Abstract
The “Spanish” influenza pandemic of 1918–19 caused acute illness in 25–30 percent of the world’s population and resulted in the death of up to an estimated 40 million people. Using fixed and frozen lung tissue of 1918 influenza victims, the complete genomic sequence of the 1918 influenza virus has been deduced. Sequence and phylogenetic analysis of the completed 1918 influenza virus genes shows them to be the most avian-like among the mammalian-adapted viruses. This finding supports the hypotheses that (1) the pandemic virus contains genes derived from avian-like influenza virus strains and that (2) the 1918 virus is the common ancestor of human and classical swine H1N1 influenza viruses. The relationship of the 1918 virus with avian influenza viruses is further supported by recent work in which the 1918 hemagglutinin (HA) protein crystal structure was resolved. Neither the 1918 hemagglutinin (HA) nor the neuraminidase (NA) genes possess mutations known to increase tissue tropicity that account for the virulence of other influenza virus strains like A/WSN/33 or the highly pathogenic avian influenza H5 or H7 viruses. Using reverse genetics approaches, influenza virus constructs containing the 1918 HA and NA on a modern human influenza virus background were lethal in mice. The complete 1918 virus was even more virulent in mice. The genotypic basis of this virulence has not yet been elucidated. The complete sequence of the non-structural (NS) gene segment of the 1918 virus was deduced and also tested for the hypothesis that enhanced virulence in 1918 could have been due to type I interferon inhibition by the NS1 protein. Results from these experiments suggest that in human cells the 1918 NS1 is a very effective interferon antagonist, but the 1918 NS1 gene does not have the amino acid change that correlates with virulence in the H5N1 virus strains identified in 1997 in Hong Kong. Sequence analysis of the 1918 pandemic influenza virus is allowing us to test hypotheses as to the origin and virulence of this strain. This information should help elucidate how pandemic influenza virus strains emerge and what genetic features contribute to virulence in humans.
Influenza a viruses are negative strand RNA viruses of the genus Orthomyxoviridae. They continually circulate in humans in yearly epidemics (mainly in the winter in temperate climates) and antigenically novel virus strains emerge sporadically as pandemic viruses (Cox and Subbarao 2000). In the United States, influenza is estimated to kill 30,000 people in an average year (Simonsen et al. 2000; Thompson et al. 2003). Every few years, influenza epidemics boost the annual number of deaths past the average, causing 10–15,000 additional deaths. Occasionally, and unpredictably, influenza sweeps the world, infecting 20 percent to 40 percent of the population in a single year. In these pandemic years, the numbers of deaths can be dramatically above average. In 1957–58, a pandemic was estimated to cause 66,000 excess deaths in the United States (Simonsen et al. 1998). In 1918, the worst pandemic in recorded history was associated with approximately 675,000 total deaths in the United States (United States Department of Commerce 1976), and killed an estimated 40 million people worldwide (Crosby 1989; Johnson and Mueller 2002; Patterson and Pyle 1991).
Influenza A viruses constantly evolve by the mechanisms of antigenic drift and shift (Webster et al. 1992). Consequently, they should be considered emerging infectious disease agents, perhaps “continually” emerging pathogens. The importance of predicting the emergence of new circulating influenza virus strains for subsequent annual vaccine development cannot be overestimated (Gensheimer et al. 1999). Pandemic influenza viruses have emerged three times in this century: in 1918 (“Spanish” influenza, H1N1), in 1957 (“Asian” influenza, H2N2), and in 1968 (“Hong Kong” influenza, H3N2; Cox and Subbarao 2000; Webby and Webster 2003). Recent circulation of highly pathogenic avian H5N1 viruses in Asia from 1997 to 2004 has caused a small number of human deaths (Claas et al. 1998; Peiris et al. 2004; Subbarao et al. 1998; Tran et al. 2004). How and when novel influenza viruses emerge as pandemic virus strains and how they cause disease is still not understood.
Studying the extent to which the 1918 influenza was like other pandemics may help us to understand how pandemic influenzas emerge and cause disease in general. On the other hand, if we determine what made the 1918 influenza different from other pandemics, we may use the lessons of 1918 to predict the magnitude of the public health risks a new pandemic virus might pose.
Origin of Pandemic Influenza Viruses
The predominant natural reservoir of influenza viruses is thought to be wild waterfowl (Webster et al. 1992). Periodically, genetic material from avian virus strains is transferred to virus strains infectious to humans by a process called reassortment. Human influenza virus strains with recently acquired avian surface and internal protein-encoding RNA segments were responsible for the pandemic influenza outbreaks in 1957 and 1968 (Kawaoka et al. 1989; Scholtissek et al. 1978a). The change in the hemagglutinin subtype or the hemagglutinin and the neuraminidase subtype is referred to as antigenic shift. Since pigs can be infected with both avian and human virus strains, and various reassortants have been isolated from pigs, they have been proposed as an intermediary in this process (Ludwig et al. 1995; Scholtissek 1994). Until recently there was only limited evidence that a wholly avian influenza virus could directly infect humans, but in 1997 eighteen people were infected with avian H5N1 influenza viruses in Hong Kong, and six died of complications after infection (Claas et al. 1998; Ludwig et al. 1995; Scholtissek 1994; Subbarao et al. 1998). Although these viruses were very poorly transmissible or non-transmissible (Claas et al. 1998; Katz et al. 1999; Ludwig et al. 1995; Scholtissek 1994; Subbarao et al. 1998), their isolation from infected patients indicates that humans can be infected with wholly avian influenza virus strains. In 2003–04, H5N1 outbreaks in poultry have become widespread in Asia (Tran et al. 2004), and at least 23 people have died of complications of infection in Vietnam and Thailand (WHO 2004). In 2003, a highly pathogenic H7N7 outbreak occurred in poultry farms in the Netherlands. This virus caused infections (predominantly conjunctivitis) in 86 poultry handlers and in 3 secondary contacts. One of the infected individuals died of pneumonia (Fouchier et al. 2004; Koopmans et al. 2004; WHO 2004). In 2004 an H7N3 influenza outbreak in poultry in Canada also resulted in the infection of a single individual (WHO 2004), and a patient in New York was reported to be sick following infection with an H7N2 virus (Lipsman 2004). Therefore, it may not be necessary to invoke swine as the intermediary in the formation of a pandemic virus strain since reassortment between an avian and a human influenza virus could take place directly in humans.
While reassortment involving genes encoding surface proteins appears to be a critical event for the production of a pandemic virus, a significant amount of data exists to suggest that influenza viruses must also acquire specific adaptations to spread and replicate efficiently in a new host. Among other features, there must be functional HA receptor binding and interaction between viral and host proteins (Weis et al. 1988). Defining the minimal adaptive changes needed to allow a reassortant virus to function in humans is essential to understanding how pandemic viruses emerge.
Once a new virus strain has acquired the changes that allow it to spread in humans, virulence is affected by the presence of novel surface protein(s) that allow the virus to infect an immunologically naïve population (Kilbourne 1977). This was the case in 1957 and 1968 and was almost certainly the case in 1918. While immunological novelty may explain much of the virulence of the 1918 influenza, it is likely that additional genetic features contributed to its exceptional lethality. Unfortunately, not enough is known about how genetic features of influenza viruses affect virulence. The degree of illness caused by a particular virus strain, or virulence, is complex and involves host factors like immune status, and viral factors like host adaptation, transmissibility, tissue tropism, or viral replication efficiency. The genetic basis for each of these features is not yet fully characterized, but is most likely polygenic in nature (Kilbourne 1977).
Prior to the analyses on the 1918 virus described in this review, only two pandemic influenza virus strains were available for molecular analysis: the H2N2 virus strain from 1957 and the H3N2 virus strain from 1968. The 1957 pandemic resulted from the emergence of a reassortant influenza virus in which both HA and NA had been replaced by gene segments closely related to those in avian virus strains (Schafer et al. 1993; Scholtissek et al. 1978b; Webster et al. 1995). The 1968 pandemic followed with the emergence of a virus strain in which the H2 subtype HA gene was exchanged with an avian-derived H3 HA RNA segment (Scholtissek et al. 1978b; Webster et al. 1995), while retaining the N2 gene derived in 1957. More recently it has been shown that the PB1 gene was replaced in both the 1957 and the 1968 pandemic virus strains, also with a likely avian derivation in both cases (Kawaoka et al. 1989). The remaining five RNA segments encoding the PA, PB2, nucleoprotein, matrix, and non-structural proteins, all were preserved from the H1N1 virus strains circulating before 1957. These segments were likely the direct descendants of the genes present in the 1918 virus. Since only the 1957 and 1968 influenza pandemic virus strains have been available for sequence analysis, it is not clear what changes are necessary for the emergence of a virus strain with pandemic potential. Sequence analysis of the 1918 influenza virus allows us potentially to address the genetic basis of virulence and human adaptation.
Historical Background
The influenza pandemic of 1918 was exceptional in both breadth and depth. Outbreaks of the disease not only swept North America and Europe, but also spread as far as the Alaskan wilderness and the most remote islands of the Pacific. It has been estimated that one-third of the world’s population (500 million people) may have been clinically infected during the pandemic (Burnet and Clark 1942; Frost 1920). The disease was also exceptionally severe, with mortality rates among the infected of more than 2.5 percent, compared with less than 0.1 percent in other influenza epidemics (Marks and Beatty 1976; Rosenau and Last 1980). Total mortality attributable to the 1918 pandemic was probably around 40 million (Crosby 1989; Johnson and Mueller 2002; Patterson and Pyle 1991).
Unlike most subsequent influenza virus strains that have developed in Asia, the “first wave” or “spring wave” of the 1918 pandemic seemingly arose in the United States in March 1918 (Barry 2004; Crosby 1989; Jordan 1927). However, the near-simultaneous appearance of influenza in March–April 1918 in North America, Europe, and Asia makes definitive assignment of a geographic point of origin difficult (Jordan 1927). It is possible that a mutation or reassortment occurred in the late summer of 1918, resulting in significantly enhanced virulence. The main wave of the global pandemic, the “fall wave” or “second wave,” occurred in September–November 1918. In many places, there was yet another severe wave of influenza in early 1919 (Jordan 1927).
