Abstract
Epithelial-to-mesenchymal transition (EMT) plays an important role during normal embryogenesis, and it has been implicated in cancer invasion and metastasis. Here, we report that Ladybird homeobox 1 (LBX1), a developmentally regulated homeobox gene, directs expression of the known EMT inducers ZEB1, ZEB2, Snail1, and transforming growth factor β2 (TGFB2). In mammary epithelial cells, overexpression of LBX1 leads to morphological transformation, expression of mesenchymal markers, enhanced cell migration, increased CD44high/CD24low progenitor cell population, and tumorigenic cooperation with known oncogenes. In human breast cancer, LBX1 is up-regulated in the unfavorable estrogen receptor (ER)/progesterone (PR)/HER2 triple-negative basal-like subtype. Thus, aberrant expression of LBX1 may lead to the activation of a developmentally regulated EMT pathway in human breast cancer.
Keywords: EMT, LBX1, breast cancer, cancer stem cell
Epithelial-to-mesenchymal transition (EMT) is a process in which epithelial cells lose characteristic epithelial traits while gaining some properties of mesenchymal cells. EMT is currently defined by a series of changes: a change from characteristic cobblestone-like epithelial morphology to a spindle, fibroblast-like shape with migratory protrusions; a conversion of epithelial apical–basal polarity to front–back polarity; acquisition of migratory and invasive ability; and a loss of the epithelial markers with concurrent acquisition of mesenchymal markers (Hay 2005). EMT is a major mechanism for tissue remodeling during normal embryogenesis, without which development cannot proceed past the blastula stage (Thiery and Sleeman 2006). The development of mesoderm and neural crest requires EMT to generate mesenchymal cells that can migrate to distant locations for organogenesis. EMT also accounts for proper heart valve development and palate fusion (Hay 2005; Yang and Weinberg 2008).
Recently, several studies have indicated that EMT might play an important role in cancer invasion and metastasis. Embryogenesis and metastasis share a major obstacle that must be overcome by epithelial cells: survival following detachment from basement membrane and migration through the extracellular matrix (ECM). EMT enables epithelial cells to overcome these restrictions, setting the stage for physiologically programmed migration or for cancer invasion. Factors regulating developmental EMT have been demonstrated to play roles in tumor progression, including the TGFβ, WNT, and Notch signaling pathways and the Snail1 (SNAI1), Slug (SNAI2), ZEB1, SIP1 (ZEB2), TWIST1, FOXC2, and Goosecoid (GSC) transcription factors (Yang and Weinberg 2008). The TGFβ pathway plays a crucial role in inducing EMT in almost all major developmental processes that depend on EMT (Thiery and Sleeman 2006). Activation of the TGFβ pathway in human cancer cell lines and in mouse tumor models can induce EMT and promote tumor cells to invade the ECM in culture and to metastasize to distant organs in mice (Oft et al. 1998; Janda et al. 2002). The zinc-finger transcription factor Snail1 also plays an important role in inducing EMT in developmental processes of mesoderm, neural crest, and cardiac valve formation (Thiery and Sleeman 2006). Expression of Snail1 is observed in various human tumors and has been correlated with tumor progression and poor prognosis (Peinado et al. 2007). Thus, EMT-associated pathways may be activated during tumorigenesis, although underlying mechanisms and functional consequences remain to be fully defined.
Ladybird homeobox 1 (LBX1) is a homeobox transcription factor implicated in normal myogenesis and neurogenesis. During fetal muscle development, LBX1 is expressed within migrating muscle precursor cells, and it is necessary for the lateral, but not ventral, migration of hypaxial muscle precursors. LBX1-deficient mice lack muscles in their limbs due to a defect in migration of muscle precursor cells along a lateral pathway to the limbs (Brohmann et al. 2000; Gross et al. 2000). In the CNS, LBX1 is required for proper differentiation and specification of neurons and interneurons in the dorsal spinal cord (Gross et al. 2002; Muller et al. 2002), for imposing a somatosensory fate on relay neurons in the hindbrain (Sieber et al. 2007), and for correct dorsal-ventral patterning of the neural tube (Kruger et al. 2002). Despite these dramatic developmental phenotypes, the mechanism of action of LBX1 is not well defined. Here, we demonstrate that LBX1 is a potent activator of EMT. In mammary epithelial cells, LBX1 induces characteristic molecular features of EMT and enhanced cellular migration, and it increases the pool of CD44high/CD24low progenitor cells, cooperating with activated HRAS to cause tumorigenesis. Significantly, LBX1 itself induces expression of multiple previously defined EMT transcriptional regulators, suggesting that under certain circumstances it may be a master regulator of this pathway. Analysis of LBX1 expression across different cancers shows a correlation with the aggressive basal-like subtype of human breast cancer.
