Abstract
The human immune system consists of a large number of T cells capable of recognizing and responding to antigens derived from various sources. The development of peptide-major histocompatibility (p/MHC) tetrameric complexes has enabled the direct detection of these antigen-specific T cells. With the goal of increasing throughput and multiplexing of T cell detection, protein microarrays spotted with defined p/MHC complexes have been reported, but studies have been limited due to the inherent instability and reproducibility of arrays produced via conventional spotted methods. Herein, we report on a platform for the detection of antigen-specific T cells on glass substrates that offers significant advantages over existing surface-bound schemes. In this approach, called “Nucleic Acid Cell Sorting (NACS)”, single-stranded DNA oligomers conjugated site-specifically to p/MHC tetramers are employed to immobilize p/MHC tetramers via hybridization to a complementary-printed substrate. Fully assembled p/MHC arrays are used to detect and enumerate T cells captured from cellular suspensions, including primary human T cells collected from cancer patients. NACS arrays outperform conventional spotted arrays assessed in key criteria such as repeatability and homogeneity. The versatility of employing DNA sequences for cell sorting is exploited to enable the programmed, selective release of target populations of immobilized T cells with restriction endonucleases for downstream analysis. Because of the performance, facile and modular assembly of p/MHC tetramer arrays, NACS holds promise as a versatile platform for multiplexed T cell detection.
Introduction
T cells constitute an important part of the acquired immune system. They recognize a diversity of antigens through the highly variable, hetero-dimeric T cell receptor protein (TCR), with approximately 107 different antigen specificities (1). The initiation of the T cell immune response is triggered by the engagement of the TCR with processed antigenic peptides (e.g. from a bacterial pathogen) that are bound to Major Histocompatibility Complex (p/MHC) molecules presented on the surface of antigen presenting cells (APCs), leading to downstream T cell proliferation and maturation into effector populations. After the pathogen has been cleared, a subset of the activated T cells transition into memory cells, providing the immune system with the capacity for rapid response towards previously encountered pathogens. As a consequence, an individual’s collection of T cells and their antigen specificities, collectively called the T cell repertoire, is an evolving, extensive repository of cellular immune responses against self and foreign antigens. It is of fundamental and therapeutic importance to detect and survey these T cell populations.
The development of soluble p/MHC tetramers for labeling antigen-specific T cells has enabled the direct phenotypic analysis of antigen-specific T cell populations with flow cytometry (2). Conventionally, p/MHC tetramers are prepared by mixing enzymatically biotinylated p/MHC molecules with preparations of streptavidin(SA)-fluorophore conjugates. While p/MHC monomers have low affinities (2,3), their tetramer counterparts exhibit much higher avidity, permitting T cell detection via flow cytometry to become a standard assay. However, because p/MHC tetramer-stained T cell populations are encoded optically (i.e. one unique fluorophore required per p/MHC specificity), the number of antigen-specificities that can be interrogated simultaneously within a population is limited by spectral overlap. In addition, serial flow cytometry detection of distinct antigen-specific T cells is generally restricted by sample size. Efforts to increase the degree of multiplexing generally revolve around polychromatic flow cytometry utilizing quantum dots (4,5). However, cost, sample preparation time, and color compensation complexity also increase correspondingly.
As an alternative to flow cytometry, several groups have reported microarray-based T cell detection schemes, in which collections of p/MHC complexes are printed on a supporting substrate (6–9). A population of cells is applied directly to the p/MHC array where target antigen-specific T cells bind to regions spotted with the cognate p/MHC and are detected optically. Analogous to DNA and protein microarrays, the readout of such assays is dependent on location rather than distinct fluorescent signals, thus potentially increasing the degree of multiplexing.
A factor that needs to be addressed before p/MHC arrays are used for broader studies and applications concerns the reproducibility and robustness of p/MHC arrays produced by spotting onto treated and/or derivatized surfaces. Ideally p/MHC complexes should be immobilized such that their functional conformations are preserved. Analogous protein arrays produced via antibody adsorption to unmodified and derivatized surfaces can suffer from surface induced effects including protein denaturation and protein adsorption in inactive orientations (10–12). To circumvent such problems, customized surfaces and relatively mild chemistries for protein immobilization have been developed (13–18). However, often the surface that meets the demands of the application requires a high level of technical expertise and/or is limited in accessibility (19).
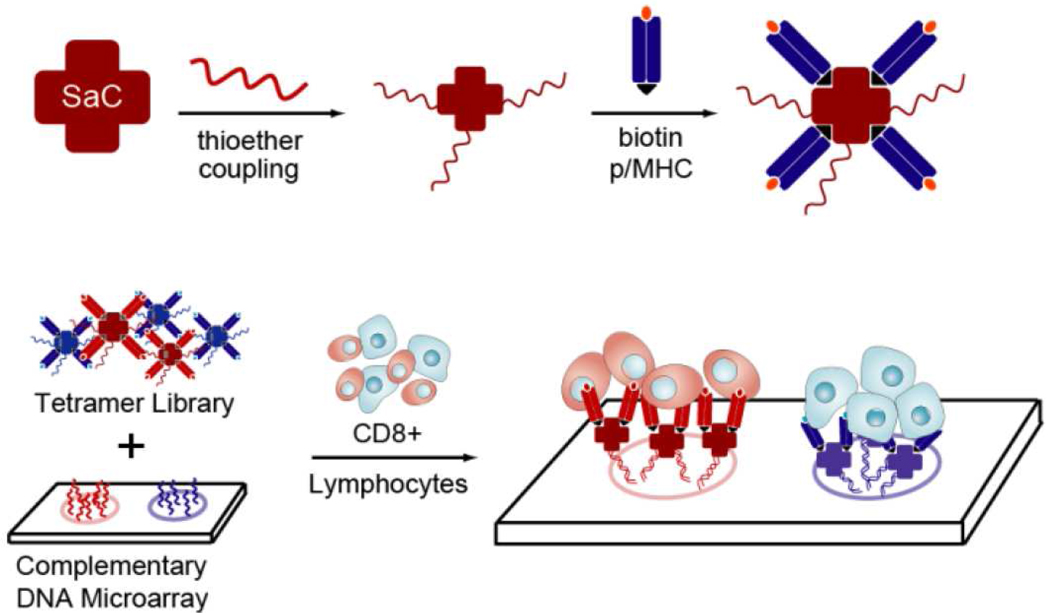
We report here on the method of Nucleic Acid Cell Sorting (NACS), which is based upon the design and application of nucleic acid assembled p/MHC tetramer arrays for multiplexed sorting of antigen-specific T cells. For NACS, p/MHC tetramers of distinct specificities are conjugated to unique sequences of ssDNA in a site-specific fashion. A collection of ssDNA-tagged p/MHC complexes is then self-assembled by DNA hybridization onto a glass slide printed with the complementary DNA sequences. Fully assembled p/MHC tetramer arrays are used to sort mixed populations of antigen-specific T cells (Figure 1). This strategy of using DNA pendants as molecular linkages (20–25) is simple and highly modular. Most importantly, T cell array binding is optimized by utilizing cysteine-engineered streptavidin (SAC) for ssDNA-p/MHC tetramer production, resulting in NACS p/MHC arrays that outperform conventional spotted arrays assessed by performance criteria such as reproducibility and homogeneity. The versatility of using DNA tags is also exploited to enable selective detachment of T cells with restriction endonucleases. Demonstrative experiments regarding NACS sensitivity, multiplexing and limit of detection are performed with cell lines and finally with T cells isolated from cancer patients.