Three extensive outbreaks of influenza within one year is unusual, and may point to unique features of the 1918 virus that could be revealed in its sequence. Interpandemic influenza outbreaks generally occur in a single annual wave in the late winter. The severity of annual outbreaks is affected by antigenic drift, with an antigenically modified virus strain emerging every two to three years. Even in pandemic influenza, while the normal late winter seasonality may be violated, the successive occurrence of distinct waves within a year is unusual. The 1890 pandemic began in the late spring of 1889 and took several months to spread throughout the world, peaking in northern Europe and the United States in late 1889 or early 1890. The second wave peaked in spring 1891 (more than a year after the first wave) and the third wave in early 1892 (Jordan 1927). As in 1918, subsequent waves seemed to produce more severe illness, so that the peak mortality was reached in the third wave of the pandemic. The three waves, however, were spread over more than three years, in contrast with less than one year in 1918. It is unclear what gave the 1918 virus this unusual ability to generate repeated waves of illness. Perhaps the surface proteins of the virus drifted more rapidly than other influenza virus strains, or perhaps the virus had an unusually effective mechanism for evading the human immune system.
It has been estimated that the influenza epidemic of 1918 killed 675,000 Americans, including 43,000 servicemen mobilized for World War I (Crosby 1989). The impact was so profound as to depress average life expectancy in the U.S. by more than ten years (fig. 1; Grove and Hetzel 1968), and may have played a significant role in ending World War I (Crosby 1989; Ludendorff 1919).
Figure 1.
Life expectancy in the United States, 1900–60, showing the impact of the 1918 influenza pandemic (Grove and Hetzel 1968; Linder and Grove 1943; United States Department of Commerce 1976).
The majority of individuals who died during the pandemic succumbed to secondary bacterial pneumonia (Jordan 1927; LeCount 1919; Wolbach 1919), since no antibiotics were available in 1918. However, a subset died rapidly after the onset of symptoms, often with either massive acute pulmonary hemorrhage or pulmonary edema, often in less than five days (LeCount 1919; Winternitz et al. 1920; Wolbach 1919). In the hundreds of autopsies performed in 1918, the primary pathologic findings were confined to the respiratory tree and death was due to pneumonia and respiratory failure (Winternitz et al. 1920). These findings are consistent with infection by a well-adapted influenza virus capable of rapid replication throughout the entire respiratory tree (Reid and Taubenberger 1999; Taubenberger et al. 2000). There was no clinical or pathological evidence for systemic circulation of the virus (Winternitz et al. 1920).
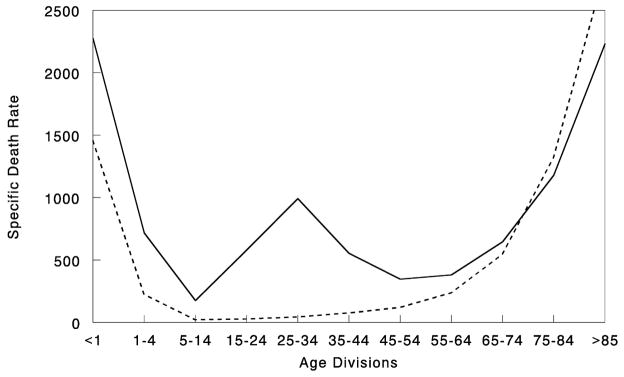
Furthermore, in the 1918 pandemic most deaths occurred among young adults, a group that usually has a very low death rate from influenza. Influenza and pneumonia death rates for 15-to-34-year-olds were more than twenty times higher in 1918 than in previous years (fig. 2; Linder and Grove 1943; Simonsen et al. 1998). The 1918 pandemic is also unique among influenza pandemics in that absolute risk of influenza mortality was higher in those less than 65 years of age than in those older than 65. Strikingly, persons less than 65 years old accounted for more than 99 percent of all excess influenza-related deaths in 1918–19 (Simonsen et al. 1998). In contrast, the under-65 age group accounted for only 36 percent of all excess influenza-related mortality in the 1957 H2N2 pandemic and 48 percent in the 1968 H3N2 pandemic. Overall, nearly half of the influenza-related deaths in the 1918 influenza pandemic were young adults, age 20–40 (fig. 2; Simonsen et al. 1998). Why this particular age group suffered such extreme mortality is not fully understood (see below).
Figure 2.
Influenza and pneumonia mortality by age, United States. Influenza and pneumonia specific mortality by age, including an average of the inter-pandemic years 1911–15 (dashed line), and the pandemic year 1918 (solid line). Specific death rate is per 100,000 of the population in each age division (Grove and Hetzel 1968; Linder and Grove 1943; United States Department of Commerce 1976).
The 1918 influenza had as another unique feature the simultaneous infection of both humans and swine. Interestingly, swine influenza was first recognized as a clinical entity in that species in the fall of 1918 (Koen 1919), concurrently with the spread of the second wave of the pandemic in humans (Dorset et al. 1922–23). Investigators were impressed with the clinical and pathological similarities of human and swine influenza in 1918 (Koen 1919; Murray and Biester 1930). An extensive review by the veterinarian W. W. Dimoch of the diseases of swine, published in August 1918, makes no mention of any swine disease resembling influenza (Dimoch 1918–19). Thus, contemporary investigators were convinced that influenza virus had not circulated as an epizootic disease in swine before 1918 and that the virus spread from humans to pigs because of the appearance of illness in pigs after the first wave of the 1918 influenza in humans (Shope 1936).
Thereafter, the disease became widespread among swine herds in the U.S. Midwest. The epizootic of 1919–20 was as extensive as in 1918–19. The disease then appeared among swine in the Midwest every year, leading to Shope’s isolation of the first influenza virus in 1930, A/swine/Iowa/30 (Shope and Lewis 1931), three years before the isolation of the first human influenza virus, A/WS/33 by Smith, Andrewes, and Laidlaw (Smith et al. 1933). Classical swine viruses have continued to circulate not only in North American pigs, but also in swine populations in Europe and Asia (Brown et al. 1995; Kupradinun et al. 1991; Nerome et al. 1982).
During the fall and winter of 1918–19, severe influenza-like outbreaks were noted in swine not only in the United States, but also in Europe and China (Beveridge 1977; Chun 1919; Koen 1919). Since 1918 there have been many examples of both H1N1 and H3N2 human influenza A virus strains’ becoming established in swine (Brown et al. 1998; Castrucci et al. 1993; Zhou et al. 2000) while swine influenza A virus strains have been isolated only sporadically from humans (Gaydos et al. 1977; Woods et al. 1981).
The unusual severity of the 1918 pandemic and the exceptionally high mortality it caused among young adults have stimulated great interest in the influenza virus strain responsible for the 1918 outbreak (Crosby 1989; Kolata 1999; Monto et al. 1997). Since the first human and swine influenza A viruses were not isolated until the early 1930s (Shope and Lewis 1931; Smith et al. 1933), characterization of the 1918 virus strain has had previously to rely on indirect evidence (Kanegae et al. 1994; Shope 1958).
Serology and Epidemiology of the 1918 Influenza Virus
Analyses of antibody titers of 1918 influenza survivors from the late 1930s suggested correctly that the 1918 virus strain was an H1N1-subtype influenza A virus, closely related to what is now known as “classic swine” influenza virus (Dowdle 1999; Philip and Lackman 1962; Shope 1936). The relationship to swine influenza is also reflected in the simultaneous influenza outbreaks in humans and pigs around the world (Beveridge 1977; Chun 1919; Koen 1919). While historical accounts described above suggest that the virus spread from humans to pigs in the fall of 1918, the relationship of these two species in the development of the 1918 influenza has not been resolved.
It is not known for certain what influenza A subtype(s) circulated before the 1918 pandemic. In a recent review of the existing archaeoserologic and epidemiologic data, Walter Dowdle concluded that an H3 subtype influenza A virus strain circulated from the 1889–91 pandemic to 1918, when it was replaced by the novel H1N1 virus strain of the 1918 pandemic (Dowdle 1999).
It is reasonable to conclude that the 1918 virus strain must have contained a hemagglutinin gene encoding a novel subtype such that large portions of the population did not have protective immunity (Kilbourne 1977; Reid and Taubenberger 1999). In fact, epidemiological data on influenza prevalence by age in the population collected between 1900 and 1918 provide good evidence for the emergence of an antigenically novel influenza virus in 1918 (Jordan 1927). Jordan showed that from 1900 to 1917, the 5–15 age group accounted for 11 percent of total influenza cases in this series, while the >65 age group similarly accounted for 6 percent of influenza cases. In 1918 the 5-to-15-year-old group jumped to 25 percent of influenza cases, compatible with exposure to an antigenically novel virus strain. The >65 age group accounted for only 0.6 percent of the influenza cases in 1918. It is likely that this age group accounted for a significantly lower percentage of influenza cases because younger people were so susceptible to the novel virus strain (as seen in the 1957 pandemic; Ministry of Health 1960; Simonsen et al. 1998), but it is also possible that this age group had pre-existing H1 antibodies. Further evidence for pre-existing H1 immunity can be derived from the age-adjusted mortality data in fig. 2. Those individuals >75 years had a lower influenza and pneumonia case mortality rate in 1918 than they had for the pre-pandemic period of 1911–17.