Results and Discussion
LBX1 induces EMT in mammary epithelial cells
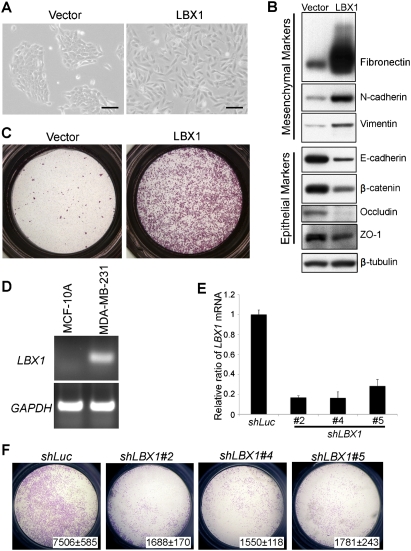
We screened a set of developmentally regulated genes expressed in various embryonic migratory mesenchymal tissues to identify novel physiological inducers of EMT (see the Supplemental Table). Among six candidate genes tested for their ability to induce EMT in cultured mammary epithelial cells, we identified LBX1 as the most potent inducer of this transformation. Stable expression of LBX1 was accomplished using pools of retrovirally infected MCF-10A cells, to avoid clonal selection bias. MCF-10A are immortalized, nontransformed human mammary epithelial cells, lacking endogenous LBX1 expression (transgene expression shown in Supplemental Fig. S1). Compared with vector-infected cells, LBX1-expressing cells demonstrated a dramatic change of cell morphology, with transformation of the cobblestone-like epithelial cells to an elongated fibroblast-like morphology with pronounced cellular scattering (Fig. 1A). The EMT phenotype was confirmed by expression of characteristic molecular markers using immunoblotting (Fig. 1B) and immunofluorescence analyses (Supplemental Fig. S2). LBX1-expressing cells have significant up-regulation of the mesenchymal markers fibronectin, N-cadherin, and vimentin, whereas the epithelial markers E-cadherin, β-catenin, occludin, and zona occludens 1 (ZO-1) are down-regulated. Cellular migration is also dramatically up-regulated, as demonstrated by transwell chamber assays in media lacking EGF (Fig. 1C). Thus, LBX1 can initiate the characteristic EMT process in mammary epithelial cells.
Figure 1.
LBX1 induces EMT in mammary epithelial cells. (A) LBX1 induces morphological changes in MCF-10A cells. MCF-10A cells were infected with vector or LBX1 retroviral expression constructs and cultured in plastic dishes. Bar, 100 μm. (B) LBX1 induces changes of EMT markers in MCF-10A cells. Expression of various epithelial and mesenchymal markers were analyzed by Western blotting. (C) LBX1 enhances the migration of MCF-10A cells. Vector- or LBX1-expressing cells were plated on top of a transwell chamber in media without EGF for 2 d. Images show cells migrated to the bottom of the chamber. (D) LBX1 mRNA expression was evaluated in MCF-10A and MDA-MB-231 cells by RT–PCR. (E) Efficiency of knocking down LBX1 level in MDA-MB-231 cells by shRNAs. ShRNA constructs against luciferase (shLuc) and LBX1 (shLBX1) were introduced into MDA-MB-231 cells by lentiviral infection and relative expression of LBX1 mRNA was measured by real-time qRT–PCR. Error bars represent mean ± SD of triplicate experiments. (F) Suppression of LBX1 in MDA-MB-231 cells inhibits migration. Migration assay of MDA-MB-231 cells with shLuc or shLBX1. Cell numbers migrated to the bottom of the chambers ± SD of triplicate experiments are indicated.
Suppression of LBX1 in MDA-MB-231 cells inhibits migration
While endogenous LBX1 mRNA level is not detectable in parental MCF-10A cells, it is expressed in several breast cancer cell lines (Supplemental Fig. S3), including a metastatic cell line MDA-MB-231 (Fig. 1D; Supplemental Fig. S3), which has mesenchymal cell characteristics, including a strong migratory ability. To test whether constitutive LBX1 expression in MDA-MB-231 cells contributes to their migratory phenotype, we knocked down endogenous LBX1 using lentivirus-driven shRNA constructs (shLBX1). Three shRNA constructs were identified as producing efficient knockdown of endogenous LBX1 mRNA, as confirmed by quantitative RT–PCR (qRT–PCR) (Fig. 1E). Inhibition of LBX1 expression in MDA-MB-231 cells using each of the three shRNA constructs resulted in significantly reduced cell migration in transwell assays (P < 0.05), whereas no effect was observed with shRNA against luciferase (shLuc) (Fig. 1F). Moreover, the LBX1 knockdown constructs also resulted in increased E-cadherin expression (for two shRNA constructs) and reduced fibronectin expression (for one shRNA construct) (Supplemental Fig. S4). Thus, suppression of LBX1 in MDA-MB-231 cells partially reverts the EMT process. In addition to MDA-MB-231 cells, overexpression of LBX1 also causes similar mesenchymal morphological changes and an increase in fibronectin in the luminal type breast cancer cell line T47D (Supplemental Fig. S5).
