To examine a possible mechanism to explain the precise expression of KNU, we examined epigenetic marks in the KNU locus. A genome-wide analysis of a well-established repressive histone mark, histone H3 Lys 27 trimethylation (H3K27me3), showed that the KNU locus is covered by H3K27me3 in vegetative seedlings (Zhang et al. 2007a). We used ap1 cal 35S∷AP1-GR inflorescences and performed time-course analysis of histone modification of the KNU locus during flower development (Fig. 6A,B). We found that the regions around the transcriptional start site and gene body are heavily covered by the repressive mark H3K27me3 at the initial stages of flower development on days 0 and 1 (primer sets P4 and P6). In contrast, we did not detect any enrichment of H3K27me3 around the AG-binding site (P2 in Fig. 6B). On day 2, the enrichment ratio for the repressive mark dropped >30% and further reduced on day 6. Another histone modification, H3K4me2, levels on KNU chromatin remained constant on days 0 and 6 (Supplemental Fig. 6), indicating that not histone density but H3K27me3 levels change during flower development. The greater reduction of H3K27me3 levels at later timing corresponds with broader KNU expression in reproductive organs in addition to floral meristems (Fig. 4C–E). This indicates that the change in H3K27me3 levels may occur only in cells where the transcriptional change is induced. Interestingly, on day 2, KNU was not yet transcriptionally induced in the ap1 cal 35S∷AP1-GR inflorescences (Fig. 1K), showing that this change in H3K27me3 at the KNU locus precedes the transcriptional change.
Figure 6.
The repressive histone modification H3K27me3 plays a role in timing control of KNU expression. (A) Positions of the primer sets in the KNU locus used for the ChIP assay in B and C. (B,C) ChIP assays using ap1 cal 35S∷AP1-GR (B) and ag-1 ap1 cal 35S∷AP1-GR (C) inflorescences 0, 1, 2, and 6 d after single DEX treatment. The Y-axis shows the ratio of bound DNAs after IP to input DNAs. Error bars represent SD of three PCR replicates. (B) The transcription start site (P4) and gene body (P6) contained higher levels of the repressive mark, which were reduced 2 d after DEX treatment in ap1 cal 35S∷AP1-GR inflorescences. (C) However, in the ag-1 background, high levels of H3K27me3 were maintained. (D–G) GUS staining in tfl2 pKNU∷KNU-GUS plants. (D,E) KNU was ectopically expressed in inflorescence stems in addition to strong GUS staining in developing anthers. (F,G) Ectopic staining was also observed in the lateral meristems (arrowhead in F) subtended by cauline leaves. (H) Diagram of KNU promoter constructs and ratio of primary transformants showing the precocious expression. (I) GUS staining of an inflorescence of pKNU∷ΔKNU-GUS plants. IM and numbers represent the inflorescence meristem and floral stages, respectively. Bars: D–F, 1 mm; G, 50 μm; I, 100 μm.
To examine whether the change in chromatin modification is dependent on AG activity, we performed the ChIP assay using ag-1 ap1 cal 35S∷AP1-GR plants (Fig. 6C). High levels of H3K27me3 repressive mark (indicated by primer sets P4 and P6) were maintained throughout days 0–6. This shows that the reduction in H3K27me3 levels at the KNU locus requires AG activity.
To examine if H3K27me3, which is mediated by polycomb group (PcG) complexes, is involved in the developmental regulation of KNU transcription, we used quantitative PCR to determine whether KNU is ectopically expressed in PcG mutants (Supplemental Fig. 7). CURLY LEAF (CLF) and SWINGER (SWN) are partially redundant histone methyltransferase E(Z) homologs and subunits of polycomb-repressive complex 2 (PRC2) (Schubert et al. 2006). Another key PRC2 component is FERTILIZATION-INDEPENDENT ENDOSPERM (FIE), an ESC homolog (Katz et al. 2004). PRC2 complexes are necessary to deposit and maintain the repressive mark H3K27me3 (Katz et al. 2004; Schubert et al. 2006). In the clf swn double-mutant and 35S∷FIE cosuppression lines (n > 20 each mixed as a pool), ectopic expression of KNU was detected in vegetative leaves (Supplemental Fig. 7), although the increase was not very strong (about twofold compared with the wild-type background, in contrast to the ectopic expression of AG [∼30-fold increase] and AP3 [threefold to sixfold increase]). KNU was not ectopically induced simply by ectopic expression of AG in early-stage floral buds or vegetative leaves (Fig. 4L; data not shown). Therefore, these results indicate that loss of PRC2 activity may lead to derepression of KNU.