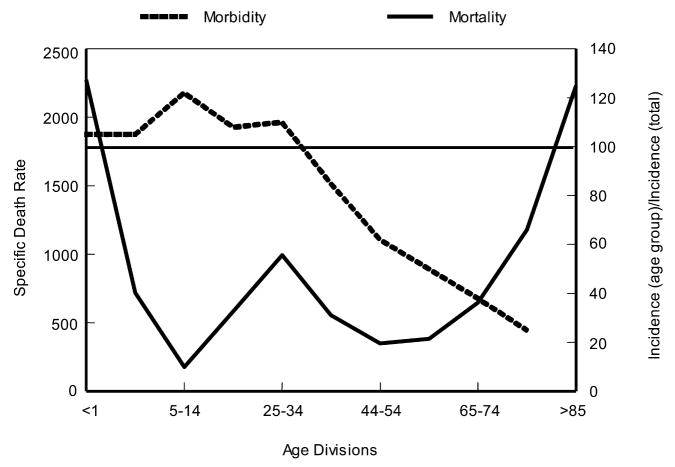
When 1918 influenza case rates by age (Jordan 1927) are superimposed on the familiar “W” shaped mortality curve (seen in fig. 2), a different perspective emerges (fig. 3). As shown, those <35 years of age in 1918 accounted for a disproportionately high influenza incidence by age. Interestingly, the 5–14 age group accounted for a large fraction of 1918 influenza cases, but had an extremely low case mortality rate compared with other age groups (fig. 3). Why this age group had such a low case fatality rate cannot yet be fully explained. Conversely, why the 25–34 age group had such a high influenza and pneumonia mortality rate in 1918 remains enigmatic, but it is one of the truly unique features of the 1918 influenza pandemic.
Figure 3.
Influenza and pneumonia mortality by age (solid line), with influenza morbidity by age (dashed line) superimposed. Influenza and pneumonia mortality by age as in figure 2. Specific death rate per age group, left ordinal axis. Influenza morbidity presented as ratio of incidence in persons of each group to incidence in persons of all ages (=100), right ordinal axis. Horizontal line at 100 (right ordinal axis) represents average influenza incidence in the total population (Taubenberger et al. 2001; adapted from Jordan 1927).
One theory that may explain these data concerns the possibility that the virus had an intrinsically high virulence that was tempered only in those patients who had been born before 1889. It can be speculated that the virus circulating prior to 1889 was an H1-like virus strain that provided partial protection against the 1918 virus strain (Ministry of Health 1960; Simonsen et al. 1998; Taubenberger et al. 2001).
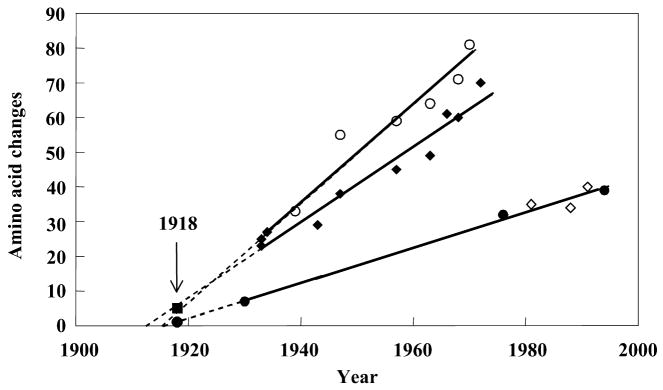
Thus, it seems clear that the H1N1 virus of the 1918 pandemic contained an antigenically novel hemagglutinin to which most humans and swine were susceptible in 1918. Given the severity of the pandemic, it is also reasonable to suggest that the other dominant surface protein, NA, would also have been replaced by antigenic shift before the start of the pandemic (Reid and Taubenberger 1999; Taubenberger et al. 2000). In fact, sequence and phylogenetic analyses suggest that the genes encoding these two surface proteins were derived from an avian-like influenza virus shortly before the start of the 1918 pandemic, and that the precursor virus did not circulate widely in either humans or swine before 1918 (Fanning et al. 2002; Reid et al. 1999; Reid et al. 2000; fig. 4). It is currently unclear what other influenza gene segments were novel in the 1918 pandemic virus in comparison with the previously circulating virus strain. It is possible that sequence and phylogenetic analyses of the gene segments of the 1918 virus may help elucidate this question.
Figure 4.
Change in hemagglutinin (HA) and neuraminidase (NA) proteins over time (Reid et al. 1999; Reid et al. 2000; Taubenberger et al. 2000). The number of amino acid changes from a hypothetical ancestor was plotted versus the date of viral isolation for viruses isolated from 1930 to 1993. Open circles, human HA; closed diamonds, human NA; closed circles, swine HA; open diamonds, swine NA. Regression lines were drawn and extrapolated to the x-intercept and then the 1918 data points (closed square, 1918 HA; closed circle, 1918 NA) were added to the graph (arrow).
Genetic Characterization of the 1918 Virus
Sequence and functional analysis of the hemagglutinin and neuraminidase gene segments
Frozen and fixed lung tissue from five fall-wave 1918 influenza victims has been used to examine directly the genetic structure of the 1918 influenza virus. Two of the cases analyzed were U.S. Army soldiers who died in September 1918, one in Camp Upton, New York, and the other in Fort Jackson, South Carolina. The available material consists of formalin-fixed, paraffin-embedded (FFPE) autopsy tissue, hematoxylin- and eosin-stained microscopic sections, and the clinical histories of these patients. A third sample was obtained from an Alaskan Inuit woman who had been interred in permafrost in Brevig Mission, Alaska, since her death from influenza in November 1918. The influenza virus sequences derived from these three cases have been called A/South Carolina/1/18 (H1N1), A/New York/1/18 (H1N1), and A/Brevig Mission/1/18 (H1N1), respectively. To date, all eight RNA segment sequences have been published (Basler et al. 2001; Reid et al. 1999; Reid et al. 2004; Reid et al. 2002; Reid et al. 2000; Taubenberger et al. 2005). The HA sequences of two additional fixed autopsy cases of 1918 influenza victims from the Royal London Hospital were determined (Reid et al. 2003). The HA sequences from these five cases show >99 percent sequence identity, but differ at amino acid residue 225 (see below).
The sequence of the 1918 HA is most closely related to that of the A/swine/Iowa/30 virus. However, despite this similarity the sequence has many avian features. Of the 41 amino acids that have been shown to be targets of the immune system and subject to antigenic drift pressure in humans, 37 match the avian sequence consensus, suggesting that there was little immunologic pressure on the HA protein before the fall of 1918 (Reid et al. 1999). Another mechanism by which influenza viruses evade the human immune system is the acquisition of glycosylation sites to mask antigenic epitopes. The HAs from modern H1N1 viruses have up to five glycosylation sites in addition to the four found in all avian HAs. The HA of the 1918 virus has only the four conserved avian sites (Reid et al. 1999).
Influenza virus infection requires binding of the HA protein to sialic acid receptors on the host cell surface. The HA receptor-binding site consists of a subset of amino acids that are invariant in all avian HAs but vary in mammalian-adapted HAs. Human-adapted influenza viruses preferentially bind sialic acid receptors with α(2–6) linkages. Those viral strains adapted to birds preferentially bind α (2–3) linked sugars (Gambaryan et al. 1997; Matrosovich et al. 1997; Weis et al. 1988). To shift from the proposed avian-adapted receptor-binding site configuration (with a preference for α(2–3) sialic acids) to that of swine H1s (which can bind both α(2–3) and α(2–6)) requires only one amino acid change, E190D. The HA sequences of all five 1918 cases have the E190D change (Reid et al. 2003). In fact, the critical amino acids in the receptor-binding site of two of the 1918 cases are identical with that of the A/swine/Iowa/30 HA. The other three 1918 cases have an additional change from the avian consensus, G225D. Since swine viruses with the same receptor site as A/swine/Iowa/30 bind both avian and mammalian-type receptors (Gambaryan et al. 1997), A/New York/1/18 virus probably also had the capacity to bind both. The change at residue 190 may represent the minimal change necessary to allow an avian H1-subtype HA to bind mammalian-type receptors (Gamblin et al. 2004; Reid et al. 1999; Reid et al. 2003; Stevens et al. 2004), a critical step in host adaptation.
The crystal structure analysis of the 1918 HA (Gamblin et al. 2004; Stevens et al. 2004) suggests that the overall structure of the receptor-binding site is akin to that of an avian H5 HA in terms of its having a narrower pocket than that identified for the human H3 HA (Wilson et al. 1981). This provides an additional clue for the avian derivation of the 1918 HA. The four antigenic sites that have been identified for another H1 HA, the A/PR/8/34 virus HA (Caton et al. 1982) also appear to be the major antigenic determinants on the 1918 HA. The X-ray analyses suggest that these sites are exposed on the 1918 HA; thus, they could be readily recognized by the human immune system.
The principal biological role of NA is the cleavage of the terminal sialic acid residues that are receptors for the virus’s HA protein (Palese and Compans 1976). The active site of the enzyme consists of 15 invariant amino acids that are conserved in the 1918 NA. The functional NA protein is configured as a homotetramer in which the active sites are found on a terminal knob carried on a thin stalk (Colman et al. 1983). Some early human virus strains have short (11–16 amino acids) deletions in the stalk region, as do many virus strains isolated from chickens. The 1918 NA has a full-length stalk and has only the glycosylation sites shared by avian N1 virus strains (Schulze 1997). Although the antigenic sites on human-adapted N1 neuraminidases have not been definitively mapped, it is possible to align the N1 sequences with N2 subtype NAs and examine the N2 antigenic sites for evidence of drift in N1. There are 22 amino acids on the N2 protein that may function in antigenic epitopes (Colman et al. 1983). The 1918 NA matches the avian consensus at 21 of these sites (Reid et al. 2000). This finding suggests that the 1918 NA, like the 1918 HA, had not circulated long in humans before the pandemic and very possibly had an avian origin (Reid and Taubenberger 2003).
Neither the 1918 HA nor the 1918 NA genes have obvious genetic features that can be related directly to virulence. Two known mutations that can dramatically affect the virulence of influenza virus strains have been described. For viral activation HA must be cleaved into two pieces, HA1 and HA2, by a host protease (Lazarowitz and Choppin 1975; Rott et al. 1995). Some avian H5 and H7 subtype viruses acquire a mutation that involves the addition of one or more basic amino acids to the cleavage site, allowing HA activation by ubiquitous proteases (Kawaoka and Webster 1988; Webster and Rott 1987). Infection with such a pantropic virus strain can cause systemic disease in birds with high mortality. This mutation was not observed in the 1918 virus (Reid et al. 1999; Taubenberger et al. 1997).