LBX1 directly up-regulates other EMT inducers
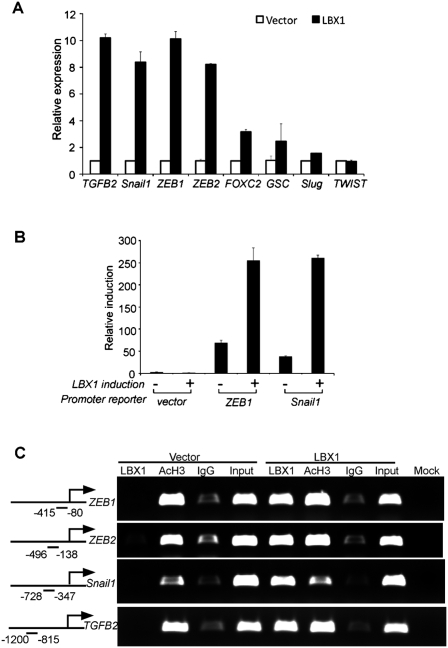
To address the potential relationship between LBX1 and other transcription factors and signaling molecules implicated in EMT, we tested whether LBX1 itself enhances the expression of known EMT inducers. LBX1 dramatically increased endogenous mRNA levels of transforming growth factor β2 (TGFB2), Snail1, ZEB1, and ZEB2 in MCF-10A cells, up to eightfold to 10-fold higher than vector-infected cells (Fig. 2A). No detectable change was observed in the level of TWIST1 mRNA. To test for a direct transcriptional effect on the promoters of the responsive EMT inducers, we transiently transfected luciferase reporter constructs containing the presumed proximal promoters of ZEB1 and Snail1 into HEK293 cells. Compared with the empty vector, LBX1 induced increased promoter activity of threefold to fourfold and fivefold to sixfold for ZEB1 and Snail1, respectively (Fig. 2B). We then used chromatin immunoprecipitation (ChIP) assays to determine whether LBX1 binds in vivo to the promoter region of these putative transcriptional targets. For each of the four potential target genes ZEB1, ZEB2, Snail1, and TGFB2, we used five overlapping primer sets, covering 2-kb regions upstream of the transcription start sites, to PCR-amplify genomic sequences from immunoprecipitated formalin cross-linked chromatin. For all four candidates, a promoter sequence containing the presumed LBX1-binding sequence was readily amplified with one or more sets of primers from the LBX1 immunoprecipitate, but not from IgG control (Fig. 2C). The full set of ChIP analyses for all five ZEB1 and TGFB2 promoter fragments, performed in duplicate, is shown in Supplemental Figure S6. To evaluate whether these EMT inducers play an important role downstream from LBX1, we performed individual and combination siRNA knockdown experiments in LBX1-expressing cells (Supplemental Fig. S7). Suppression of ZEB1, ZEB2, Snail1, and TGFB2 together, but not individually, significantly inhibited LBX1-induced migration and partially reverted expression of EMT marker. Taken together, these results suggest that LBX1 may be a direct transcriptional activator of ZEB1, ZEB2, Snail1, and TGFB2, four critical factors that have been implicated in the regulation of EMT.
Figure 2.
LBX1 directly up-regulates the expression of other EMT inducers. (A) qRT–PCR showing relative mRNA expression levels of known EMT inducers in MCF-10A cells expressing vector or LBX1. Error bars represent mean ± SD of triplicate experiments. (B) LBX1 acts as transcriptional inducers of ZEB1 and Snail1. HEK293T cells were transiently cotransfected pWPI vector or pWPI-LBX1 construct together with pGL4.15 backbone, or ZEB1 and Snail1 promoters. Relative luciferase activity is shown. Error bars represent mean ± SD of triplicate experiments. (C) LBX1 binds to the promoters of ZEB1, ZEB2, Snail1, and TGFB2 evaluated by ChIP. Antibodies against LBX1, acetylated histone H3 (AcH3), and corresponding IgG were used in vector- and LBX1-expressing MCF-10A cells to immunoprecipitate the corresponding protein-bound genomic DNA. PCR products corresponding to the promoter region are illustrated in the left panel.