To examine whether KNU expression is affected in the mutant for a factor necessary to maintain the repressive status associated with the H3K27me3 mark, we crossed the pKNU∷KNU-GUS line with terminal flower2 (tlf2) plants (Fig. 6D–G). TFL2 encodes A. thaliana LIKE HETEROCHROMATIN PROTEIN 1 (LHP1), which epigenetically represses FLOWERING LOCUS C (FLC), AG, and AP3 through the H3K27me3 repressive mark (Sung et al. 2006; Turck et al. 2007). Genome-wide analysis of LHP1-binding sites showed that the KNU locus is bound by LHP1 in seedlings (Zhang et al. 2007b). In tfl2 mutants, the main inflorescence shoots are terminated after three to four flowers are created, and lateral shoots are often terminated as a single flower (Fig. 6D; Takada and Goto 2003; Turck et al. 2007; Zhang et al. 2007b). In tfl2 mutants (n = 20), KNU-GUS was strongly expressed in flowers, and occasionally the strong ectopic expression was detected in the vasculature of inflorescence stems (Fig. 6D–F; Supplemental Fig. 8) (for the control in the wild-type background). The KNU ectopic staining was also observed in the lateral meristems subtended by cauline leaves (arrowhead in Fig. 6F,G). These results indicate that PcG complexes are involved in preventing precocious KNU expression.
We further confirmed these results through a KNU promoter analysis (Fig. 6H). Deletion of part of the KNU coding region (represented by the P6 primer set in Fig. 6A, pKNU∷ΔKNU-GUS) but with intact upstream and downstream sequences resulted in ectopic expression in the vegetative tissues, SAMs, young floral buds, and developing flowers in the majority of T1 transgenic plants (Fig. 6I; Table 1). In pKNU∷ΔKNU-GUS inflorescences, the GUS reporter RNA level was ∼14-fold higher than in pKNU∷KNU-GUS inflorescences (Supplemental Fig. 9), showing that the ectopic expression occurs at the transcriptional level. This shows that the full KNU coding region covered by the H3K27me3 repressive mark is necessary for developmental regulation of KNU expression. These data indicate that KNU expression is suppressed by repressive histone modification of the KNU locus, and it is induced in a proper developmental timing through the AG-dependent removal of the repressive mark.
Discussion
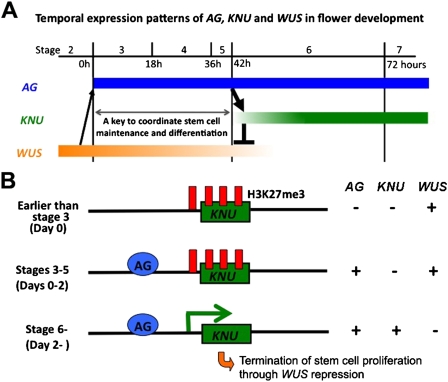
In flower development, stem cell proliferation is precisely terminated to ensure floral organ differentiation and to prevent extra organ formation. Here we showed that the C2H2-type zinc finger repressor protein KNU is a key link in the feedback regulation of WUS by AG, and the precise balance between proliferation and differentiation is executed at the level of KNU transcriptional regulation (Fig. 7A). Overexpression of AG does not terminate the floral meristem precociously (Mizukami and Ma 1992), but the overexpression of KNU was sufficient to shut off WUS expression and prematurely terminate the stem cell population. Furthermore, we showed that AG-dependent histone demethylation is involved in controlling the timing of KNU expression.
Figure 7.
Model of stem cell regulation in flower development. (A) Timing of AG, KNU, and WUS expression in the control of stem cell maintenance and differentiation in flower development. (B) Models of developmental timing control. Earlier than stage 3, corresponding to day 0 of ap1 cal 35S∷AP1 floral buds, the KNU locus is heavily covered by the H3K27me3 repressive mark. At stage 3, AG expression starts, and it binds to the upstream promoter of KNU. The KNU transcript cannot be induced due to the repressive marks. At stage 6, the repressive mark is removed in an AG-dependent manner, allowing the basic transcriptional machinery to access the KNU locus and induce transcription. Thereafter, WUS transcription is repressed and stem cell activity is terminated.
AG directly induces KNU with a 2-d time lag in flower development
We showed that AG directly binds to the KNU promoter and activates transcription. Why does AG induce KNU after ∼2 d delay even though AG binds directly to the KNU promoter earlier? One possible explanation is that higher levels of AG activity may be necessary to induce KNU (quantitative model). This is unlikely, however, because between days 1 and 3, the binding of AG to the KNU promoter (measured by ChIP enrichment rate) did not increase substantially. Moreover, in the 35S∷AG-GR plants, AG is expressed under a constitutive ubiquitous promoter, but KNU induction still took ∼2 d. Another explanation for the time lag is that AG induces KNU with proper timing according to qualitative differences in AG (e.g., post-translational modifications), other AG-interacting proteins, and/or modification of the KNU genomic region itself (qualitative model).