The second mutation with a significant effect on virulence through pantropism has been identified in the NA gene of two mouse-adapted influenza virus strains, A/WSN/33 and A/NWS/33. Mutations at a single codon (N146R or N146Y, leading to the loss of a glycosylation site) appear, like the HA cleavage site mutation, to allow the virus to replicate in many tissues outside the respiratory tract (Li et al. 1993). This mutation was also not observed in the NA of the 1918 virus (Reid et al. 2000).
Therefore, neither surface protein-encoding gene has known mutations that would allow the 1918 virus to become pantropic. Since clinical and pathological findings in 1918 showed no evidence of replication outside the respiratory system (Winternitz et al. 1920; Wolbach 1919), mutations allowing the 1918 virus to replicate systemically would not have been expected. However, the relationship of other structural features of these proteins (aside from their presumed antigenic novelty) to virulence remains unknown. In their overall structural and functional characteristics, the 1918 HA and NA are avian-like, but they also have mammalian-adapted characteristics.
Interestingly, recombinant influenza viruses containing the 1918 HA and NA and up to five additional genes derived from the 1918 virus (the other genes being derived from either the A/WSN/33 virus or a modern human H1N1 virus) were all highly virulent in mice (Tumpey et al. 2004; Tumpey et al. 2005). Furthermore, expression microarray analysis performed on whole lung tissue of mice infected with the 1918 HA/NA recombinant showed increased upregulation of genes involved in apoptosis, tissue injury, and oxidative damage (Kash et al. 2004). These findings were unusual because the viruses with the 1918 genes had not been adapted to mice. On the other hand replacement of only a single gene, the NS gene, in the A/WSN/33 virus background led to a dramatic decrease in the LD50 value for mice, suggesting that the lack of mouse adaptation by the interferon antagonist (coded for by the 1918 NS gene) is associated with a decrease in virulence (Basler et al. 2001). Again, the 1918 NS gene in the context of additional 1918 genes (the HA, NA, NP, and M genes) appears to result in recombinant viruses with high virulence in mice (see above). One explanation is that the combination of the genes/proteins of the 1918 virus was “optimal,” and that this virus—unfortunately for humans and pigs—was a “lucky” winner. As a consequence, viruses with many (or most) genes derived from the 1918 virus are highly pathogenic in different species because the 1918 genes possibly work synergistically in terms of virulence. The completion of the sequence of the entire genome of the 1918 virus (Taubenberger et al. 2005) and the reconstruction and characterization of viruses with 1918 genes under appropriate biosafety conditions (Tumpey et al. 2005) have shed more light on this hypothesis and should allow a definitive examination of this explanation.
Antigenic analysis of recombinant viruses possessing the 1918 HA and NA by hemagglutination inhibition tests using ferret and chicken antisera suggested a close relationship with the A/swine/Iowa/30 virus and H1N1 viruses isolated in the 1930s (Tumpey et al. 2004), further supporting data of Shope from the 1930s (Shope 1936). Interestingly, when mice were immunized with different H1N1 virus strains, challenge studies using the 1918-like viruses revealed partial protection by this treatment, suggesting that current vaccination strategies are adequate against a 1918-like virus (Tumpey et al. 2004). In fact, the data may even allow us to suggest that the human population, having experienced a long period of exposure to H1N1 viruses, may be partially protected against a 1918-like virus (Tumpey et al. 2004).
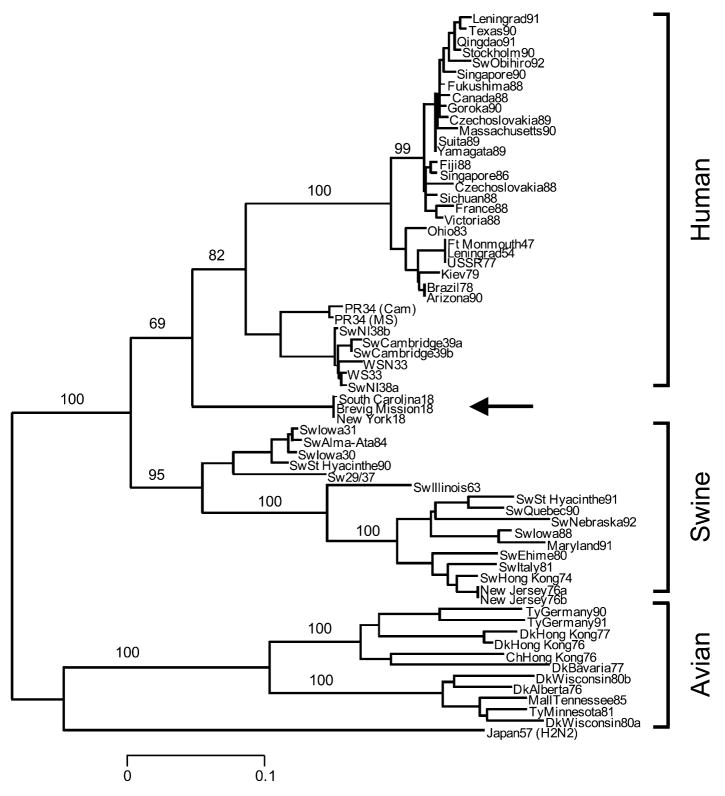
Since virulence (in the immunologically naïve person) has not yet been mapped to particular sequence motifs of the 1918 HA and NA genes, what can gene sequencing tell us about the origin of the 1918 virus? The best approach to analyzing the relationships among influenza viruses is phylogenetics, whereby hypothetical family trees are constructed. These family trees take available sequence data and use them to make assumptions about the ancestral relationships between current and historical influenza virus strains (Fitch et al. 1991; Gammelin et al. 1990; Scholtissek et al. 1993; fig. 5). Since influenza viruses possess eight discrete RNA segments that can move independently between virus strains by the process of reassortment, these evolutionary studies must be performed independently for each gene segment.
Figure 5.
Phylogenetic tree of the influenza virus hemagglutinin gene segment. Amino acid changes in three lineages of the influenza virus hemagglutinin protein segment, HA1. The tree shows the numbers of unambiguous changes between these sequences, with branch lengths being proportional to number of changes.
A comparison of the complete 1918 HA (fig. 5) and NA genes with those of numerous human, swine, and avian sequences demonstrates the following. Phylogenetic analyses based upon HA nucleotide changes (whether total, synonymous, or nonsynonymous) or HA amino acid changes always place the 1918 HA with the mammalian viruses, not with the avian viruses (Reid et al. 1999). In fact, both synonymous and nonsynonymous changes place the 1918 HA in the human clade. Phylogenetic analyses of total or synonymous NA nucleotide changes also place the 1918 NA sequence with the mammalian viruses, but analysis of nonsynonymous changes or amino acid changes places the 1918 NA with the avian viruses (Reid et al. 2000). Since the 1918 HA and NA have avian features and most analyses place HA and NA near the root of the mammalian clade (close to an ancestor of the avian genes), it is likely that both genes emerged from an avian-like influenza reservoir just prior to 1918 (Fanning et al. 2000; Fanning and Taubenberger 1999; Reid et al. 1999; Reid et al. 2000; Reid and Taubenberger 2003; fig. 4). Clearly, by 1918 the virus had acquired enough mammalian-adaptive changes to function as a human pandemic virus and to form a stable lineage in swine.
Sequence and functional analysis of the non-structural gene segment
The complete coding sequence of the 1918 non-structural (NS) segment was completed (Basler et al. 2001). The functions of the two proteins, NS1 and NS2 (NEP), encoded by overlapping reading frames (Lamb and Lai 1980) of the NS segment are still being elucidated (García-Sastre 2002; García-Sastre et al. 1998; Krug et al. 2003; Li et al. 1998; O’Neill et al. 1998). The NS1 protein has been shown to prevent type I interferon (IFN) production, by preventing activation of the latent transcription factors IRF-3 (Talon et al. 2000) and NF-κB (Wang et al. 2000). One of the distinctive clinical characteristics of the 1918 influenza was its ability to produce rapid and extensive damage to both the upper and lower respiratory epithelium (Winternitz et al. 1920). Such a clinical course suggests a virus that replicated to a high titer and spread quickly from cell to cell. Thus, an NS1 protein that was especially effective at blocking the type I IFN system might have contributed to the exceptional virulence of the 1918 virus strain (García-Sastre et al. 1998; Talon et al. 2000; Wang et al. 2000). To address this possibility, transfectant A/WSN/33 influenza viruses were constructed with the 1918 NS1 gene or with the entire 1918 NS segment (coding for both NS1 and NS2 [NEP] proteins; Basler et al. 2001). In both cases, viruses containing 1918 NS genes were attenuated in mice, compared with wild-type A/WSN/33 controls. The attenuation demonstrates that NS1 is critical for the virulence of A/WSN/33 in mice. On the other hand, transcriptional profiling (microarray analysis) of infected human lung epithelial cells showed that a virus with the 1918 NS1 gene was more effective at blocking the expression of IFN-regulated genes than the isogenic parental mouse-adapted A/WSN/33 virus (Geiss et al. 2002), suggesting that the 1918 NS1 contributes virulence characteristics in human cells but not murine ones. The 1918 NS1 protein varies from that of the WSN virus at 10 amino acid positions. The amino acid differences between the 1918 and A/WSN/33 NS segments may be important in the adaptation of the latter virus strain to mice, and likely account for the observed differences in virulence in these experiments. Recently, a single amino acid change (D92E) in the NS1 protein was associated with increased virulence of the 1997 Hong Kong H5N1 viruses in a swine model (Seo et al. 2002). This amino acid change was not found in the 1918 NS1 protein.
Sequence and functional analysis of the matrix gene segment
The coding region of influenza A RNA segment 7 from the 1918 pandemic virus, consisting of the open reading frames of the two matrix genes, M1 and M2, has been sequenced (Reid et al. 2002). While this segment is highly conserved among influenza virus strains, the 1918 sequence does not match any previously sequenced influenza virus strains. The 1918 sequence matches the consensus over the M1 RNA-binding domains and nuclear localization signal and the highly conserved transmembrane domain of M2. Amino acid changes that correlate with high yield and pathogenicity in animal models were not found in the 1918 virus strain.