Figure 1. Self-assembled ssDNA-p/MHC tetramer arrays for multiplexed sorting of antigen-specific cells.
ssDNA-tagged p/MHC tetramers are produced by coupling ssDNA site-specifically to SAC prior to exposure to molar excess of biotinylated p/MHC monomers. p/MHC tetramer arrays are formed by pooling ssDNA-p/MHC tetramers of select specificity and hybridization to a complementary printed ssDNA microarray. T cells expressing the cognate TCR are detected by binding to the surface confined tetramer.
RESULTS AND DISCUSSION
Rational design of ssDNA-encoded p/MHC tetramers
The standard scaffold most frequently used to assemble p/MHC monomers into tetramers is a SA-phycobiliprotein (using the protein fluorophores phycoerythrin (PE) or allophycocyanain (APC)) conjugate. SA-phycobiliprotein conjugates are produced via chemical cross-linking, which means that most functional groups are exhausted and/or altered, which prohibits the conjugation of ssDNA. In addition, the attachment of molecular fluorophores to native SA reduces the binding capacity for biotin, an effect attributed to the modification of lysine121 that occurs with amide coupling strategies. This residue is in close proximity to the ligand binding pockets (26, 27). To circumvent this, Altman and co-workers (26) employed a recombinant mutant of SA for fluorescent p/MHC tetramer preparations. This variant incorporates a cysteine residue at the carboxy-terminus (28), a site removed from the biotin binding pocket. The conjugation of cysteine-reactive maleimide derivatives is restricted to the C-terminus because cysteine residues are absent in native SA.
We expressed SAC, coupled the protein with 5’-maleimide ssDNA, and verified the formation of conjugates with mobility shift assays (Figure S1A,B). In parallel, ssDNA was coupled to native SA for direct comparison. To test biotin binding capacity, SAC-oligo conjugates were probed with 2-(4′-Hydroxyazobenzene) benzoic acid (HABA) (29), a molecular mimic of biotin with distinct optical density coefficients dependent on whether biotin is bound to SA or not. A biotin:SA molar ratio of association significantly below 4 in the assay would indicate a reduction in biotin binding capacity. Conjugates derived from native SA were greater than one full unit below the expected value (2.86 versus 4.0), while conjugates formed with SAC maintained near optimal (3.7) binding capacity (Figure S1C). These conjugates were then tested across 4 different monoclonal T cell populations (2 human TCR-transduced cell lines and 2 murine TCR-transgenic splenocyte cell suspensions). ssDNA-tagged SAC constructs had markedly higher cell capture efficiencies compared with p/MHC tetramers prepared with native SA (Figure S2). All subsequent NACS tetramers were prepared with the SAC variant.
Performance of p/MHC arrays produced via DNA immobilization and direct spotting
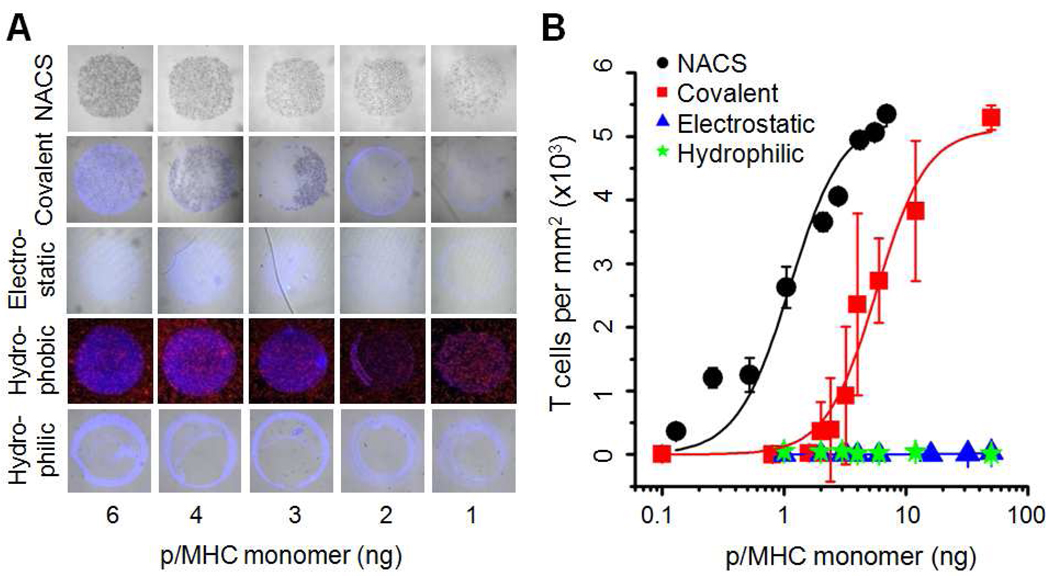
We compared the performance of NACS with conventional spotting strategies on various model substrates. The substrates were selected to represent the spectrum of surface chemistries typically used to immobilize proteins (covalent, electrostatic, hydrophobic, and hydrophilic adsorption). Serial dilutions of a fluorescent p/MHC tetramer, MART-1, were spotted on the substrates according to manufacturer’s instructions. Jurkatα-MART-1 T cells (Jurkat human T cells transduced with the TCR (30) specific for MART-1) were applied to the array and representative images collected (Figure 2A) and quantified (Figure 2B). We observed little to no T cell capture (electrostatic, hydrophilic) or significant noise (hydrophobic) on the majority of the surfaces investigated compared to NACS arrays immobilized with identical concentrations of p/MHC tetramers. T cell binding was observed on one surface (covalent) but cell capture was highly variable as evidenced by both intra-spot and inter-spot heterogeneity and cross experimental variation (Figure S3). Moreover, to achieve equivalent T cell capture densities, NACS p/MHC arrays required >5 times less material than covalent immobilization. (p/MHC monomer at half max ≡ K1/2 = 1.1 ng for NACS and 5.7 ng for covalent immobilization).