Ectopic expression of LBX1 in mammary epithelial cells increases their CD44high/CD24low progenitor subpopulation
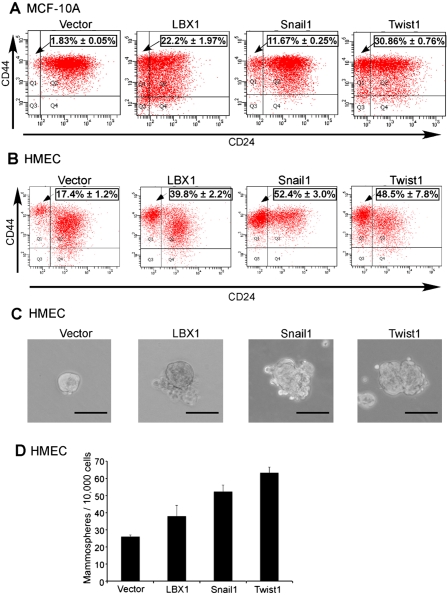
Mammary epithelial cells that have undergone EMT have been shown recently to have an increased CD44high/CD24low population, a phenotype linked to both normal breast stem cells and potential breast cancer progenitors (Al-Hajj et al. 2003; Dontu et al. 2003; Sleeman et al. 2006). This is associated with an increased capacity for mammosphere formation, an anchorage-independent growth characteristic correlated with pluripotent progenitors (Mani et al. 2008). To test the effect of LBX1 on these features of breast cancer “stemness,” we compared the effect of LBX1 on MCF-10A cells with that of the prototype EMT regulators Snail1 and Twist1. Flow cytometric analysis of LBX1-expressing cells demonstrated a 10-fold increase in the CD44high/CD24low population, compared with vector-transduced cells (P < 0.001), comparable with the effect of Snail1 and Twist1 (Fig. 3A). Breast progenitor cell populations have been defined most carefully in the human mammary epithelial cell (HMEC) assay (Elenbaas et al. 2001; Mani et al. 2008). Consistent with our observations in MCF-10A cells, we detected a significant increase in the CD44high/CD24low population within LBX1-expressing HMECs (Fig. 3B). Mammosphere formation assays also showed an increase in both the size and number of mammospheres in LBX1-expressing cells, in both MCF-10A cells and HMECs (Figs. 3C,D; Supplemental Fig. S9). Conversely, suppression of endogenous LBX1 in MDA-MB-231 breast cancer cells led to reduced growth in the specialized mammosphere media, but not under standard in vitro culture conditions (Supplemental Fig. S10). The bFGF- and EGF-supplemented, serum-free mammosphere media has been shown to selectively enhance proliferation of stem cell-like subpopulation of cancer cells (Lee et al. 2006). Collectively, these results indicate that LBX1 triggers expression of markers and functional characteristics associated with breast cancer progenitors, a feature recently defined for inducers of EMT (Mani et al. 2008).
Figure 3.
LBX1 overexpression in mammary epithelial cells enhances stemness. (A) Flow cytometric evaluation of CD44high/CD24low population in MCF-10A cells expressing vector, LBX1, Snail1, or Twist1. Percentages of mean CD44high/CD24low population ± SD of triplicate experiments are indicated. (B) Flow cytometric evaluation of CD44high/CD24low population in HMEC cells expressing vector, LBX1, Snail1, or Twist1. Percentages of mean CD44high/CD24low population ± SD of triplicate experiments are indicated. (C) Representative images of mammospheres formed by HMEC cells. Bar, 100 μm. (D) Quantification of mammosphere numbers for HMEC cells. Error bars, mean ± SD of triplicate experiments.
Oncogenic cooperation by LBX1
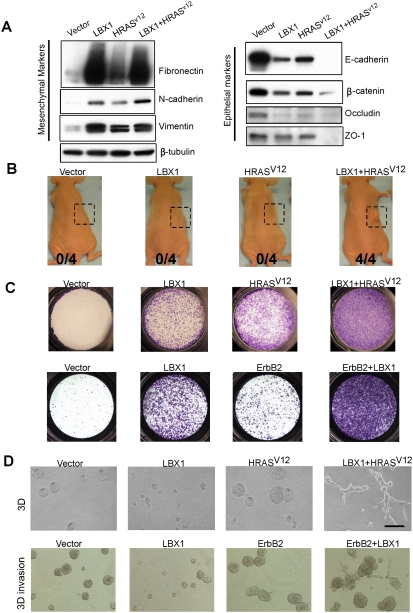
Twist proteins have been shown recently to cooperate with activation of the Ras pathway in promoting even more profound characteristics of EMT and initiation of tumorigenesis (Ansieau et al. 2008). To investigate the functional properties of LBX1 in a tumorigenesis model, we coexpressed it with an activated form of HRAS (HRASV12) in MCF-10A cells. Cell morphology in LBX1 + HRASV12-infected cells was dramatically different than seen with either construct alone, with prominent elongated spindle-shaped cells (Supplemental Fig. S11). Analysis of mesenchymal and epithelial cell markers showed a cooperative effect of LBX1 and HRASV12 in inducing characteristic features of EMT (Fig. 4A): N-cadherin induction was higher in LBX1 + HRASV12 cells relative to either LBX1 alone or HRASV12 alone, and most striking was the complete loss of epithelial markers E-cadherin and ZO-1 in LBX1 + HRASV12 cells. Thus, these two constructs appear to work together in inducing a more complete EMT phenotype. We further tested their ability to cooperate in generating tumors following subcutaneous inoculation into nude mice (Fig. 4B). None of four mice injected with MCF-10A cells expressing vector, LBX1, or HRASV12 alone developed tumors at 3 mo. In contrast, four out of four mice inoculated with cells expressing LBX1 + HRASV12 cells gave rise to tumors within 1 mo. LBX1 + HRASV12 cells demonstrated even more migratory activity in transwell assays than cells expressing either construct alone (Fig. 4C). Similar cooperative effect with LBX1 was also evident with ErbB2, a growth factor receptor commonly overexpressed in human breast cancer (Fig. 4C; Supplemental Fig. S12). For instance, in three-dimensional (3D) Matrigel culture, dramatic differences were evident: LBX1-expressing MCF-10A cells formed smaller acinar structures than vector-infected cells, whereas HRASV12-expressing cells formed larger 3D structures with preserved boundaries and failure to invade into the Matrigel (Fig. 4D). Remarkably, MCF-10A cells expressing both LBX1 and HRASV12 showed a prominent invasive phenotype, forming multiple cellular protrusions extending into the Matrigel (Fig. 4D). The invasive phenotype displayed by cells coexpressing LBX1 and ErbB2 was further evident in Matrigel/collagen 3D assay (Fig. 4D), which highlights the ability of cancer cells to invade through collagen (Xiang and Muthuswamy 2006). Taken together, these results demonstrate potent cooperation between LBX1 and characteristic oncogenes in promoting EMT and invasive properties correlated with tumorigenesis.