We showed that the repressive mark H3k27me3 covers the nucleosomes specifically around the transcription start site and the gene body of KNU. The following results strongly suggest that H3K27me3 mediated by the PRC is necessary to control the timing of KNU expression: (1) H3K27me3 levels at the KNU locus were reduced in an AG-dependent manner on day 2 prior to the transcriptional induction of KNU in ap1 cal 35S∷AP1-GR inflorescences; (2) in PcG mutants, KNU was expressed ectopically; and (3) deletion of the KNU coding region from the KNU reporter construct marked by H3K27me3 resulted in ectopic and precocious KNU expression. Based on these data, we propose a model for how AG regulates KNU (Fig. 7B). AG is induced at stage 3, and AG directly binds to the KNU promoter. At that time, the transcription start site and gene body of KNU are covered by the repressive mark, which prevents transcription of KNU. During stages 3–5, the repressive mark in the transcriptional start site and the gene body is removed in an AG-dependent manner. Thereafter, the basic transcriptional machinery, including RNA polymerase II, can access the KNU promoter and trigger KNU transcription at stage 6. When KNU is up-regulated, WUS transcription is rapidly repressed.
The KNU reporter assay with the deletion of the coding region also showed that KNU can be ectopically expressed in cells where AG is not expressed. This indicates that the KNU upstream region contains an AG-independent promoter activity and that requirement of AG is for timely removal of repressive modification from the KNU locus. Why are 2 d necessary to remove the H3K27me3 at the KNU locus? H3K27me3 is inherited through cell division by the actions of PcG complexes (Hansen et al. 2008). Live imaging analysis in Arabidopsis shoot apical and floral meristems showed that cells divide once in 1–2 d (Reddy et al. 2004). These indicate that 2-d timing could be correlated with one to two cell division cycles. The next challenge is to reveal how AG is involved in the dynamics of epigenetic marks and to understand a possible link between chromatin modification and cell cycle progression (Dominguez and Berger 2008).
The negative regulation of WUS by KNU
WUS is a central regulator of meristem identity. We showed that KNU is the key mediator of AG to repress WUS to terminate stem cell activity in the floral meristem by the following results: (1) KNU and WUS showed temporally anti-correlated expression patterns; (2) WUS expression was prolonged in the meristematic region of knu flowers; (3) overexpression of KNU led to precocious termination of WUS expression, resulting in wus-like phenotypes; (4) KNU induction resulted in WUS suppression rapidly; and (5) overexpression of KNU was sufficient to rescue the indeterminate phenotype of ag. As KNU contains a repression motif, these results suggest that KNU may directly regulate WUS. We used ChIP to examine whether KNU directly binds the WUS genomic region, but we could not obtain a definitive result (data not shown). We also tried to test WUS expression in the inducible line of KNU with a protein synthesis inhibitor, but we could not detect clear WUS repression within the executed time range of several hours, possibly due to the RNA stability (data not shown). Thus, we do not know currently whether KNU directly represses WUS transcription.
AG coordinates proliferation and differentiation
One of the intriguing questions in developmental biology is how homeotic proteins coordinate growth and differentiation. The floral homeotic protein AG controls various target genes in a stage- and tissue-specific manner to specify reproductive organs (Ito et al. 2004, 2007; Gomez-Mena et al. 2005). In addition to promoting stamen and carpel identity, AG controls the determinate nature of the floral meristem by negatively affecting the transcription of the homeobox gene WUS (Lenhard et al. 2001; Lohmann et al. 2001). However, how AG-dependent transcription is developmentally regulated is largely unknown. Here we showed that AG activity from stage 3 is necessary for floral meristem determinacy through KNU activation from stage 6. In contrast, AG activity starting from stage 6 is necessary and sufficient to induce stamen identity and produce fully mature stamens (Ito et al. 2007). Stamen identity requires the direct induction of SPL by AG at organ primordia in whorl 3 at stage 6 (Ito et al. 2004). Thus, the requirement for AG activity in the activation of SPL and KNU is different. SPL is rapidly induced within a couple of hours after induction of AG, suggesting that SPL induction appears to require the context of stage 6, which is induced AG-independently. In contrast, KNU induction in the floral meristem was not observed until 2 d after AG was activated. We showed that induction timing of KNU involves AG-dependent changes in the levels of H3K27me3. KNU induction by AG in developing stamens was further delayed. These results indicate that different AG target genes are regulated in different ways—some depend on stage-specific cofactors, whereas others require precedent epigenetic modification of target loci.
Our results indicate that balancing stem cell maintenance and differentiation via AG is partly achieved by the direct induction of KNU, which represses WUS to terminate the floral meristem. Delayed activation of KNU by AG allows initial activation of AG by WUS, before WUS is switched off by KNU. Such a negative feedback loop is a common theme in biology, but not much clear mechanistic understanding has been made in multicellular organisms, except for the study of the circadian clock (Wijnen and Young 2006; Pruneda-Paz et al. 2009). The timing mechanism we uncovered in plant stem cell regulation may well be present in animals, as it has been shown previously that mammalian stem cell pluripotency is maintained via repression of differentiation-promoting genes by the same repressive mark—H3K27me3 (Bernstein et al. 2006; Boyer et al. 2006).