Figure 2. Comparison of NACS versus spotted p/MHC arrays.
(A) Brightfield and fluorescent images of Jurkatα-MART-1 T cells captured on various model substrates. (B) Quantification of T cell capture efficiencies (hydrophobic surface was excluded because signal:noise ≤ 1). Each data point was derived from three representative spots.
The performance and reproducibility of NACS p/MHC arrays is markedly improved and represents an integral step towards expanding array-based T cell detection schemes for broader applications. This likely has a few causes. First, surface-tethered ssDNA-p/MHC tetramers may enjoy greater orientational freedom at the surface/solution interface compared with adsorbed proteins which are required to conform to the surface. This effect may increase the density of functional protein and consequently reduce the amount of material required for array production. Second, the hydration state of the environment during the production and subsequent storage of protein arrays is an important factor for array reproducibility (10,13,18). This effect is minimized with NACS because DNA chips can be printed and stored dry for extended periods of time and ssDNA-tagged p/MHC tetramer arrays are self-assembled in solution immediately prior to an experiment.
NACS specificity and limit of detection
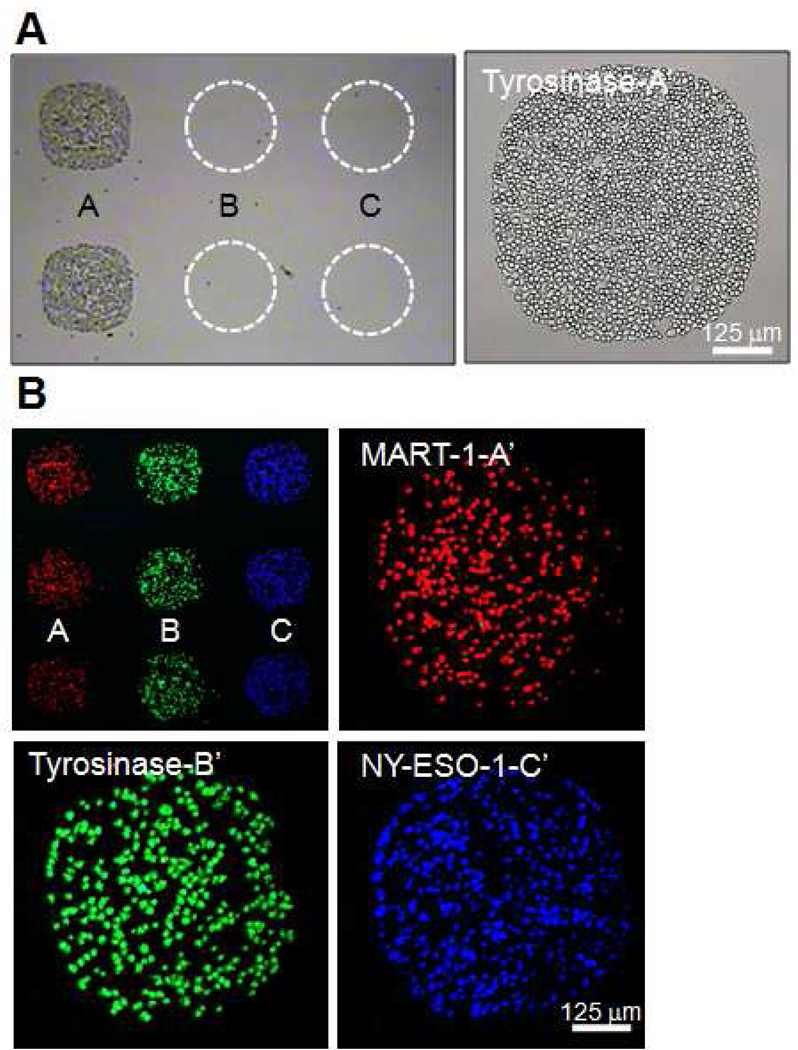
To evaluate the specificity of p/MHC array assembly and T cell sorting, a ssDNA-p/MHC tetramer, tyrosinase, with pendant DNA sequence A’ was hybridized to a DNA microarray printed with the complementary strand (A) along with two additional distinct sequences (designated B and C). A homogeneous population of Jurkatα-Tyr cells (Jurkat cells transduced with a TCR specific for tyrosinase) (31) was then applied to the array. Jurkat α-Tyr T cells localized to the complementary spots (A) containing the hybridized cognate p/MHC but not to spots printed with the non-complementary sequences B and C (Figure 3A). The mean binding capacity calculated from three spots (~600 µm) was ~1486 ± 62 Jurkat α-Tyr T cells.
Figure 3. Mulitplexed nucleic acid cell sorting of antigen-specific T cells.
(A) Tyrosinase p/MHC tetramer conjugated to ssDNA sequence A’ was hybridized to an array printed with DNA complement strand A and non-complement strands B and C (dashed circles). Jurkat α −Tyr cells were localized to spot A only. (B) A 1:1:1 mixture of Jurkatα-a-MART-1 (red), Jurkatα -Tyr (green) and Jurkatα–NY-ESO-1 cells was selectively sorted on an array hybridized with MART-1, tyrosinase, and NY-ESO-1 p/MHC ssDNA-tetramers (top left). The remaining three panels are representative images of spots A, B and C.