Figure 4.
Oncogenic cooperation by LBX1. Cell lines were generated by sequentially infecting MCF-10A cells with retroviral pBabe vector (vector), pBabe-LBX1 (LBX1), and lentiviral pMSCV-HRASV12 construct (HRASV12 or LBX1 + HRASV12). ErbB2-overexpressing MCF-10A cells were infected with pBabe-vector (ErbB2) or pBabe-LBX1 (ErbB2 + LBX1). (A) Immunoblotting of cell lysates for different EMT markers. (B) Cells (5 × 106) were subcutaneously grafted into Swiss nude mice. The number of mice developing tumors is indicated. The boxes illustrate the injection area at 3 mo. (C) Transwell migration assays (2 d). (D) Cells were cultured in 3D culture (3D) on Matrigel for 4 d or 3D invassion assay on Matrigel/collagen mix for 6 d. Bars, 100 μm.
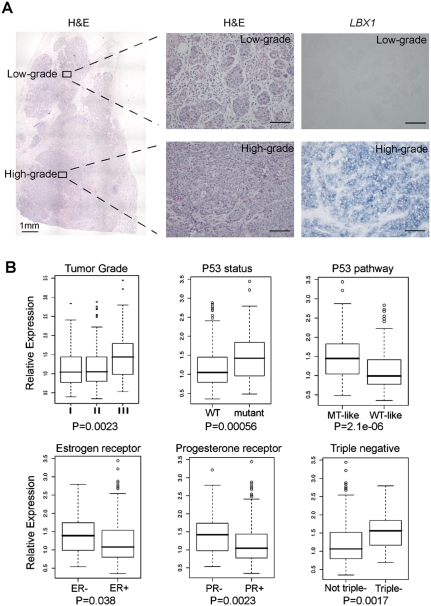
LBX1 expression is associated with invasive human breast cancer
Although LBX1 is physiologically expressed during neural and muscle differentiation, database screening indicated that its expression is more prevalent in breast cancer than in cancers of neural or muscle origin. Indeed, RNA in situ hybridization for LBX1 in 14 human breast cancers demonstrated high-level expression in nine tumors, but not in surrounding normal breast epithelium (Supplemental Fig. S13). Interestingly, in one breast tumor of the triple-negative subtype (negative for estrogen receptor [ER], progesterone [PR], and Her2), which had adjacent foci of histologically high-grade and low-grade tumor, LBX1 expression was only present within the invasive lesion (Fig. 5A). To address a possible correlation between LBX1 expression and other known prognostic markers in breast cancer, we interrogated a large published data set, including 251 clinically annotated cases of breast cancer for which RNA microarray and protein immunohistochemistry (IHC) data are available (Miller et al. 2005). LBX1 expression in these breast cancers was significantly correlated with advanced tumor grade (grade III) (P = 0.0023), mutant p53 status (P = 0.00056), negative ER (P = 0.038), and PR status (P = 0.0023) (Fig. 5B). The correlation between LBX1 expression and adverse prognostic markers was also evident when analyzing p53 mutant-like tumors defined by their microarray expression profile analysis (P = 2.1e − 06). Since no ERBB2/HER2 protein expression data are available for this breast cancer data set, we analyzed mRNA expression of ERBB2/HER2, together with the IHC-verified ER/PR status, to define the subset of cancers characterized as ER/PR/HER2 “triple negative.” Clinically, this breast cancer subset is known to be associated with higher tumor grades, poor response to chemotherapy, and an unfavorable prognosis. Indeed, higher LBX1 mRNA levels were present in triple-negative tumors (P = 0.0017). Taken together, these data suggest that aberrant expression of the developmentally regulated EMT inducer LBX1 is associated with more invasive subtypes of human breast cancer.
Figure 5.