Influenza A virus RNA segment 7 encodes two proteins, the matrix proteins M1 and M2. The M1 mRNA is colinear with the viral RNA, while the M2 mRNA is encoded by a spliced transcript (Lamb and Krug 2001). The proteins encoded by these mRNAs share their initial nine amino acids and also have a stretch of 14 amino acids in overlapping reading frames. The M1 protein is a highly conserved 252 amino acid protein. It is the most abundant protein in the viral particle, lining the inner layer of the viral membrane and contacting the ribonucleoprotein core. M1 has been shown to have several functions (Lamb and Krug 2001) including regulation of nuclear export of vRNPs, both permitting the transport of vRNP particles into the nucleus upon infection and preventing newly exported vRNP particles from re-entering the nucleus. The 97 amino acid M2 protein is a homotetrameric integral membrane protein that exhibits ion channel activity and is the target of the drug amantadine (Hay et al. 1985). The ion channel activity of M2 is important both during virion uncoating and during viral budding (Lamb and Krug 2001).
In the transmembrane region of the M2 protein, five amino acid sites have been identified that are involved in resistance to the antiviral drug amantadine: sites 26, 27, 30, 31, and 34 (Holsinger et al. 1994). The 1918 influenza M2 sequence is identical at these positions with that of the amantadine-sensitive influenza virus strains. Thus, it was predicted that the M2 protein of the 1918 influenza virus would be sensitive to amantadine. This was recently demonstrated experimentally. A recombinant virus possessing the 1918 matrix segment was inhibited effectively both in tissue culture and in vivo by the M2 ion-channel inhibitors amantadine and rimantadine (Tumpey et al. 2002).
The phylogenetic analyses suggest that the 1918 matrix genes, while more avian-like than those of other mammalian influenza viruses, were mammalian-adapted (Reid et al. 2002). For example, the extra-cellular domain of the M2 protein contains 4 amino acids that differ consistently between the avian and mammalian clades (M2 residues #14, 16, 18, and 20). The 1918 sequence matches the mammalian sequence at all four of these residues (Reid et al. 2002), suggesting that the matrix segment may have been circulating in human virus strains for at least several years before 1918.
Sequence and functional analysis of the nucleoprotein gene segment
The nucleoprotein gene (NP) of the 1918 pandemic influenza A virus has been amplified and sequenced from archival material (Reid et al. 2004). The NP gene is known to be involved in many aspects of viral function and to interact with host proteins, thereby playing a role in host specificity (Portela and Digard 2002). NP is highly conserved, with a maximum amino acid difference of 11 percent among virus strains, probably because it must bind to multiple proteins, both viral and cellular. Numerous studies suggest that NP is a major determinant of host specificity (Scholtissek et al. 1985; Scholtissek et al. 1978a). The 1918 NP amino acid sequence differs at only 6 amino acids from avian consensus sequences, consistent with reassortment from an avian source shortly before 1918. However, the 1918 NP nucleotide sequence has more than 170 differences from avian consensus sequences, suggesting substantial evolutionary distance from known avian sequences. Both the 1918 NP gene and protein sequences fall within the mammalian clade upon phylogenetic analysis.
Phylogenetic analyses of NP sequences from many virus strains result in trees with two main branches, one consisting of mammalian-adapted virus strains and one of avian-adapted virus strains (Gammelin et al. 1990; Gorman et al. 1991; Shu et al. 1993). The NP gene segment was not replaced in the pandemics of 1957 and 1968, so it is likely that the sequences in the mammalian clade are descended from the 1918 NP segment. The mammalian branches, unlike the avian branch, show a slow but steady accumulation of changes over time. Extrapolation of the rate of change along the human branch back to a putative common ancestor suggests that this NP entered the mammalian lineage sometime after 1900 (Gammelin et al. 1990; Gorman et al. 1991; Shu et al. 1993). Separate analyses of synonymous and nonsynonymous substitutions also placed the 1918 virus NP gene in the mammalian clade (Reid et al. 2004). When synonymous substitutions were analyzed, the 1918 virus gene was placed within and near the root of swine viruses. When nonsynonymous viruses were analyzed, the 1918 virus gene was placed within and near the root of the human viruses.
The evolutionary distance of the 1918 NP from avian and mammalian sequences was examined using several different parameters. There are at least three possibilities for the origin of the 1918 NP gene segment (Reid et al. 2004). First, it could have been retained from the previously circulating human virus, as was the case with the 1957 and 1968 pandemic virus strains, whose NP segments are descendants of the 1918 NP. The large number of nucleotide changes from the avian consensus and the placement of the 1918 sequence in the mammalian clade are consistent with this hypothesis. NJ analyses of nonsynonymous nucleotide sequences or of amino acid sequences place the 1918 sequence within and near the root of the human clade. The 1918 NP has only a few amino acid differences from most bird virus strains, but this consistent group of amino acid changes is shared by the 1918 NP and its subsequent mammalian descendants and is not found in any birds, resulting in the 1918 sequence’s being placed outside the avian clade (Reid et al. 2004). One or more of these amino acid substitutions may be important for adaptation of the protein to humans. However, the very small number of amino acid differences from the avian consensus argues for recent introduction from birds—eighty years after 1918, the NP genes of human influenza virus strains have accumulated more than 30 additional amino acid differences from the avian consensus (a rate of 2.3 amino acid changes per year). Thus, it seems unlikely that the 1918 NP, with only 6 amino acid differences from the avian consensus, could have been in humans for many years before 1918. This conclusion is supported by the regression analysis that suggests that the progenitor of the 1918 virus probably entered the human population around 1915 (Reid et al. 2004).
A second possible origin for the 1918 NP segment is direct reassortment from an avian virus. The small number of amino acid differences between 1918 and the avian consensus supports this hypothesis. While 1918 varies at many nucleotides from the nearest avian virus strain, avian virus strains are quite diverse at the nucleotide level. Synonymous/non-synonymous ratios between 1918 and avian virus strains are similar to the ratios between avian virus strains, opening the possibility that avian virus strains may exist that are more closely related to 1918. The great evolutionary distance between the 1918 sequence and the avian consensus suggests that no avian virus strain similar to those in the currently identified clades could have provided the 1918 virus strain with its NP segment.
A final possibility is that the 1918 gene segment was acquired shortly before 1918 from a source not currently represented in the database of influenza sequences. There may be a currently unknown influenza host that, while similar to currently characterized avian virus strains at the amino acid level, is quite different at the nucleotide level. It is possible that such a host was the source of the 1918 NP segment (Reid et al. 2004).
Future Work
All eight RNA segments of the 1918 influenza virus have been sequenced and analyzed. Their characterization has shed light on the origin of the virus and strongly supports the hypothesis that the 1918 virus was the common ancestor of both subsequent human and swine H1N1 lineages. Sequence analysis of the genes to date offers no definitive clue as to the genotypic basis of the exceptional virulence of the 1918 virus strain. Thus, experiments testing models of virulence using reverse genetics approaches with 1918 influenza genes have begun.
It is hoped that in future work the 1918 pandemic virus strain can be placed in the context of influenza virus strains that preceded it and followed it. The direct precursor of the pandemic virus, the first or “spring” wave virus strain, lacked the exceptional virulence of the “fall” wave virus strain. Identification of an influenza RNA-positive case from the first wave would have tremendous value in deciphering the genetic basis for virulence, by allowing differences in the sequences to be highlighted. Identification of pre-1918 human influenza RNA samples would clarify which gene segments were novel in the 1918 virus.
In many respects, the 1918 influenza pandemic was similar to other influenza pandemics. In its epidemiology, disease course, and pathology, the pandemic generally was different in degree but not in kind from previous and subsequent pandemics. Furthermore, laboratory experiments using recombinant influenza viruses containing genes from the 1918 virus suggest that the 1918 and 1918-like viruses would be as sensitive to the FDA-approved anti-influenza drugs rimantadine and oseltamivir as other virus strains (Tumpey et al. 2002).
However, there are some characteristics of the pandemic that appear to be unique. Mortality was exceptionally high, ranging from five to twenty times higher than normal. Clinically and pathologically, the high mortality appears to be the result of a higher proportion of severe and complicated infections of the respiratory tract, not with systemic infection or involvement of organ systems outside the influenza virus’s normal targets. The mortality was concentrated in an unusually young age group. Finally, the waves of influenza activity followed on each other unusually rapidly, resulting in three major outbreaks within a year’s time. Each of these unique characteristics may find its explanation in genetic features of the 1918 virus. The challenge will be in determining the links between the biological capabilities of the virus and the known history of the pandemic.
Acknowledgments
This work has been partially supported by NIH grants to JKT, and by grants from the Veterans Administration and the American Registry of Pathology (JKT). I also thank the following colleagues and their groups for wonderful and stimulating collaborations during the last several years: Chris Basler, Roger Bumgarner, Thomas Fanning, Adolfo García-Sastre, Michael Katze, Peter Palese, David Swayne, Terry Tumpey, Ian Wilson, and especially Ann Reid.
Footnotes
Read 9 November 2002, as part of the symposium “Archaeology: New Techniques and Methods.”