To illustrate the multiplexing capability of NACS, MART-1, tyrosinase, and NY-ESO-1 ssDNA-p/MHC tetramers encoded to DNA sequences A, B and C respectively were combined and assembled simultaneously to a three element DNA microarray (strands A, B, and C). A 1:1:1 mixed population of Jurkatα-MART-1, Jurkatα-Tyr and Jurkat α-NY-ESO-1 (Jurkat human T cells transduced with the TCR (32) specific for NY-ESO-1) cells prestained with lipophilic dyes (red, green and blue respectively) was applied to the array and localized into alternating columns (Figure 3B). Minimal cross-reactivity was observed. The average density of spots was about a factor of three less than homogeneous sorting (440 ± 28 T cells/spot).
To determine the limit of detection, target populations of Jurkat α-Tyr cells were spiked in at 10%, 1% and 0.1% into wild type (w.t.) Jurkat cells and sorted (Figure 4A). The T cell capture density per spot per species for each mixture was enumerated and averaged (Figure 4B). The number of non-specific w.t. Jurkat cells that adhered to the array was constant throughout all dilutions while the number of Jurkat α-Tyr T cells captured per spot decreased linearly in relation to the fractional composition of Jurkatα-Tyr cells with a detection limit that was ~ 1 in 1000 cells – a limit that corresponds well to the total number of cells that can be captured per spot. Thus, the sensitivity of this approach is strictly a geometric constraint since antigen-specific T cells that settle on inert areas cannot sample and bind to their cognate p/MHC tetramer. The sensitivity can be improved by increasing the size of the capture region (i.e. increase spot diameter and/or incorporate spot redundancy) or by reducing inert regions (i.e. increase printing density). It should be noted, however, that the sensitivity of this approach cannot be increased without limiting scalability. Since spot diameters are required to be sufficiently large to detect a population at low frequency, this sets an upper bound on the number of distinct spots that can be patterned on a substrate. In the current instance, 600 µm spots are printed in 12 by 12 grids (~1 in2), enabling the potential identification of 144 distinct antigen-specificities ≥ 0.1% from 106 T cells (106 cells is typically required to cover a 1 in2 region). In order to detect target populations below 0.1% without decreasing multiplexing, p/MHC-specific enrichment strategies (e.g. magnetic bead-based schemes (33)) can be implemented prior to cell sorting. Lastly, the recovery of this technique—defined as the total number of antigen-specific T cells captured as a fraction of the number of T cells applied to the array—is low relative to other sorting technologies like FACS and bead-based schemes. Typical T cell recovery for NACS is ≤ 20% (34). In comparison, fluorescent-based sorting techniques like FACS have high recoveries, since each stained T cell can be identified individually by the cytometer and sorted from the null population. The recovery (as well as sensitivity) of NACS will likely be improved by circulating T cells over the p/MHC array with agitation or with integrated microfluidic devices to allow T cells to sample the entire array.
Figure 4. NACS limit of detection.
(A) Jurkatα-Tyr cells (red) were serially diluted in wild type (WT) Jurkat cells (green) at frequencies 50%, 10%, 1% and 0.1% and detected by NACS. (B) The average number of Jurkatα-Tyr cells and WT Jurkat cells per dilution per spot plotted in a histogram.
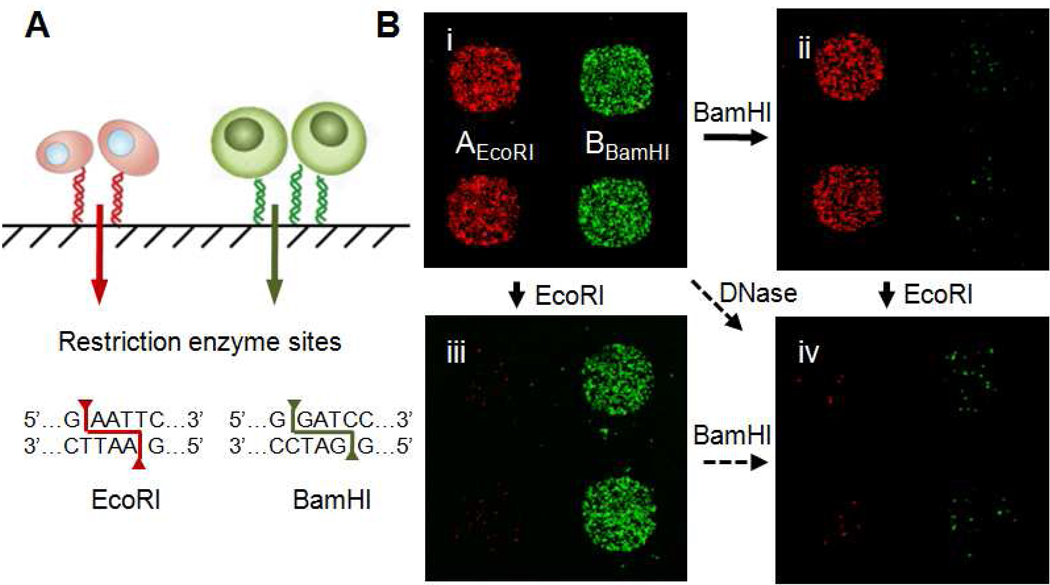
Selective Release of Immobilized T cells with Restriction Endonucleases
Antigen-specific T cells immobilized onto glass are immediately available for secondary assays, since many assays such as immunohistochemistry (IHC), fluorescent in situ hybrization (FISH) and cytokine secretion assays (6,8) are traditionally performed or are compatible with cells localized to a substrate. However, several other relevant assays, such as those designed to assess T cell phenotype or functional status like genomic/mRNA analysis or simply, further culture for phenotypic enrichment would require a method for releasing the captured cells. Any release scheme should ideally be selective for given cell types. For NACS, we explored whether the DNA tethers could be selectively cleaved by exploiting the sequence specificity of restriction endonucleases. We integrated unique restriction sites to each DNA sequence employed for cell sorting, and found that the adhesion of different populations of antigen-specific T cells could, in fact, be independently controlled (Figure 5A). Jurkatα-MART-1 and Jurkat α-Tyr cells prestained with lipophilic dyes (red and green respectivey) were sorted on an array printed with DNA sequences AEcoRI and BBamHI (Figure 5Bi). These oligonucleotides were modified by incorporating 6 bp restriction sites specific for endonucleases EcoRI and BamHI respectively. After T cell immobilization, the array was treated with BamHI which cleaved the BBamHI spots and selectively released the bound Jurkat α-Tyr cells (Figure 5Bii). Conversely, on a separate but identical array, Jurkatα-MART-1 cells were released after treatment with EcoRI (Figure 5Biii). A second round of enzymatic treatment with the complementary endonuclease (EcoRI to state (ii) or BamHI to state (iii)) removed the remaining adherent cells (Figure 5Biv). Alternatively all captured cells (i) could be released non-selectively in a single step with the addition of DNase.