Materials and methods
Plant materials and chemical treatments
All plants carrying the temperature-sensitive knu-1 (Payne et al. 2004) mutation were grown at 25°C (nonpermissive condition) or 16°C (permissive condition). All other plants were of the Landsberg erecta (Ler) background and were grown at 22°C under continuous light. knu-1 (Wassilewskija background) was backcrossed into the Ler background three times and used for genetic analyses. Plant photographs were taken by a Nikon SMZ 1500 stereoscopic microscope attached to a digital camera SIGHT DS-U1. Scanning electron microscopic images were taken by an Electron Microscope JEOL JSM-6360LV.
DEX and 5α-androstan-17β-ol-3-one (DHT) treatments were conducted by inverting the plants and submerging the inflorescences for 1 min in solution containing either 1 μM DEX (for 35S∷AP1-GR) or 10 μM DEX (for 35S∷AG-GR and 35S∷KNU-GR), or 100 nM DHT (for 35S∷KNU-AR) together with 0.015% Silwet L-77. The day of initial DEX or DHT treatment was designated as day 0. Repeated treatments were done at 1-d intervals.
Genetic analysis
The mutant combinations ag-1 knu-1, ag-4 knu-1, and knu-1 wus-1 were identified in the F2 segregants self-pollinated from F1 plants by crossing parental heterozygous mutants. Genotyping of the desired mutant alleles was confirmed by PCR amplification using primer sets KNUgenF and KNUgenR or WUSgenF and WUSgenR (Supplemental Table 2), followed by restriction digestion with HpyCH4III or Asp718 to identify knu-1 or wus-1, respectively.
RNA extraction and expression analysis
Total RNA was isolated from flower bud clusters younger than stage 10, when each bud is smaller than 0.5 mm in diameter, by the RNeasy plant mini kit (Qiagen). Reverse transcription was conducted using the ThermoScriptIII RT–PCR system (Invitrogen). Quantitative real-time PCR assays were performed in triplicate by the 7900HT fast real-time PCR system (Applied Biosystems) using the SYBR Green PCR master mix (Applied Biosystems). Supplemental Table 2 lists all the primer sequences used in expression analyses. Expression assays were repeated at least twice and confirmed to show reproductive trends. One representative set of data is shown in the figures.
In situ hybridization
Nonradioactive in situ hybridizations were performed as described (Carles et al. 2005). To produce a WUS-specific antisense probe, a pMHwus16 (Carles et al. 2005) clone carrying WUS cDNA was used as a template for in vitro transcription.
GUS staining
GUS staining was performed as described (Ito et al. 2003) and observed in whole mount in clearing solution using a stereomicroscope (Figs. 3D–G, 4F–K; Supplemental Fig. 8), on paraffin-sections (Fig. 4B–E) or on glass slides mounted in clearing solution (insets in Figs. 4J–L, 5C,D, 6D–G,I). Photographs were taken using a Nikon Eclipse 80i binocular and a Nikon digital sight DS-U1 camera.
Vector constructs and plant transformation
pKNU∷KNU-GUS and pKNU∷KNU-GR constructs were produced as follows: The 3980-bp full-length KNU genomic region (containing a 2918-bp promoter, 486-bp protein-coding sequence, and a 576-bp downstream sequence) was amplified by high-fidelity Pfu Ultra DNA polymerase (Stratagene), sequenced, and then cloned into cloning vector pCR8 TOPO (Invitrogen). The restriction site SfoI was introduced to replace the KNU stop codon with the primers KNUmutF and KNUmutR using the QuickChange II Site-Directed Mutagenesis kit (Stratagene). Next, blunt-ended GUS or GR fragments were cloned into the SfoI site of the pCR8-KNU vector. Finally, the entire cassettes of pKNU∷KNU-GUS or pKNU∷KNU-GR in pCR8 were recombined into CD3-694, pEarleyGate303 (Earley et al. 2006) by LR-recombinase (Invitrogen).
The construct pMutatedKNU∷KNU-GUS with the mutation of three putative AG-binding half-sites was prepared as follows: The NdeI restriction fragment containing three putative AG-binding sites in the KNU promoter region was amplified with the primers KNUNdeIF and KNUNdeIR and cloned into Topo pCRII vector (Invitrogen). The resulting plasmid was used for PCR-based site-directed mutagenesis to introduce GG-to-AA mutations sequentially with three pair of primers: KNUmAG1F, KNUmAG1R, KNUmAG2F, KNUmAG2R, KNUmAG3F, and KNUmAG3R. Then the NdeI fragment with the mutations in three putative AG-binding sites was subcloned into pKNU∷KNU-GUS to replace the original NdeI restriction fragment. The construct with the deletion of a part of the KNU coding region was prepared based on the pKNU∷KNU-GUS construct. A portion of the KNU coding region was amplified by the primer set PpuMI-del-KNU-F and SgrAI-del-KNU-R and subcloned into the pKNU∷KNU-GUS construct to replace the original PpuMI-SgrAI restriction fragment to delete a 231-bp fragment of the latter part of the coding region. To construct 35S∷KNU-AR, the DNA sequence containing the 5′ untranslated region and the full-length KNU protein-coding sequence was amplified by the primer set XhoI-KNU-F and Bsp120I-KNU-R, and subsequently cloned into vectors pGreen-35S-GR and pGreen-35S-AR (Chang et al. 1988; T. Ito, unpubl.), which are modified versions of pGreen (Hellens et al. 2000). Supplemental Table 2 lists all primer sequences used for plant vector construction.