References
- Barry JM. The great influenza: The epic story of the deadliest plague in history. New York: Viking Press; 2004. p. 560. [Google Scholar]
- Basler CF, Reid AH, Dybing JK, Janczewski TA, Fanning TG, Zheng H, Salvatore M, Perdue ML, Swayne DE, García-Sastre A, Palese P, Taubenberger JK. Sequence of the 1918 pandemic influenza virus non-structural gene (NS) segment and characterization of recombinant viruses bearing the 1918 NS genes. Proc Natl Acad Sci U S A. 2001;98:2746–51. doi: 10.1073/pnas.031575198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beveridge W. Influenza: The last great plague, an unfinished story of discovery. New York: Prodist; 1977. [Google Scholar]
- Brown IH, Chakraverty P, Harris PA, Alexander DJ. Disease outbreaks in pigs in Great Britain due to an influenza A virus of H1N2 subtype. Vet Rec. 1995;136:328–29. doi: 10.1136/vr.136.13.328. [DOI] [PubMed] [Google Scholar]
- Brown IH, Harris PA, McCauley JW, Alexander DJ. Multiple genetic reassortment of avian and human influenza A viruses in European pigs, resulting in the emergence of an H1N2 virus of novel genotype. J Gen Virol. 1998;79:2947–55. doi: 10.1099/0022-1317-79-12-2947. [DOI] [PubMed] [Google Scholar]
- Burnet F, Clark E. Influenza: A survey of the last 50 years in the light of modern work on the virus of epidemic influenza. Melbourne: MacMillan; 1942. p. 188. [Google Scholar]
- Castrucci MR, Donatelli I, Sidoli L, Barigazzi G, Kawaoka Y, Webster RG. Genetic reassortment between avian and human influenza A viruses in Italian pigs. Virology. 1993;193:503–06. doi: 10.1006/viro.1993.1155. [DOI] [PubMed] [Google Scholar]
- Caton AJ, Brownlee GG, Yewdell JW, Gerhard W. The antigenic structure of the influenza virus A/PR/8/34 hemagglutinin (H1 subtype) Cell. 1982;31:417–27. doi: 10.1016/0092-8674(82)90135-0. [DOI] [PubMed] [Google Scholar]
- Chun J. Influenza including its infection among pigs. Natl Med J (of China) 1919;5:34–44. [Google Scholar]
- Claas EC, Osterhaus AD, van Beek R, De Jong JC, Rimmelzwaan GF, Senne DA, Krauss S, Shortridge KF, Webster RG. Human influenza A H5N1 virus related to a highly pathogenic avian influenza virus. Lancet. 1998;351:472–77. doi: 10.1016/S0140-6736(97)11212-0. [DOI] [PubMed] [Google Scholar]
- Colman PM, Varghese JN, Laver WG. Structure of the catalytic and antigenic sites in influenza virus neuraminidase. Nature. 1983;303:41–44. doi: 10.1038/303041a0. [DOI] [PubMed] [Google Scholar]
- Cox NJ, Subbarao K. Global epidemiology of influenza: past and present. Annu Rev Med. 2000;51:407–21. doi: 10.1146/annurev.med.51.1.407. [DOI] [PubMed] [Google Scholar]
- Crosby A. America’s forgotten pandemic. Cambridge: Cambridge University Press; 1989. [Google Scholar]
- Dimoch WW. Diseases of swine. J Am Vet Med Assn. 1918–19;54:321–40. [Google Scholar]
- Dorset M, McBryde CN, Niles WB. Remarks on “hog” flu. J Am Vet Med Assn. 1922–23;62:162–71. [Google Scholar]
- Dowdle WR. Influenza A virus recycling revisited. Bull World Health Organ. 1999;77:820–28. [PMC free article] [PubMed] [Google Scholar]
- Fanning TG, Reid AH, Taubenberger JK. Influenza A virus neuraminidase: regions of the protein potentially involved in virus-host interactions. Virology. 2000;276:417–23. doi: 10.1006/viro.2000.0578. [DOI] [PubMed] [Google Scholar]
- Fanning TG, Slemons RD, Reid AH, Janczewski TA, Dean J, Taubenberger JK. 1917 avian influenza virus sequences suggest that the 1918 pandemic virus did not acquire its hemagglutinin directly from birds. J Virol. 2002;76:7860–62. doi: 10.1128/JVI.76.15.7860-7862.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fanning TG, Taubenberger JK. Phylogenetically important regions of the influenza A H1 hemagglutinin protein. Virus Res. 1999;65:33–42. doi: 10.1016/s0168-1702(99)00098-2. [DOI] [PubMed] [Google Scholar]
- Fitch W, Leiter J, Li X, Palese P. Positive Darwinian evolution in human influenza A viruses. Proc Natl Acad Sci U S A. 1991;88:4270–74. doi: 10.1073/pnas.88.10.4270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fouchier RA, Schneeberger PM, Rozendaal FW, Broekman JM, Kemink SA, Munster V, Kuiken T, Rimmelzwaan GF, Schutten M, Van Doornum GJ, Koch G, Bosman A, Koopmans M, Osterhaus AD. Avian influenza A virus (H7N7) associated with human conjunctivitis and a fatal case of acute respiratory distress syndrome. Proc Natl Acad Sci U S A. 2004;101:1356–61. doi: 10.1073/pnas.0308352100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frost W. Statistics of influenza morbidity. Public Health Reports. 1920;35:584–97. [Google Scholar]
- Gambaryan A, Tuzikov A, Piskarev V, Yamnikova S, Lvov D, Robertson J, Bovin N, Matrosovich M. Specification of receptor-binding phenotypes of influenza virus isolates from different hosts using synthetic sialylglyco-polymers: Non-egg-adapted human H1 and H3 influenza A and influenza B viruses share a common high binding affinity for 6′-sialyl(N-acetyllactosamine) Virology. 1997;232:345–50. doi: 10.1006/viro.1997.8572. [DOI] [PubMed] [Google Scholar]
- Gamblin SJ, Haire LF, Russell RJ, Stevens DJ, Xiao B, Ha Y, Vasisht N, Steinhauer DA, Daniels RS, Elliot A, Wiley DC, Skehel JJ. The structure and receptor binding properties of the 1918 influenza hemagglutinin. Science. 2004;303:1838–42. doi: 10.1126/science.1093155. [DOI] [PubMed] [Google Scholar]
- Gammelin M, Altmuller A, Reinhardt U, Mandler J, Harley V, Hudson P, Fitch W, Scholtissek C. Phylogenetic analysis of nucleoproteins suggests that human influenza A viruses emerged from a 19th-century avian ancestor. Mol Biol Evol. 1990;7:194–200. doi: 10.1093/oxfordjournals.molbev.a040594. [DOI] [PubMed] [Google Scholar]
- García-Sastre A. Mechanisms of inhibition of the host interferon alpha/beta-mediated antiviral responses by viruses. Microbes Infect. 2002;4:647–55. doi: 10.1016/s1286-4579(02)01583-6. [DOI] [PubMed] [Google Scholar]
- García-Sastre A, Egorov A, Matassov D, Brandt S, Levy DE, Durbin JE, Palese P, Muster T. Influenza A virus lacking the NS1 gene replicates in interferon-deficient systems. Virology. 1998;252:324–30. doi: 10.1006/viro.1998.9508. [DOI] [PubMed] [Google Scholar]
- Gaydos J, Hodder R, Top FJ, Soden V, Allen R, Bartley J, Zabkar J, Nowosiwsky T, Russell P. Swine influenza A at Fort Dix, New Jersey (January–February 1976). I. Case finding and clinical study of cases. J Infect Dis. 1977;136:S356–62. doi: 10.1093/infdis/136.supplement_3.s356. [DOI] [PubMed] [Google Scholar]
- Geiss GK, Salvatore M, Tumpey TM, Carter VS, Wang X, Basler CF, Taubenberger JK, Bumgarner RE, Palese P, Katze MG, García-Sastre A. Cellular transcriptional profiling in influenza A virus-infected lung epithelial cells: the role of the nonstructural NS1 protein in the evasion of the host innate defense and its potential contribution to pandemic influenza. Proc Natl Acad Sci U S A. 2002;99:10736–41. doi: 10.1073/pnas.112338099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gensheimer KF, Fukuda K, Brammer L, Cox N, Patriarca PA, Strikas RA. Preparing for pandemic influenza: the need for enhanced surveillance. Emerg Infect Dis. 1999;5:297–99. doi: 10.3201/eid0502.990219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorman O, Bean W, Kawaoka Y, Donatelli I, Guo Y, Webster R. Evolution of influenza A virus nucleoprotein genes: implications for the origins of H1N1 human and classical swine viruses. J Virol. 1991;65:3704–14. doi: 10.1128/jvi.65.7.3704-3714.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grove RD, Hetzel AM. Vital statistics rates in the United States: 1940–1960. Washington, DC: US Government Printing Office; 1968. [Google Scholar]
- Hay A, Wolstenholme A, Skehel J, Smith M. The molecular basis of the specific anti-influenza action of amantadine. EMBO. 1985;4:3021–24. doi: 10.1002/j.1460-2075.1985.tb04038.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holsinger LJ, Nichani D, Pinto LH, Lamb RA. Influenza A virus M2 ion channel protein: a structure-function analysis. J Virol. 1994;68:1551–63. doi: 10.1128/jvi.68.3.1551-1563.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson NP, Mueller J. Updating the accounts: global mortality of the 1918–1920 “Spanish” influenza pandemic. Bull Hist Med. 2002;76:105–15. doi: 10.1353/bhm.2002.0022. [DOI] [PubMed] [Google Scholar]
- Jordan E. Epidemic influenza: A survey. Chicago: American Medical Association; 1927. p. 355. [Google Scholar]
- Kanegae Y, Sugita S, Sortridge K, Yoshioka Y, Nerome K. Origin and evolutionary pathways of the H1 hemagglutinin gene of avian, swine and human influenza viruses: Cocirculation of two distinct lineages of swine viruses. Arch Virol. 1994;134:17–28. doi: 10.1007/BF01379103. [DOI] [PubMed] [Google Scholar]