Figure 5. Programmed release of sorted T cells by endonuclease cleavage.
(A) DNA microarrays were printed with orthogonal sequences containing EcoRI and BamHI restriction sites. (B) Fluorescent images of Jurkatα-MART-1 (red) and Jurkatα-Tyro (green) cells captured on p/MHC array (i) and after treatment with BamHI (ii) or EcoRI (iii). Only cells localized to DNA spots containing the target restriction sequence were released. A second round of enzymatic treatment released the remaining bound cells (iv, image representative of BamHI to EcoRI cleavage).
NACS sorting of endogenous primary human T cells
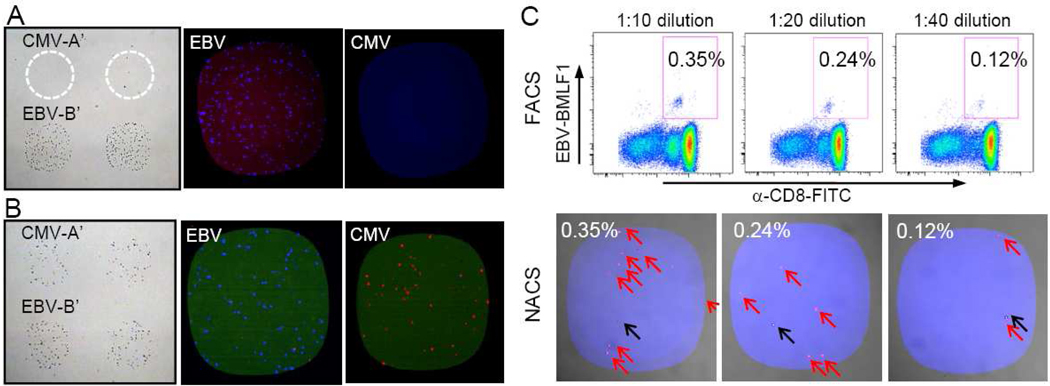
Detection of primary human T cells isolated from peripheral blood is generally more demanding than cultured cell lines because a single population of antigen-specific T cells is present within a large background of differing blood cells and of T cells expressing monoclonal and polyclonal TCRs of diverse specificities. In addition, these T cells would be expressing endogenous levels of TCR. We explored whether the same attributes of NACS that were found in the above examples would apply equally to endogenous primary human T cells. Frozen white blood cell samples from patient NRA13 were CD8+ enriched and applied to a cytomegalovirus (CMV) and Epstein-barr virus (EBV) p/MHC array. T cells were captured only within the EBV regions only (Figure 6A). NRA13 CD8+ cells were verified by flow cytometry to be ~5% EBV-specific and ~0% CMV-specific (Figure S4A). The registry of the array was determined with the addition of A’-cy3 (red) and B’-cy5 (blue) conjugates.
Figure 6. NACS sorting of endogenous primary human T cells specific for Epstein-Barr virus and Cytomegalovirus.
(A) CD8+ T cells from patient NRA 13 were captured on EBV p/MHC spots and no T cells were captured on CMV spots (left panel). The right two panels are representative images after the cells were stained with fluorescent EBV (blue) and CMV p/MHC tetramers (red). (B) T cells detected from a 1:1 mixture of NRA11 and NRA 13 (left panel) were verified to be specific for EBV and CMV (right panels). (C) Mixtures of ~0.4%, 0.2% and 0.1% EBV-specific T cell populations (upper panels) were detected via NACS (bottom panels). Populations of EBV-specific T cells are marked with red arrows and non-specific cells are marked with black arrows.
For multiplexed detection, a 1:1 mixture of EBV-specific and CMV-specific CD8+ T cells was produced by combining NRA13 T cells with CMV-specific T cells from patient NRA11 (Figure S4B). Following cell sorting and fluorescent p/MHC tetramer staining, the populations were complementary stained for the appropriate antigen-specificity (Figure 6B). The detection limit of antigen-specific T cells was evaluated from using serially diluted mixtures of EBV-specific T cells (~0.4%, 0.2%, and 0.1% by FACS) that were probed on an array. Isolated hits were resolved in frequencies as low as ~0.1% (Figure 6C, red arrows). The number of unstained cells within the capture regions (black arrow) was constant throughout all dilutions (~1–2 cells/spot) and likely represents the level of background from non-specific interactions. It should be noted that while we incorporated fluorescent p/MHC tetramer staining after T cell immobilization for illustrative purposes, the specificity of the captured cells could be determined solely from the registry of the array.