Except for pKNU∷KNU-GR, which was introduced into knu-1, pKNU∷KNU-GUS, 35S∷KNU-GR, and 35S∷KNU-AR were introduced into wild-type Ler plants, using the Agrobacterium-mediated floral dip method (Clough and Bent 1998). All transgenic plants were selected by the herbicide Basta (Bayer; 0.002% of the commercial solution).
ChIP
ChIP experiments were performed as described (Ito et al. 1997; Liu et al. 2007) with slight modification. Inflorescences from ap1 cal 35S∷AP1-GR were ground in liquid nitrogen and post-fixed with 1% formaldehyde for 10 min. Chromatin was isolated and solubilized by sonication to generate DNA fragments with an average length of 400 bp. After incubation with salmon sperm DNA/protein-A agarose beads (Upstate Biotechnologies), the solubilized chromatin was incubated overnight with anti-AG serum, normal rabbit IgG (Santa Cruz Biotechnologies, for AG ChIP experiment as a control), or anti-H3K27me3 and anti-H3K4me2 antibodies (Upstate Biotechnologies; for histone modification ChIP experiments). DNA fragments were recovered from the purified DNA–protein complexes and then used for enrichment tests by real-time PCR analysis in triplicates. The primary ratio between the input DNA before IP and the bound DNA after IP was calculated for all the representative primer sets spanning the KNU genomic region, and the ratios were plotted to show the relative changes in the levels of epigenetic marks. The relative enrichment for AG on the KNU locus was the secondary ratio generated from the primary ratio of AG over that of IgG. For all ChIP experiments, primers for the Mu-like transposon, TUB, or ACT gene were included as negative controls. All ChIP assays were repeated at least twice and confirmed to show reproductive trends. One representative set of data is shown in the figures.
Accession numbers
Sequence information can be found in the Arabidopsis Genome Initiative database under the following accession numbers: AGAMOUS (AG, At4g18960); KNUCKLES (KNU, At5g14010); SPOROCYTELESS (SPL, At4g27330); WUSCHEL (WUS, At2g17950); ACTIN 2 (ACT, At3g18780); and TUBULIN2 (TUB, At5g62690).
Acknowledgments
We appreciate the continuous support of K. Okada and E.M. Meyerowitz. We thank F. Wellmer for providing ap1 cal 35S::AP1-GR and ag-1/+ ap1 cal 35S::AP1-GR seeds, A.M. Koltunow for knu-1 seeds, T. Laux for pWUS::GUS seeds, and N. Ohad for 35S::GFP-FIE seeds. We also thank T.I.'s laboratory members and my colleagues—Y. Hao, F. Berger, J. Dinneny, H. Yuehui, S. Cohen, and K. Oyama—for valuable comments on the manuscript. This work was supported by a research grant to T.I. from Temasek Life Sciences Laboratory and from JST. N.K. is supported by the Singapore Millennium Foundation.
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1800409.
Supplemental material is available at http://www.genesdev.org.