- Kash JC, Basler CF, García-Sastre A, Carter V, Billharz R, Swayne DE, Przygodzki RM, Taubenberger JK, Palese P, Katze MG, Tumpey TM. The global host immune response: Contribution of HA and NA genes from the 1918 Spanish influenza to viral pathogenesis. Journal of Virology. 2004;78:9499–511. doi: 10.1128/JVI.78.17.9499-9511.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz J, Lim W, Bridges C, Rowe T, Hu-Primmer J, Lu X, Abernathy R, Clarke M, Conn L, Kwong H, Lee M, Au G, Ho Y, Mak K, Cox N, Fukuda K. Antibody response in individuals infected with avian influenza A (H5N1) viruses and detection of anti-H5 antibody among household and social contacts. J Infect Dis. 1999;180:1763–70. doi: 10.1086/315137. [DOI] [PubMed] [Google Scholar]
- Kawaoka Y, Krauss S, Webster RG. Avian-to-human transmission of the PB1 gene of influenza A viruses in the 1957 and 1968 pandemics. J Virol. 1989;63:4603–08. doi: 10.1128/jvi.63.11.4603-4608.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawaoka Y, Webster RG. Molecular mechanism of acquisition of virulence in influenza virus in nature. Microb Pathog. 1988;5:311–18. doi: 10.1016/0882-4010(88)90032-0. [DOI] [PubMed] [Google Scholar]
- Kilbourne E. Influenza pandemics in perspective. JAMA. 1977;237:1225–28. [PubMed] [Google Scholar]
- Koen JS. A practical method for field diagnoses of swine diseases. Am J Vet Med. 1919;14:468–70. [Google Scholar]
- Kolata GB. Flu: The story of the great influenza pandemic of 1918 and the search for the virus that caused it. New York: Farrar Straus & Giroux; 1999. [Google Scholar]
- Koopmans M, Wilbrink B, Conyn M, Natrop G, van der Nat H, Vennema H, Meijer A, van Steenbergen J, Fouchier R, Osterhaus A, Bosman A. Transmission of H7N7 avian influenza A virus to human beings during a large outbreak in commercial poultry farms in the Netherlands. Lancet. 2004;363:587–93. doi: 10.1016/S0140-6736(04)15589-X. [DOI] [PubMed] [Google Scholar]
- Krug RM, Yuan W, Noah DL, Latham AG. Intracellular warfare between human influenza viruses and human cells: the roles of the viral NS1 protein. Virology. 2003;309:181–89. doi: 10.1016/s0042-6822(03)00119-3. [DOI] [PubMed] [Google Scholar]
- Kupradinun S, Peanpijit P, Bhodhikosoom C, Yoshioka Y, Endo A, Nerome K. The first isolation of swine H1N1 influenza viruses from pigs in Thailand. Arch Virol. 1991;118:289–97. doi: 10.1007/BF01314040. [DOI] [PubMed] [Google Scholar]
- Lamb R, Krug R. Orthomyxovirdiae: The viruses and their replication. In: Knipe D, Howley P, editors. Fields virology. 4. Philadelphia: Lippincott Williams & Wilkins; 2001. pp. 1487–1531. [Google Scholar]
- Lamb RA, Lai CJ. Sequence of interrupted and uninterrupted mRNAs and cloned DNA coding for the two overlapping nonstructural proteins of influenza virus. Cell. 1980;21:475–85. doi: 10.1016/0092-8674(80)90484-5. [DOI] [PubMed] [Google Scholar]
- Lazarowitz SG, Choppin PW. Enhancement of the infectivity of influenza A and B viruses by proteolytic cleavage of the hemagglutinin polypeptide. Virology. 1975;68:440–54. doi: 10.1016/0042-6822(75)90285-8. [DOI] [PubMed] [Google Scholar]
- LeCount ER. The pathologic anatomy of influenzal bronchopneumonia. JAMA. 1919;72:650–52. [Google Scholar]
- Li S, Schulman J, Itamura S, Palese P. Glycosylation of neuraminidase determines the neurovirulence of influenza A/WSN/33 virus. J Virol. 1993;67:6667–73. doi: 10.1128/jvi.67.11.6667-6673.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Yamakita Y, Krug R. Regulation of a nuclear export signal by an adjacent inhibitory sequence: the effector domain of the influenza virus NS1 protein. Proc Natl Acad Sci U S A. 1998;95:4864–69. doi: 10.1073/pnas.95.9.4864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linder FE, Grove RD. Vital statistics rates in the United States: 1900–1940. Washington, DC: Government Printing Office; 1943. [Google Scholar]
- Lipsman J. H7N2 avian influenza identified in Westchester resident. Westchester County Department of Health; 2004. [Google Scholar]
- Ludendorff E. Meine Kriegserinnerungen 1914–1918. Berlin: Ernst Siegfried Mittler und Sohn Verlagsbuchhandlung; 1919. p. 514. [Google Scholar]
- Ludwig S, Stitz L, Planz O, Van H, Fitch W, Scholtissek C. European swine virus as a possible source for the next influenza pandemic? Virology. 1995;212:551–61. doi: 10.1006/viro.1995.1513. [DOI] [PubMed] [Google Scholar]
- Marks G, Beatty WK. Epidemics. New York: Scribner; 1976. [Google Scholar]
- Matrosovich M, Gambaryan A, Teneberg S, Piskarev V, Yamnikova S, Lvov D, Robertson J, Karlsson K. Avian influenza A viruses differ from human viruses by recognition of sialyloigosaccharides and gangliosides and by a higher conservation of the HA receptor-binding site. Virology. 1997;233:224–34. doi: 10.1006/viro.1997.8580. [DOI] [PubMed] [Google Scholar]
- Ministry of Health, U. K. Reports on public health and medical subjects. London: Ministry of Health; 1960. The influenza epidemic in England and Wales 1957–1958. [Google Scholar]
- Monto AS, Iacuzio DA, La Montaigne JR. Pandemic influenza: Confronting a re-emergent threat. J Infect Dis. 1997;176:S1–3. [PubMed] [Google Scholar]
- Murray C, Biester HE. Swine influenza. J Am Vet Med Assn. 1930;76:349–55. [Google Scholar]
- Nerome K, Ishida M, Oya A, Oda K. The possible origin H1N1 (Hsw1N1) virus in the swine population of Japan and antigenic analysis of the isolates. J Gen Virol. 1982;62:171–75. doi: 10.1099/0022-1317-62-1-171. [DOI] [PubMed] [Google Scholar]
- O’Neill RE, Talon J, Palese P. The influenza virus NEP (NS2 protein) mediates the nuclear export of viral ribonucleoproteins. EMBO J. 1998;17:288–96. doi: 10.1093/emboj/17.1.288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palese P, Compans RW. Inhibition of influenza virus replication in tissue culture by 2-deoxy- 2,3-dehydro-N-trifluoroacetylneuraminic acid (FANA): mechanism of action. J Gen Virol. 1976;33:159–63. doi: 10.1099/0022-1317-33-1-159. [DOI] [PubMed] [Google Scholar]
- Patterson KD, Pyle GF. The geography and mortality of the 1918 influenza pandemic. Bull Hist Med. 1991;65:4–21. [PubMed] [Google Scholar]
- Peiris JS, Yu WC, Leung CW, Cheung CY, Ng WF, Nicholls JM, Ng TK, Chan KH, Lai ST, Lim WL, Yuen KY, Guan Y. Re-emergence of fatal human influenza A subtype H5N1 disease. Lancet. 2004;363:617–19. doi: 10.1016/S0140-6736(04)15595-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philip RN, Lackman DB. Observations on the present distribution of influenza A/swine antibodies among Alaskan natives relative to the occurrence of influenza in 1918–1919. Am J Hygiene. 1962;75:322–34. doi: 10.1093/oxfordjournals.aje.a120253. [DOI] [PubMed] [Google Scholar]
- Portela A, Digard P. The influenza virus nucleoprotein: a multifunctional RNA-binding protein pivotal to virus replication. J Gen Virol. 2002;83:723–34. doi: 10.1099/0022-1317-83-4-723. [DOI] [PubMed] [Google Scholar]
- Reid AH, Fanning TG, Hultin JV, Taubenberger JK. Origin and evolution of the 1918 “Spanish” influenza virus hemagglutinin gene. Proc Natl Acad Sci U S A. 1999;96:1651–56. doi: 10.1073/pnas.96.4.1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid AH, Fanning TG, Janczewski TA, Lourens R, Taubenberger JK. Novel origin of the 1918 pandemic influenza virus nucleoprotein gene segment. J Virol. 2004 doi: 10.1128/JVI.78.22.12462-12470.2004. forthcoming. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid AH, Fanning TG, Janczewski TA, McCall S, Taubenberger JK. Characterization of the 1918 “Spanish” influenza virus matrix gene segment. J Virol. 2002;76:10717–23. doi: 10.1128/JVI.76.21.10717-10723.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid AH, Fanning TG, Janczewski TA, Taubenberger JK. Characterization of the 1918 “Spanish” influenza virus neuraminidase gene. Proc Natl Acad Sci U S A. 2000;97:6785–90. doi: 10.1073/pnas.100140097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid AH, Janczewski TA, Lourens RM, Elliot AJ, Daniels RS, Berry CL, Oxford JS, Taubenberger JK. 1918 Influenza pandemic caused by highly conserved viruses with two receptor-binding variants. Emerg Infect Dis. 2003;9:1249–53. doi: 10.3201/eid0910.020789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid AH, Taubenberger JK. The 1918 flu and other influenza pandemics: “Over there” and back again. Lab Invest. 1999;79:95–101. [PubMed] [Google Scholar]
- Reid AH, Taubenberger JK. The origin of the 1918 pandemic influenza virus: a continuing enigma. J Gen Virol. 2003;84:2285–92. doi: 10.1099/vir.0.19302-0. [DOI] [PubMed] [Google Scholar]
- Rosenau MJ, Last JM. Maxcy-Rosenau preventative medicine and public health. New York: Appleton-Century-Crofts; 1980. [Google Scholar]
- Rott R, Klenk HD, Nagai Y, Tashiro M. Influenza viruses, cell enzymes, and pathogenicity. Am J Respir Crit Care Med. 1995;152:S16–19. doi: 10.1164/ajrccm/152.4_Pt_2.S16. [DOI] [PubMed] [Google Scholar]
- Schafer JR, Kawaoka Y, Bean WJ, Suss J, Senne D, Webster RG. Origin of the pandemic 1957 H2 influenza A virus and the persistence of its possible progenitors in the avian reservoir. Virology. 1993;194:781–88. doi: 10.1006/viro.1993.1319. [DOI] [PubMed] [Google Scholar]