Persistence of MART-1 specific, TCR-engineered human T cells in vivo
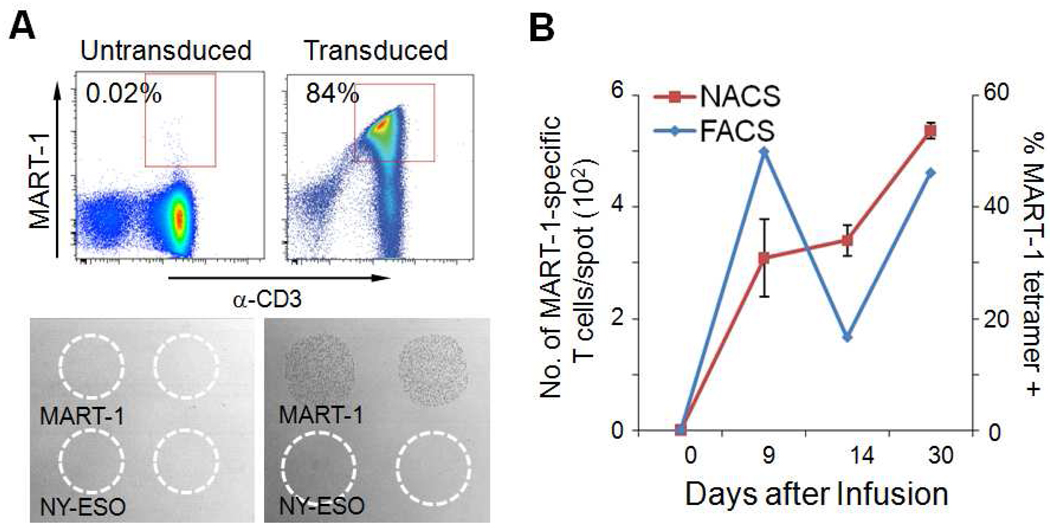
One important potential application of NACS is to utilize the technique for monitoring cancer patients that are undergoing a particular type of immunotherapy that involves TCR engineering of peripheral blood mononuclear cells (PBMCs). This is an emerging clinical approach to rapidly generate large numbers of tumor antigen-specific T cells. In particular, a cancer patient’s T cells are collected, genetically modified to express a TCR specific for a desired cancer antigen, and then they are introduced back into the patient as a cancer therapy. After re-infusion into the patient, the modified T cells can subsequently traffic to and engage with cancer cells, promoting tumor regression in a subset of patients with metastatic cancers. This type of directed cell therapy is utilized primarily in patients with melanoma (skin cancer), but also for other cancers (35, 36). This approach is called adoptive cell transfer (ACT). We monitored the presence and abundance of TCR-engineered T cells in peripheral blood of a patient undergoing ACT. White blood cells were collected from patient F5-1 with metastatic melanoma. After in vitro expansion and TCR transduction, the cells were > 80% specific for the MART-1 p/MHC tetramer (Figure 7A). These cells were subsequently infused into patient F5-1 and the presence and persistence of MART-1-specific T cells in the peripheral blood was monitored by NACS at days 0 (prior to infusion), 9, 14 and 30 (Figure 7B). MART-1-specific T cells were not detectable in pre-infusion samples from the patient but they were detectable at all subsequent time points. This is represented as a gradual rise in the abundance of MART-1-specific T cells until day 30 (left axis). In comparison, the frequency of MART-1-specific T cells detected by flow cytometry spiked initially before decreasing and finally increasing by day 30 (right axis).
Figure 7. Monitoring the presence of infused MART-1-specific, TCR-engineered T cells.
(A) Flow cytometry plots (top panels) and NACS (bottom panels) monitoring the transduction of T cells with a MART-1-specific TCR. (B) Blood cells from patient F5-1 were collected at days 0, 9, 14 and 30 after infusion. The abundance of MART-1-specific T cells was monitored by NACS (left axis) and flow cytometry (right axis).
The abundance of TCR-engineered T cells, as measured by NACS, was generally correlated with parallel measurements using flow cytometry. The prospect of array-based T cell detection schemes for experimental cancer immuno-therapies such as ACT will likely increase as more cancer associated antigens are identified and targeted. For example, more than 50 different melanoma associated antigens have thus far been characterized (37, 38), and this means that more T cells types are likely to be employed for future therapies (36, 39, 40). However, the desire to probe larger sets of cancer associated antigens will be compounded by the amount of sample that can be practically collected from a patient. NACS appears to provide a feasible approach towards carrying out the highly multiplexed cellular measurements that will eventually be required as ACT-like therapies move forward.
CONCLUSION
We have described a method for generating robust and modular p/MHC arrays for high efficiency T cell sorting. The inclusion of a larger set of orthogonal DNA sequences (20,25) will enable the modular assembly of higher order p/MHC arrays for T cell screening experiments (e.g. one working set of DNA sequences can be used interchangeably to generate any combination of p/MHC arrays). This would find immediate utility in the field of TCR peptide epitope discovery where recently, novel antigen peptides were discovered via high-throughput CD8+ screening experiments utilizing multi-color flow cytometry in mice and humans (41, 42) (as many as 2,000 distinct p/MHC tetramers were prepared and tested). NACS arrays have the potential to streamline such experiments. Although traditional methods of producing single p/MHC monomers are time and labor intensive, recent reports using conditional peptide exchange technology enables the relatively straightforward construction of 1000 element p/MHC libraries rapidly (41–43). The integration of NACS with these peptide exchange technologies is a realistic option.
We have demonstrated a number of advantages of the NACS platform. It significantly outperforms literature approaches that utilize surface-bound p/MHC tetramers to capture cells. It is simple and inexpensive to implement since cell sorting is performed on glass substrates prepared via traditional DNA printing technologies. In addition, sorted cells may be selectively released, which should permit for the deployment of a host of bioanalytical methods on NACS sorted cells. We envision that NACS will find uses beyond multiplexed sorting of T cells based on TCR specificity. The principal components of this platform—streptavidin-cysteine core and orthogonal single stranded DNA sequences—were rationally developed to enable oriented coupling and spatial addressing. Thus this platform is amenable to any family of binding proteins or small molecule binders labeled with biotin. The increase in avidity of p/MHC tetramers over monomers as a consequence of the valency of SA should likewise extend to other capture agents, making it feasible to generate cellular arrays with probes ranging from high to moderate affinities like antibodies, aptamers or peptides.
Experimental Procedures
Microarray Fabrication
All DNA strands were purchased from IDT. DNA microarrays were printed by the microarray facility at the Institute for Systems Biology (ISB--Seattle, WA) on amine-coated glass slides (GAPS II, Corning) in identical triplicate 12×12 arrays containing alternative rows of A, B and C spots, or AEcoRI and BBamHI with a SMPXB15 pin (Arrayit). Sequences for all strands can be found at Supporting Information Table 1 online.