References
- Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- Bowman JL, Smyth DR, Meyerowitz EM. Genes directing flower development in Arabidopsis. Plant Cell. 1989;1:37–52. doi: 10.1105/tpc.1.1.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman JL, Drews GN, Meyerowitz EM. Expression of the Arabidopsis floral homeotic gene AGAMOUS is restricted to specific cell types late in flower development. Plant Cell. 1991;3:749–758. doi: 10.1105/tpc.3.8.749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer LA, Plath K, Zeitlinger J, Brambrink T, Medeiros LA, Lee TI, Levine SS, Wernig M, Tajonar A, Ray MK, et al. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature. 2006;441:349–353. doi: 10.1038/nature04733. [DOI] [PubMed] [Google Scholar]
- Brand U, Fletcher JC, Hobe M, Meyerowitz EM, Simon R. Dependence of stem cell fate in Arabidopsis on a feedback loop regulated by CLV3 activity. Science. 2000;289:617–619. doi: 10.1126/science.289.5479.617. [DOI] [PubMed] [Google Scholar]
- Busch MA, Bomblies K, Weigel D. Activation of a floral homeotic gene in Arabidopsis. Science. 1999;285:585–587. doi: 10.1126/science.285.5427.585. [DOI] [PubMed] [Google Scholar]
- Carles CC, Choffnes-Inada D, Reville K, Lertpiriyapong K, Fletcher JC. ULTRAPETALA1 encodes a SAND domain putative transcriptional regulator that controls shoot and floral meristem activity in Arabidopsis. Development. 2005;132:897–911. doi: 10.1242/dev.01642. [DOI] [PubMed] [Google Scholar]
- Chang CS, Kokontis J, Liao ST. Structural analysis of complementary DNA and amino acid sequences of human and rat androgen receptors. Proc Natl Acad Sci. 1988;85:7211–7215. doi: 10.1073/pnas.85.19.7211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark SE, Running MP, Meyerowitz EM. CLAVATA1, a regulator of meristem and flower development in Arabidopsis. Development. 1993;119:397–418. doi: 10.1242/dev.119.2.397. [DOI] [PubMed] [Google Scholar]
- Clark SE, Williams RW, Meyerowitz EM. The CLAVATA1 gene encodes a putative receptor kinase that controls shoot and floral meristem size in Arabidopsis. Cell. 1997;89:575–585. doi: 10.1016/s0092-8674(00)80239-1. [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- Deyhle F, Sarkar AK, Tucker EJ, Laux T. WUSCHEL regulates cell differentiation during anther development. Dev Biol. 2007;302:154–159. doi: 10.1016/j.ydbio.2006.09.013. [DOI] [PubMed] [Google Scholar]
- Dominguez M, Berger F. Chromatin and the cell cycle meet in Madrid. Development. 2008;135:3475–3480. doi: 10.1242/dev.000844. [DOI] [PubMed] [Google Scholar]
- Earley KW, Haag JR, Pontes O, Opper K, Juehne T, Song K, Pikaard CS. Gateway-compatible vectors for plant functional genomics and proteomics. Plant J. 2006;45:616–629. doi: 10.1111/j.1365-313X.2005.02617.x. [DOI] [PubMed] [Google Scholar]
- Fletcher JC. The ULTRAPETALA gene controls shoot and floral meristem size in Arabidopsis. Development. 2001;128:1323–1333. doi: 10.1242/dev.128.8.1323. [DOI] [PubMed] [Google Scholar]
- Fletcher JC, Brand U, Running MP, Simon R, Meyerowitz EM. Signaling of cell fate decisions by CLAVATA3 in Arabidopsis shoot meristems. Science. 1999;283:1911–1914. doi: 10.1126/science.283.5409.1911. [DOI] [PubMed] [Google Scholar]
- Gallois JL, Nora FR, Mizukami Y, Sablowski R. WUSCHEL induces shoot stem cell activity and developmental plasticity in the root meristem. Genes & Dev. 2004;18:375–380. doi: 10.1101/gad.291204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Mena C, de Folter S, Costa MM, Angenent GC, Sablowski R. Transcriptional program controlled by the floral homeotic gene AGAMOUS during early organogenesis. Development. 2005;132:429–438. doi: 10.1242/dev.01600. [DOI] [PubMed] [Google Scholar]
- Gross-Hardt R, Lenhard M, Laux T. WUSCHEL signaling functions in interregional communication during Arabidopsis ovule development. Genes & Dev. 2002;16:1129–1138. doi: 10.1101/gad.225202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen KH, Bracken AP, Pasini D, Dietrich N, Gehani SS, Monrad A, Rappsilber J, Lerdrup M, Helin K. A model for transmission of the H3K27me3 epigenetic mark. Nat Cell Biol. 2008;10:1291–1300. doi: 10.1038/ncb1787. [DOI] [PubMed] [Google Scholar]
- Hellens RP, Edwards EA, Leyland NR, Bean S, Mullineaux PM. pGreen: A versatile and flexible binary Ti vector for Agrobacterium-mediated plant transformation. Plant Mol Biol. 2000;42:819–832. doi: 10.1023/a:1006496308160. [DOI] [PubMed] [Google Scholar]