- Scholtissek C. Source for influenza pandemics. Eur J Epidemiol. 1994;10:455–58. doi: 10.1007/BF01719674. [DOI] [PubMed] [Google Scholar]
- Scholtissek C, Burger H, Kistner O, Shortridge KF. The nucleoprotein as a possible major factor in determining host specificity of influenza H3N2 viruses. Virology. 1985;147:287–94. doi: 10.1016/0042-6822(85)90131-x. [DOI] [PubMed] [Google Scholar]
- Scholtissek C, Koennecke I, Rott R. Host range recombinants of fowl plague (influenza A) virus. Virology. 1978a;91:79–85. doi: 10.1016/0042-6822(78)90356-2. [DOI] [PubMed] [Google Scholar]
- Scholtissek C, Ludwig S, Fitch W. Analysis of influenza A virus nucleoproteins for the assessment of molecular genetic mechanisms leading to new phylogenetic virus lineages. Arch Virol. 1993;131:237–50. doi: 10.1007/BF01378629. [DOI] [PubMed] [Google Scholar]
- Scholtissek C, Rohde W, Von Hoyningen V, Rott R. On the origin of the human influenza virus subtypes H2N2 and H3N2. Virology. 1978b;87:13–20. doi: 10.1016/0042-6822(78)90153-8. [DOI] [PubMed] [Google Scholar]
- Schulze IT. Effects of glycosylation on the properties and functions of influenza virus hemagglutinin. J Infect Dis. 1997;176 (Suppl 1):S24–28. doi: 10.1086/514170. [DOI] [PubMed] [Google Scholar]
- Seo SH, Hoffmann E, Webster RG. Lethal H5N1 influenza viruses escape host anti-viral cytokine responses. Nat Med. 2002;8:950–54. doi: 10.1038/nm757. [DOI] [PubMed] [Google Scholar]
- Shope R. Influenza: History, epidemiology, and speculation. Public Health Reports. 1958;73:165–78. [PMC free article] [PubMed] [Google Scholar]
- Shope RE. The incidence of neutralizing antibodies for swine influenza virus in the sera of human beings of different ages. J Exp Med. 1936;63:669–84. doi: 10.1084/jem.63.5.669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shope RE, Lewis PA. Swine influenza. J Exp Med. 1931:54. doi: 10.1084/jem.54.3.361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu L, Bean W, Webster R. Analysis of the evolution and variation of the human influenza A virus nucleoprotein gene from 1933 to 1900. J Virol. 1993;67:2723–29. doi: 10.1128/jvi.67.5.2723-2729.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonsen L, Clarke MJ, Schonberger LB, Arden NH, Cox NJ, Fukuda K. Pandemic versus epidemic influenza mortality: A pattern of changing age distribution. J Infect Dis. 1998;178:53–60. doi: 10.1086/515616. [DOI] [PubMed] [Google Scholar]
- Simonsen L, Fukuda K, Schonberger LB, Cox NJ. The impact of influenza epidemics on hospitalizations. J Infect Dis. 2000;181:831–37. doi: 10.1086/315320. [DOI] [PubMed] [Google Scholar]
- Smith W, Andrewes C, Laidlaw P. A virus obtained from influenza patients. Lancet. 1933;225:66–68. [Google Scholar]
- Stevens J, Corper AL, Basler CF, Taubenberger JK, Palese P, Wilson IA. Structure of the uncleaved human H1 hemagglutinin from the extinct 1918 influenza virus. Science. 2004;303:1866–70. doi: 10.1126/science.1093373. [DOI] [PubMed] [Google Scholar]
- Subbarao K, Klimov A, Katz J, Regnery H, Lim W, Hall H, Perdue M, Swayne D, Bender C, Huang J, Hemphill M, Rowe T, Shaw M, Xu X, Fukuda K, Cox N. Characterization of an avian influenza A (H5N1) virus isolated from a child with a fatal respiratory illness. Science. 1998;279:393–96. doi: 10.1126/science.279.5349.393. [DOI] [PubMed] [Google Scholar]
- Talon J, Horvath CM, Polley R, Basler CF, Muster T, Palese P, García-Sastre A. Activation of interferon regulatory factor 3 is inhibited by the influenza A virus NS1 protein. J Virol. 2000;74:7989–96. doi: 10.1128/jvi.74.17.7989-7996.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taubenberger JK, Reid A, Fanning T. The 1918 influenza virus: A killer comes into view. Virology. 2000;274:241–45. doi: 10.1006/viro.2000.0495. [DOI] [PubMed] [Google Scholar]
- Taubenberger JK, Reid AH, Janczewski TA, Fanning TG. Integrating historical, clinical and molecular genetic data in order to explain the origin and virulence of the 1918 Spanish influenza virus. Philos Trans R Soc Lond B Biol Sci. 2001;356:1829–39. doi: 10.1098/rstb.2001.1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taubenberger JK, Reid AH, Krafft AE, Bijwaard KE, Fanning TG. Initial genetic characterization of the 1918 “Spanish” influenza virus. Science. 1997;275:1793–96. doi: 10.1126/science.275.5307.1793. [DOI] [PubMed] [Google Scholar]
- Taubenberger JK, Reid AH, Lourens RM, Wang R, Jin G, Fanning TG. Characterization of the 1918 influenza virus polymerase genes. Nature. 2005;437:889–93. doi: 10.1038/nature04230. [DOI] [PubMed] [Google Scholar]
- Thompson WW, Shay DK, Weintraub E, Brammer L, Cox N, Anderson LJ, Fukuda K. Mortality associated with influenza and respiratory syncytial virus in the United States. JAMA. 2003;289:179–86. doi: 10.1001/jama.289.2.179. [DOI] [PubMed] [Google Scholar]
- Tran TH, Nguyen TL, Nguyen TD, Luong TS, Pham PM, Nguyen VC, Pham TS, Vo CD, Le TQ, Ngo TT, Dao BK, Le PP, Nguyen TT, Hoang TL, Cao VT, Le TG, Nguyen DT, Le HN, Nguyen KT, Le HS, Le VT, Christiane D, Tran TT, Menno de J, Schultsz C, Cheng P, Lim W, Horby P, Farrar J. Avian influenza A (H5N1) in 10 patients in Vietnam. N Engl J Med. 2004;350:1179–88. doi: 10.1056/NEJMoa040419. [DOI] [PubMed] [Google Scholar]
- Tumpey TM, Basler CF, Aguilar PV, Zeng H, Solorzano A, Swayne DE, Cox NJ, Katz JM, Taubenberger JK, Palese P, García-Sastre A. Characterization of the reconstructed 1918 Spanish influenza pandemic virus. Science. 2005;310:77–80. doi: 10.1126/science.1119392. [DOI] [PubMed] [Google Scholar]
- Tumpey TM, García-Sastre A, Mikulasova A, Taubenberger JK, Swayne DE, Palese P, Basler CF. Existing antivirals are effective against influenza viruses with genes from the 1918 pandemic virus. Proc Natl Acad Sci U S A. 2002;99:13849–54. doi: 10.1073/pnas.212519699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tumpey TM, García-Sastre A, Taubenberger JK, Palese P, Swayne DE, Basler CF. Pathogenicity and immunogenicity of influenza viruses with genes from the 1918 pandemic virus. Proc Natl Acad Sci U S A. 2004;101:3166–71. doi: 10.1073/pnas.0308391100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- United States Department of Commerce. Historical statistics of the United States: Colonial times to 1970. Washington, DC: Government Printing Office; 1976. [Google Scholar]
- WHO. Avian influenza A(H7) human infections in Canada. WHO; 2004. [Google Scholar]
- Wang X, Li M, Zheng H, Muster T, Palese P, Beg AA, García-Sastre A. Influenza A virus NS1 protein prevents activation of NF-kappaB and induction of alpha/beta interferon. J Virol. 2000;74:11566–73. doi: 10.1128/jvi.74.24.11566-11573.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webby RJ, Webster RG. Are we ready for pandemic influenza? Science. 2003;302:1519–22. doi: 10.1126/science.1090350. [DOI] [PubMed] [Google Scholar]
- Webster R, Rott R. Influenza virus A pathogenicity: The pivotal role of hemagglutinin. Cell. 1987;50:665–66. doi: 10.1016/0092-8674(87)90321-7. [DOI] [PubMed] [Google Scholar]
- Webster RG, Bean WJ, Gorman OT, Chambers TM, Kawaoka Y. Evolution and ecology of influenza A viruses. Microbiol Rev. 1992;56:152–79. doi: 10.1128/mr.56.1.152-179.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webster RG, Sharp GB, Claas EC. Interspecies transmission of influenza viruses. Am J Respir Crit Care Med. 1995;152:S25–30. doi: 10.1164/ajrccm/152.4_Pt_2.S25. [DOI] [PubMed] [Google Scholar]
- Weis W, Brown JH, Cusack S, Paulson JC, Skehel JJ, Wiley DC. Structure of the influenza virus haemagglutinin complexed with its receptor, sialic acid. Nature. 1988;333:426–31. doi: 10.1038/333426a0. [DOI] [PubMed] [Google Scholar]
- Wilson IA, Skehel JJ, Wiley DC. Structure of the haemagglutinin membrane glycoprotein of influenza virus at 3 A resolution. Nature. 1981;289:366–73. doi: 10.1038/289366a0. [DOI] [PubMed] [Google Scholar]
- Winternitz MC, Wason IM, McNamara FP. The pathology of influenza. New Haven: Yale University Press; 1920. [Google Scholar]
- Wolbach SB. Comments on the pathology and bacteriology of fatal influenza cases, as observed at Camp Devens, Mass. Johns Hopkins Hospital Bulletin. 1919;30:104. [Google Scholar]
- Woods GT, Schnurrenberger PR, Martin RJ, Tompkins WA. Swine influenza virus in swine and man in Illinois. J Occup Med. 1981;23:263–67. [PubMed] [Google Scholar]
- Zamarin D, Palese P. Influenza virus: Lessons learned. In: Kowalski JB, Morissey JB, editors. International Kilmer Conference Proceedings. Champlain, NY: Polyscience Publications, Inc; 2004. [Google Scholar]
- Zhou NN, Senne DA, Landgraf JS, Swenson SL, Erickson G, Rossow K, Liu L, Yoon KJ, Krauss S, Webster RG. Emergence of H3N2 reassortant influenza A viruses in North American pigs. Vet Microbiol. 2000;74:47–58. doi: 10.1016/s0378-1135(00)00165-6. [DOI] [PubMed] [Google Scholar]