Production of ssDNA-SAC conjugates
The pET-3a plasmid containing the SAC gene was a kind gift from Takeshi Sano (Harvard Medical School). The expression of SAC was performed according to previously published protocols (44). Prior to conjugation, stock SAC was buffer exchanged to PBS containing 5mM Tris(2-Carboxyethyl) phosphine Hydrochloride (TCEP) using zeba desalting columns (Pierce). MHPH (3-N-Maleimido-6-hydraziniumpyridine hydrochloride, Solulink) in DMF was added to SAC at a molar excess of 300:1. In parallel, SFB in DMF (succinimidyl 4-formylbenzoate, Solulink) was added in a 40:1 molar excess to 5’aminated oligos. The mixtures were reacted at room temperature (RT) for 3–4 hours before buffer exchanged to citrate (50mM sodium citrate, 150 mM NaCl, pH 6.0) using zeba columns. The SFB-labled oligos were combined in a 20:1 molar excess with the derivatized SAC and incubated overnight at RT. Unreacted oligos were removed using a Pharmacia Superdex 200 gel filtration column at 0.5 ml/min isocratic flow of PBS. Fractions containing the SAC-oligo conjugates were concentrated using 10K mwco concentration filters (Millipore).
Preparation of T cells
The cDNA from the alpha and beta chains of a TCR specific for tyrosinase368–376 was a kind gift from Michael I. Nishimura (Medical University of South Carolina, Charleston, SC). The alpha and beta chains were cloned into a lentiviral vector where both transgenes were linked by a 2A self-cleaving sequence as described (45). Concentrated supernatant from this lentiviral vector was used to infect Jurkat cells to generate Jurkatα-Tyro cells. The MSGV1-F5AfT2AB retroviral vector expressing the F5 MART-1 TCR was a kind gift from Steven A. Rosenberg and Richard Morgan (Surgery Branch, National Cancer Institute Bethesda, MD). The MSGV1-F5AfT2AB retroviral supernatant was used to infect Jurkat cells to generate the Jurkatα-MART-1 cell line. The Jurkatα-NY-ESO-1 cell line was a generous gift from Robert Prins (UCLA). To generate primary human T cell cultures expressing the F5 MART-1 TCR, PBMCs obtained from leukapheresis were activated for 48 hours with 50 ng/ml of OKT3 (muromonab anti-human CD3 antibody, Ortho-Biotech, Bridgewater, NJ) and 300 U/ml of IL-2 (adesleukin, Novartis, Emeryville, CA). MSGV1-F5AfT2AB retrovirus supernatant was applied to retronectin-coated wells (Takara Bio Inc., Japan). Then activated PBMC in RPMI plus 5% human AB serum supplemented by 300 IU of IL-2 were added to these wells and incubated at 37°C overnight at 5% CO2. On the following day, PBMC are transferred to a second set of pre-coated retronectin retroviral vector tissue culture plates and incubated at 37°C overnight at 5% CO2. Cells were subsequently washed and re-suspended in culture media described above. Frozen leukapheresis fractions from patients NRA11, NRA 13 (UCLA IRB#03–12–023 ) and F5-1 (UCLA IRB #08-02–020-02A) were thawed and incubated overnight in RPMI supplemented with 10% human AB serum and 1% penicillin, streptomycin, and amphotericin (Omega Scientific). F5-1 cells were used immediately following incubation. NRA11 and NRA13 samples were CD8+ enriched (anti-CD8 microbeads, Miltenyi Biotech) using an AutoMACS machine according to the manufacturer’s instructions. Following separation, the cells were kept at in RPMI-humanAB media containing 30 U IL2/mL.
Cell Sorting Methods
The HLA-A*0201 restricted MHC class I monomers loaded with tyrosinase369–377 (YMDGTMSQV) and MART-126-35 (ELAGIGILTV), and NY-ESO-1157–165 (SLLMWITQC) were produced in house according to previous published protocols (46). HLA-A*0201-restricted EBV BMLF1259–267 (GLCTLVAML), CMV pp65495–503 (NLVPMVATV), murine H-2Kb/-OVA257–264 (SIINFEKL), and murine H-2Db/-gp10025–33 (KVPRNQDWL) as well as all fluorescent HLA-A*0201 tetramers were purchased from Beckman Coulter. Lipophilic cell membrane staining dyes DiO, DiD, and DiL were purchased from Invitrogen.
Prior to experiments, microarray slides were blocked to prevent non-specific cell binding with 1 mg/ml PEG-NHS ester (Sunbio) in PBS for 2 hours at RT. Four-fold molar excess of p/MHC monomers were combined with ssDNA-SAC at 37°C for 20 min. ssDNA-p/MHC tetramers were hybridized to DNA arrays for 1 hour at 37°C in 200 µl media and rinsed with 3% FBS in PBS. T cells (106 /100 µl media) were incubated on the array at 37°C for 30 min. The arrays were rinsed with 3% FBS in PBS and cell capture visualized via brightfield (Nikon Eclipse TE2000) and/or confocal microscropy (Nikon E800). Post T cell capture p/MHC tetramer staining was done by incubating the array with 200 µl of media containing fluorescent p/MHC tetramer along with fluorescent cDNA (Cy5-A’ and/or Cy3-B’). The arrays were rinsed with 3% FBS in PBS prior to imaging. For selective T cell release experiments, three identical arrays were used to immobilize cells. Treatment with EcoRI, BamHI, or DNase was in RPMI media for 1–2 hours at 37°C. DNase was purchased from Sigma, all other enzymes from NEbiolabs.
For p/MHC comparative studies, SuperEpoxy and SuperProtein (representing covalent and hydrophobic surfaces respectively) were purchased from Arrayit (Sunnyvale, CA). Amine GAPS II slides (electrostatic) were purchased from Corning. Polycarboxylate hydrogel (hydrophilic) slides were purchased from XanTec (Germany). Fluorescent MART-1 tetramers were printed according to manufacturer’s instructions for each slide. Cell sorting images were quantified with ImageJ (NIH) and fitted to the Hill Function (NACS n=2, R2=0.95, Covalent n=2.1, R2 =0.97) with Origin (OriginLab, MA).