- Huang H, Mizukami Y, Hu Y, Ma H. Isolation and characterization of the binding sequences for the product of the Arabidopsis floral homeotic gene AGAMOUS. Nucleic Acids Res. 1993;21:4769–4776. doi: 10.1093/nar/21.20.4769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito T, Takahashi N, Shimura Y, Okada K. A serine/threonine protein kinase gene isolated by an in vivo binding procedure using the Arabidopsis floral homeotic gene product, AGAMOUS. Plant Cell Physiol. 1997;38:248–258. doi: 10.1093/oxfordjournals.pcp.a029160. [DOI] [PubMed] [Google Scholar]
- Ito T, Sakai H, Meyerowitz EM. Whorl-specific expression of the SUPERMAN gene of Arabidopsis is mediated by cis elements in the transcribed region. Curr Biol. 2003;13:1524–1530. doi: 10.1016/s0960-9822(03)00612-2. [DOI] [PubMed] [Google Scholar]
- Ito T, Wellmer F, Yu H, Das P, Ito N, Alves-Ferreira M, Riechmann JL, Meyerowitz EM. The homeotic protein AGAMOUS controls microsporogenesis by regulation of SPOROCYTELESS. Nature. 2004;430:356–360. doi: 10.1038/nature02733. [DOI] [PubMed] [Google Scholar]
- Ito T, Ng KH, Lim TS, Yu H, Meyerowitz EM. The homeotic protein AGAMOUS controls late stamen development by regulating a jasmonate biosynthetic gene in Arabidopsis. Plant Cell. 2007;19:3516–3529. doi: 10.1105/tpc.107.055467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz A, Oliva M, Mosquna A, Hakim O, Ohad N. FIE and CURLY LEAF polycomb proteins interact in the regulation of homeobox gene expression during sporophyte development. Plant J. 2004;37:707–719. doi: 10.1111/j.1365-313x.2003.01996.x. [DOI] [PubMed] [Google Scholar]
- Kondo T, Sawa S, Kinoshita A, Mizuno S, Kakimoto T, Fukuda H, Sakagami Y. A plant peptide encoded by CLV3 identified by in situ MALDI-TOF MS analysis. Science. 2006;313:845–848. doi: 10.1126/science.1128439. [DOI] [PubMed] [Google Scholar]
- Lenhard M, Bohnert A, Jurgens G, Laux T. Termination of stem cell maintenance in Arabidopsis floral meristems by interactions between WUSCHEL and AGAMOUS. Cell. 2001;105:805–814. doi: 10.1016/s0092-8674(01)00390-7. [DOI] [PubMed] [Google Scholar]
- Liu C, Zhou J, Bracha-Drori K, Yalovsky S, Ito T, Yu H. Specification of Arabidopsis floral meristem identity by repression of flowering time genes. Development. 2007;134:1901–1910. doi: 10.1242/dev.003103. [DOI] [PubMed] [Google Scholar]
- Lloyd AM, Schena M, Walbot V, Davis RW. Epidermal cell fate determination in Arabidopsis: Patterns defined by a steroid-inducible regulator. Science. 1994;266:436–439. doi: 10.1126/science.7939683. [DOI] [PubMed] [Google Scholar]
- Lohmann JU, Hong RL, Hobe M, Busch MA, Parcy F, Simon R, Weigel D. A molecular link between stem cell regulation and floral patterning in Arabidopsis. Cell. 2001;105:793–803. doi: 10.1016/s0092-8674(01)00384-1. [DOI] [PubMed] [Google Scholar]
- Mayer KF, Schoof H, Haecker A, Lenhard M, Jurgens G, Laux T. Role of WUSCHEL in regulating stem cell fate in the Arabidopsis shoot meristem. Cell. 1998;95:805–815. doi: 10.1016/s0092-8674(00)81703-1. [DOI] [PubMed] [Google Scholar]
- Mizukami Y, Ma H. Ectopic expression of the floral homeotic gene AGAMOUS in transgenic Arabidopsis plants alters floral organ identity. Cell. 1992;71:119–131. doi: 10.1016/0092-8674(92)90271-d. [DOI] [PubMed] [Google Scholar]
- Mizukami Y, Ma H. Determination of Arabidopsis floral meristem identity by AGAMOUS. Plant Cell. 1997;9:393–408. doi: 10.1105/tpc.9.3.393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payne T, Johnson SD, Koltunow AM. KNUCKLES (KNU) encodes a C2H2 zinc-finger protein that regulates development of basal pattern elements of the Arabidopsis gynoecium. Development. 2004;131:3737–3749. doi: 10.1242/dev.01216. [DOI] [PubMed] [Google Scholar]
- Pruneda-Paz JL, Breton G, Para A, Kay SA. A functional genomics approach reveals CHE as a component of the Arabidopsis circadian clock. Science. 2009;323:1481–1485. doi: 10.1126/science.1167206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prunet N, Morel P, Thierry AM, Eshed Y, Bowman JL, Negrutiu I, Trehin C. REBELOTE, SQUINT, and ULTRAPETALA1 function redundantly in the temporal regulation of floral meristem termination in Arabidopsis thaliana. Plant Cell. 2008;20:901–919. doi: 10.1105/tpc.107.053306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy GV, Meyerowitz EM. Stem-cell homeostasis and growth dynamics can be uncoupled in the Arabidopsis shoot apex. Science. 2005;310:663–667. doi: 10.1126/science.1116261. [DOI] [PubMed] [Google Scholar]