LBX1 expression level is associated with invasive subtype of human breast cancer. (A) Human breast tumor sample examined by hematoxylin and eosin stain (H&E) and in situ hybridization for LBX1 (LBX1). The left panel shows low magnification H&E-stained tumor. Higher magnification of the adjacent low-grade and high-grade tumor regions is shown in the right panel for H&E (histology) and in situ hybridization with probes against LBX1 mRNA. (B) Microarray analysis in human breast tumors for correlation of LBX1 expression level with different known prognostic factors. Relative expression of LBX1 mRNA level was compared in different groups according to tumor grade, P53 mutation status, P53 pathway status, estrogen receptor, progesterone receptor, and triple-negative status by box plotting. P-values analyzed by Kruskal-Wallis test were indicated.
Concluding remarks
During normal muscle development, LBX1 is expressed specifically in hypaxial muscle precursors derived from dermomyotomes and it is indispensible for these precursors to migrate over long distances to form most of the skeletal muscles in the limb (Mennerich et al. 1998; Gross et al. 2000). Lbx1−/− mice lack most appendicular muscles, except for some forelimb flexors and hindlimb extensors. Diaphragm and tongue muscles, which do not involve extensive precursor migration, still form in these mice (Franz et al. 1993; Gross et al. 2000). These observations have implicated LBX1 in cellular migration, but have not determined whether LBX1 expression is itself a cause or result of EMT transformation. Our results indicate that LBX1 can indeed function as a driver for EMT.
The regulation of EMT involves a complex network of signaling molecules and transcription factors, presumably linked to the precise temporal and spatial requirement of EMT in specific cell types at precisely coordinated times during development. Key environmental signals transduced through the TGFβ, WNT, Notch, and receptor tyrosine kinase pathways are thought to lead to the activation of the transcriptional regulators Snail1, Slug, ZEB1, ZEB2, TWIST1, FOXC2, and Goosecoid that in turn program suppression of epithelial proteins and up-regulation of mesenchymal proteins. The observation that LBX1 directly up-regulates expression of several EMT-inducing transcription factors raises the possibility that it may function as an upstream or “master” regulator. In this context, it is interesting that Snail1, Slug, TWIST1, and ZEB2 are all involved in neural crest formation (Yang and Weinberg 2008), while LBX1 mediates cell fate determination in specific regions of the CNS (Gross et al. 2002; Kruger et al. 2002). A similar hierarchy may exist during myogenesis where LBX1, itself regulated by the Scatter factor/hepatocyte growth factor (SF/HGF) and c-MET pathways, may integrate different environmental cues to guide and maintain the migration of the muscle precursor cells to the distal limbs (Brohmann et al. 2000; Gross et al. 2000). The integrative role proposed for LBX1 in normal neural and muscle differentiation makes it a particularly interesting candidate for a role in mediating EMT during tumorigenesis by coregulating its targets.
Of all the developmentally regulated candidate genes tested here, LBX1 was the most potent inducer of EMT in human mammary epithelial cells, with characteristic changes in cell morphology and migratory potential, down-regulation of epithelial markers, and up-regulation of mesenchymal markers. Recently, EMT in breast epithelial precursors has been linked to “stemness,” a phenomenon defined by expression of the CD44high/CD24low stem cell signature, and the ability to generate anchorage-independent colonies in serum-free media, called mammospheres (Mani et al. 2008). It is uncertain whether EMT truly reverts epithelial cells to a more primitive cell fate or whether it induces a cellular program that shares certain functional properties with stem-like progenitors. However, its potential impact on cancer is illustrated by the striking tumorigenic cooperation evident by inducing an EMT-driven cell fate, together with proliferative signals exemplified by activated HRAS. In all of these properties, LBX1 appears to function as an important driver of EMT and its tumorigenic potential.
While further studies will be required to define the contribution of LBX1 across the diverse forms of human cancer, the striking correlation between its aberrant expression and the high-grade, p53 mutant, ER/PR/HER2 triple-negative, so-called “basal-like” breast cancers is consistent with a role in this particularly invasive subtype. Basal-like carcinomas account for up to 15% of all breast cancers, most frequently affecting young women, and are noteworthy for their high rate of distant metastasis to brain and visceral organs, and their unfavorable prognosis (Nielsen et al. 2004; Calza et al. 2006; Reis-Filho and Tutt 2008). Of note, basal-like breast carcinomas have generated considerable interest as a distinct subset thought to originate from normal basal/myoepithelial cells of the breast, and EMT markers have been correlated preferentially with such cell types (Sarrio et al. 2008). The up-regulation of LBX1 and its associated EMT cellular program may therefore reflect a lineage-specific property in a partially invasive subset of breast cancer.
Materials and methods
Constructs
The human LBX1 ORF was cloned into pBabe or pWPI expression vectors as a BamHI–EcoRI fragment. The HRasV12 pMSCV-IRES-GFP construct is a generous gift from Patrick Humbert. Mouse Snail1 and Twist1 proteins were expressed in pBabe vector. One-kilobase sequence fragments upstream of human Snail1 (−1078 ∼ −79) and ZEB1 (−1079 ∼ −80) transcription start site were cloned into pGL4.15 luciferase reporter vector (Promega). shRNAs targeting the human LBX1 and control luciferase were from the RNAi consortium at the Broad Institute.