- Reddy GV, Heisler MG, Ehrhardt DW, Meyerowitz EM. Real-time lineage analysis reveals oriented cell divisions associated with morphogenesis at the shoot apex of Arabidopsis thaliana. Development. 2004;131:4225–4237. doi: 10.1242/dev.01261. [DOI] [PubMed] [Google Scholar]
- Schoof H, Lenhard M, Haecker A, Mayer KF, Jurgens G, Laux T. The stem cell population of Arabidopsis shoot meristems in maintained by a regulatory loop between the CLAVATA and WUSCHEL genes. Cell. 2000;100:635–644. doi: 10.1016/s0092-8674(00)80700-x. [DOI] [PubMed] [Google Scholar]
- Schubert D, Primavesi L, Bishopp A, Roberts G, Doonan J, Jenuwein T, Goodrich J. Silencing by plant Polycomb-group genes requires dispersed trimethylation of histone H3 at lysine 27. EMBO J. 2006;25:4638–4649. doi: 10.1038/sj.emboj.7601311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiraishi H, Okada K, Shimura Y. Nucleotide sequences recognized by the AGAMOUS MADS domain of Arabidopsis thaliana in vitro. Plant J. 1993;4:385–398. doi: 10.1046/j.1365-313x.1993.04020385.x. [DOI] [PubMed] [Google Scholar]
- Sieburth LE, Running MP, Meyerowitz EM. Genetic separation of third and fourth whorl functions of AGAMOUS. Plant Cell. 1995;7:1249–1258. doi: 10.1105/tpc.7.8.1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sieburth LE, Drews GN, Meyerowitz EM. Non-autonomy of AGAMOUS function in flower development: Use of a Cre/loxP method for mosaic analysis in Arabidopsis. Development. 1998;125:4303–4312. doi: 10.1242/dev.125.21.4303. [DOI] [PubMed] [Google Scholar]
- Smyth DR, Bowman JL, Meyerowitz EM. Early flower development in Arabidopsis. Plant Cell. 1990;2:755–767. doi: 10.1105/tpc.2.8.755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steeves TA, Sussex IM. Patterns in plant development. 2nd ed. Cambridge University Press; Cambridge, UK: 1989. [Google Scholar]
- Sung S, He Y, Eshoo TW, Tamada Y, Johnson L, Nakahigashi K, Goto K, Jacobsen SE, Amasino RM. Epigenetic maintenance of the vernalized state in Arabidopsis thaliana requires LIKE HETEROCHROMATIN PROTEIN 1. Nat Genet. 2006;38:706–710. doi: 10.1038/ng1795. [DOI] [PubMed] [Google Scholar]
- Takada S, Goto K. Terminal flower2, an Arabidopsis homolog of heterochromatin protein1, counteracts the activation of flowering locus T by constans in the vascular tissues of leaves to regulate flowering time. Plant Cell. 2003;15:2856–2865. doi: 10.1105/tpc.016345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turck F, Roudier F, Farrona S, Martin-Magniette ML, Guillaume E, Buisine N, Gagnot S, Martienssen RA, Coupland G, Colot V. Arabidopsis TFL2/LHP1 specifically associates with genes marked by trimethylation of histone H3 lysine 27. PLoS Genet. 2007;3:e86. doi: 10.1371/journal.pgen.0030086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellmer F, Alves-Ferreira M, Dubois A, Riechmann JL, Meyerowitz EM. Genome-wide analysis of gene expression during early Arabidopsis flower development. PLoS Genet. 2006;2:e117. doi: 10.1371/journal.pgen.0020117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wijnen H, Young MW. Interplay of circadian clocks and metabolic rhythms. Annu Rev Genet. 2006;40:409–448. doi: 10.1146/annurev.genet.40.110405.090603. [DOI] [PubMed] [Google Scholar]
- Yanofsky MF, Ma H, Bowman JL, Drews GN, Feldmann KA, Meyerowitz EM. The protein encoded by the Arabidopsis homeotic gene agamous resembles transcription factors. Nature. 1990;346:35–39. doi: 10.1038/346035a0. [DOI] [PubMed] [Google Scholar]
- Zhang X, Clarenz O, Cokus S, Bernatavichute YV, Pellegrini M, Goodrich J, Jacobsen SE. Whole-genome analysis of histone H3 lysine 27 trimethylation in Arabidopsis. PLoS Biol. 2007a;5:e129. doi: 10.1371/journal.pbio.0050129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Germann S, Blus BJ, Khorasanizadeh S, Gaudin V, Jacobsen SE. The Arabidopsis LHP1 protein colocalizes with histone H3 Lys27 trimethylation. Nat Struct Mol Biol. 2007b;14:869–871. doi: 10.1038/nsmb1283. [DOI] [PubMed] [Google Scholar]
- Zhao Y, Medrano L, Ohashi K, Fletcher JC, Yu H, Sakai H, Meyerowitz EM. HANABA TARANU is a GATA transcription factor that regulates shoot apical meristem and flower development in Arabidopsis. Plant Cell. 2004;16:2586–2600. doi: 10.1105/tpc.104.024869. [DOI] [PMC free article] [PubMed] [Google Scholar]