Transwell migration assay
Cells (1 × 105) were plated without EGF on 8-μm pore size Transwell filters (Corning) in 3D medium as described (Debnath et al. 2003). Assays were stained and quantified after cells migrated for 48 h. For MDA-MB-231 cells, 0.3 × 105 cells were used in the assay for 24 h.
Real-time qRT–PCR
RNA was isolated using RNeasy Mini kit (Qiagen) and used for real-time qRT–PCR using SYBR Green in an ABI PRISM 7500 sequence detection system with 96-block module and automation accessory (Applied Biosystems). GAPDH was used as an internal control gene. All samples are analyzed in triplicate. The primer sequences are listed in the Supplemental Material.
Acknowledgments
We thank Miguel N. Rivera for his help with pathological evaluation and Dennis Sgroi for discussion and technical help. This work was supported by grants from the NIH (CA-129933) and the Howard Hughes Medical Institute.
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1809309.
Supplemental material is available at http://www.genesdev.org.
References
- Al-Hajj M, Wicha MS, Benito-Hernandez A, Morrison SJ, Clarke MF. Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci. 2003;100:3983–3988. doi: 10.1073/pnas.0530291100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ansieau S, Bastid J, Doreau A, Morel AP, Bouchet BP, Thomas C, Fauvet F, Puisieux I, Doglioni C, Piccinin S, et al. Induction of EMT by twist proteins as a collateral effect of tumor-promoting inactivation of premature senescence. Cancer Cell. 2008;14:79–89. doi: 10.1016/j.ccr.2008.06.005. [DOI] [PubMed] [Google Scholar]
- Brohmann H, Jagla K, Birchmeier C. The role of Lbx1 in migration of muscle precursor cells. Development. 2000;127:437–445. doi: 10.1242/dev.127.2.437. [DOI] [PubMed] [Google Scholar]
- Calza S, Hall P, Auer G, Bjohle J, Klaar S, Kronenwett U, Liu ET, Miller L, Ploner A, Smeds J, et al. Intrinsic molecular signature of breast cancer in a population-based cohort of 412 patients. Breast Cancer Res. 2006;8:R34. doi: 10.1186/bcr1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debnath J, Muthuswamy SK, Brugge JS. Morphogenesis and oncogenesis of MCF-10A mammary epithelial acini grown in three-dimensional basement membrane cultures. Methods. 2003;30:256–268. doi: 10.1016/s1046-2023(03)00032-x. [DOI] [PubMed] [Google Scholar]
- Dontu G, Abdallah WM, Foley JM, Jackson KW, Clarke MF, Kawamura MJ, Wicha MS. In vitro propagation and transcriptional profiling of human mammary stem/progenitor cells. Genes & Dev. 2003;17:1253–1270. doi: 10.1101/gad.1061803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elenbaas B, Spirio L, Koerner F, Fleming MD, Zimonjic DB, Donaher JL, Popescu NC, Hahn WC, Weinberg RA. Human breast cancer cells generated by oncogenic transformation of primary mammary epithelial cells. Genes & Dev. 2001;15:50–65. doi: 10.1101/gad.828901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franz T, Kothary R, Surani MA, Halata Z, Grim M. The Splotch mutation interferes with muscle development in the limbs. Anat Embryol (Berl) 1993;187:153–160. doi: 10.1007/BF00171747. [DOI] [PubMed] [Google Scholar]
- Gross MK, Moran-Rivard L, Velasquez T, Nakatsu MN, Jagla K, Goulding M. Lbx1 is required for muscle precursor migration along a lateral pathway into the limb. Development. 2000;127:413–424. doi: 10.1242/dev.127.2.413. [DOI] [PubMed] [Google Scholar]
- Gross MK, Dottori M, Goulding M. Lbx1 specifies somatosensory association interneurons in the dorsal spinal cord. Neuron. 2002;34:535–549. doi: 10.1016/s0896-6273(02)00690-6. [DOI] [PubMed] [Google Scholar]
- Hay ED. The mesenchymal cell, its role in the embryo, and the remarkable signaling mechanisms that create it. Dev Dyn. 2005;233:706–720. doi: 10.1002/dvdy.20345. [DOI] [PubMed] [Google Scholar]
- Janda E, Lehmann K, Killisch I, Jechlinger M, Herzig M, Downward J, Beug H, Grunert S. Ras and TGFβ cooperatively regulate epithelial cell plasticity and metastasis: Dissection of Ras signaling pathways. J Cell Biol. 2002;156:299–313. doi: 10.1083/jcb.200109037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruger M, Schafer K, Braun T. The homeobox containing gene Lbx1 is required for correct dorsal-ventral patterning of the neural tube. J Neurochem. 2002;82:774–782. doi: 10.1046/j.1471-4159.2002.01078.x. [DOI] [PubMed] [Google Scholar]
- Lee J, Kotliarova S, Kotliarov Y, Li A, Su Q, Donin NM, Pastorino S, Purow BW, Christopher N, Zhang W, et al. Tumor stem cells derived from glioblastomas cultured in bFGF and EGF more closely mirror the phenotype and genotype of primary tumors than do serum-cultured cell lines. Cancer Cell. 2006;9:391–403. doi: 10.1016/j.ccr.2006.03.030. [DOI] [PubMed] [Google Scholar]
- Mani SA, Guo W, Liao MJ, Eaton EN, Ayyanan A, Zhou AY, Brooks M, Reinhard F, Zhang CC, Shipitsin M, et al. The epithelial–mesenchymal transition generates cells with properties of stem cells. Cell. 2008;133:704–715. doi: 10.1016/j.cell.2008.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mennerich D, Schafer K, Braun T. Pax-3 is necessary but not sufficient for lbx1 expression in myogenic precursor cells of the limb. Mech Dev. 1998;73:147–158. doi: 10.1016/s0925-4773(98)00046-x. [DOI] [PubMed] [Google Scholar]
- Miller LD, Smeds J, George J, Vega VB, Vergara L, Ploner A, Pawitan Y, Hall P, Klaar S, Liu ET, et al. An expression signature for p53 status in human breast cancer predicts mutation status, transcriptional effects, and patient survival. Proc Natl Acad Sci. 2005;102:13550–13555. doi: 10.1073/pnas.0506230102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller T, Brohmann H, Pierani A, Heppenstall PA, Lewin GR, Jessell TM, Birchmeier C. The homeodomain factor lbx1 distinguishes two major programs of neuronal differentiation in the dorsal spinal cord. Neuron. 2002;34:551–562. doi: 10.1016/s0896-6273(02)00689-x. [DOI] [PubMed] [Google Scholar]
- Nielsen TO, Hsu FD, Jensen K, Cheang M, Karaca G, Hu Z, Hernandez-Boussard T, Livasy C, Cowan D, Dressler L, et al. Immunohistochemical and clinical characterization of the basal-like subtype of invasive breast carcinoma. Clin Cancer Res. 2004;10:5367–5374. doi: 10.1158/1078-0432.CCR-04-0220. [DOI] [PubMed] [Google Scholar]
- Oft M, Heider KH, Beug H. TGFβ signaling is necessary for carcinoma cell invasiveness and metastasis. Curr Biol. 1998;8:1243–1252. doi: 10.1016/s0960-9822(07)00533-7. [DOI] [PubMed] [Google Scholar]
- Peinado H, Olmeda D, Cano A. Snail, Zeb and bHLH factors in tumour progression: An alliance against the epithelial phenotype? Nat Rev Cancer. 2007;7:415–428. doi: 10.1038/nrc2131. [DOI] [PubMed] [Google Scholar]
- Reis-Filho JS, Tutt AN. Triple negative tumours: A critical review. Histopathology. 2008;52:108–118. doi: 10.1111/j.1365-2559.2007.02889.x. [DOI] [PubMed] [Google Scholar]
- Sarrio D, Rodriguez-Pinilla SM, Hardisson D, Cano A, Moreno-Bueno G, Palacios J. Epithelial–mesenchymal transition in breast cancer relates to the basal-like phenotype. Cancer Res. 2008;68:989–997. doi: 10.1158/0008-5472.CAN-07-2017. [DOI] [PubMed] [Google Scholar]
- Sieber MA, Storm R, Martinez-de-la-Torre M, Muller T, Wende H, Reuter K, Vasyutina E, Birchmeier C. Lbx1 acts as a selector gene in the fate determination of somatosensory and viscerosensory relay neurons in the hindbrain. J Neurosci. 2007;27:4902–4909. doi: 10.1523/JNEUROSCI.0717-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sleeman KE, Kendrick H, Ashworth A, Isacke CM, Smalley MJ. CD24 staining of mouse mammary gland cells defines luminal epithelial, myoepithelial/basal and non-epithelial cells. Breast Cancer Res. 2006;8:R7. doi: 10.1186/bcr1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiery JP, Sleeman JP. Complex networks orchestrate epithelial–mesenchymal transitions. Nat Rev Mol Cell Biol. 2006;7:131–142. doi: 10.1038/nrm1835. [DOI] [PubMed] [Google Scholar]
- Xiang B, Muthuswamy SK. Using three-dimensional acinar structures for molecular and cell biological assays. Methods Enzymol. 2006;406:692–701. doi: 10.1016/S0076-6879(06)06054-X. [DOI] [PubMed] [Google Scholar]
- Yang J, Weinberg RA. Epithelial–mesenchymal transition: At the crossroads of development and tumor metastasis. Dev Cell. 2008;14:818–829. doi: 10.1016/j.devcel.2008.05.009. [DOI] [PubMed] [Google Scholar]