Supplementary Material
Acknowledgements
The authors thank Takeshi Sano at Harvard Medical School for providing the pET-3a–SAC plasmid, John Altman at Emory University for providing an initial aliquot of SAC for trial experiments. We also thank Bruz Marzolf at the Institute for Systems Biology for printing the DNA microarrays, Mireille Riedinger at UCLA for assistance with SAC expression, Lilah Morris M.D. at UCLA for assistance with primary murine cell purifications. K.H. is supported by Samsung Fellowship. O.N.W. is an Investigator of the Howard Hughes Medical Institute. This work was primarily funded by the National Cancer Institute Grant No. 5U54 CA119347 (J.R.H., P.I.). Flow cytometry was performed in the UCLA Jonsson Comprehensive Cancer Center and Center for AIDS Research Flow Cytometry Core Facility that is supported by the National Institutes of Health Awards CA-16042 and AI_28697, by the Jonsson Cancer Center, the UCLA AIDS Institute and the UCLA School of Medicine.
References
- 1.Arstila TP, Casrouge A, Baron V, Even J, Kanellopoulos J, Kourilsky P. Science. 1999;286:958–961. doi: 10.1126/science.286.5441.958. [DOI] [PubMed] [Google Scholar]
- 2.Altman JD, Moss PAH, Goulder PJR, Barouch DH, McHeyzer-Williams MG, Bell JI, McMichael AJ, Davis MM. Science. 1996;274:94–96. [PubMed] [Google Scholar]
- 3.Matsui K, Boniface JJ, Steffner P, Reay PA, Davis MM. Proc. Natl. Acad. Sci. USA. 1994;91:12862–12866. doi: 10.1073/pnas.91.26.12862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McLaughlin BE, Baumgarth N, Bigos M, Roederer M, DeRosa SC, Altman JD, Nixon DF, Ottinger J, Oxford C, Evans TG, Asmuth DM. Cytometry A. 2008;73:400–4010. doi: 10.1002/cyto.a.20555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chattopadhyay PK, Price DA, Harper TF, Betts MR, Yu J, Gostick E, Perfetto SP, Goepfert P, Koup RA, De Rosa SC, Bruchez MP, Roederer M. Nat. Med. 2006;12:972–977. doi: 10.1038/nm1371. [DOI] [PubMed] [Google Scholar]
- 6.Chen DS, Soen Y, Stuge TB, Lee PP, Weber JS, Brown PO, Davis MM. PLoS Med. 2005;2:1018–1030. doi: 10.1371/journal.pmed.0020265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Soen Y, Chen DS, Kraft DL, Davis MM, Brown PO. PLoS Biol. 2003;1:429–438. doi: 10.1371/journal.pbio.0000065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stone JD, Demkowicz WE, Stern LJ. Proc. Natl. Acad. Sci. USA. 2005;102:3744–3749. doi: 10.1073/pnas.0407019102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Deviren G, Gupta K, Paulaitis ME, Schneck JP. J. Mol. Recognit. 2007;20:32–38. doi: 10.1002/jmr.805. [DOI] [PubMed] [Google Scholar]
- 10.Haab BB, Dunham MJ, Brown PO. Genome Biology. 2001;2:1–13. doi: 10.1186/gb-2001-2-2-research0004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Butler JE, Ni L, Brown WR, Joshi KS, Chang J, Rosenberg B, Voss EW., Jr Mol Immunol. 1993;30:1165–1175. doi: 10.1016/0161-5890(93)90135-x. [DOI] [PubMed] [Google Scholar]
- 12.Butler JE, Ni L, Nessler R, Joshi KS, Suter M, Rosenberg B, Chang J, Brown WR, CantareroButler LA. J Immunol Methods. 1992;150:77–90. doi: 10.1016/0022-1759(92)90066-3. [DOI] [PubMed] [Google Scholar]
- 13.MacBeath G, Schreiber SL. Science. 2000;289:1760–1763. doi: 10.1126/science.289.5485.1760. [DOI] [PubMed] [Google Scholar]
- 14.Lesaicherre ML, Lue YPR, Chen GYJ, Zhu Q, Yao Q. J. Am. Chem. Soc. 2002;124:8786. doi: 10.1021/ja0265963. [DOI] [PubMed] [Google Scholar]
- 15.Peluso P, Wilson D, Do D, Tran H, Venkatasubbaiah M, Quincy D, Heidecker B, Poindexter K, Tolani N, Phelan M, Witte K, Jung L, Wagner P, Nock S. Anal. Biochem. 2003;312:113–124. doi: 10.1016/s0003-2697(02)00442-6. [DOI] [PubMed] [Google Scholar]
- 16.Kwon Y, Han Z, Karatan E, Mrksich M, Kay BK. Anal. Chem. 2004;76:5713–5720. doi: 10.1021/ac049731y. [DOI] [PubMed] [Google Scholar]
- 17.Arenkov P, Kukhtin A, Gemmel A, Voloshchuk S, Chupeeva V, Mirzabekov A. Anal. Biochem. 2000;278:123–131. doi: 10.1006/abio.1999.4363. [DOI] [PubMed] [Google Scholar]
- 18.Kiyonaka S, Sada K, Yoshimura I, Shinkai S, Kato N, Hamachi I. Nat. Mater. 2004;3:58–64. doi: 10.1038/nmat1034. [DOI] [PubMed] [Google Scholar]
- 19.For example, the polyacrylamide surface employed by Davis and co-workers6,7 has been discontinued from the manufacturer (Perkin Elmer)
- 20.Bailey RC, Kwong GA, Radu CG, Witte ON, Heath JR. J Am. Chem. Soc. 2007;129:1959–1967. doi: 10.1021/ja065930i. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Boozer C, Ladd J, Chen SF, Jiang ST. Anal. Chem. 2006;78:1515–1519. doi: 10.1021/ac051923l. [DOI] [PubMed] [Google Scholar]
- 22.Niemeyer CM. Nano Today. 2007;2:42–52. [Google Scholar]
- 23.Chandra RA, Douglas ES, Mathies RA, Bertozzi CR, Francis MB. Angew. Chem. Int. Engl. 2006;45:896–901. doi: 10.1002/anie.200502421. [DOI] [PubMed] [Google Scholar]
- 24.Hsiao SC, Crow AK, Lam WA, Bertozzi CR, Fletcher DA, Francis MB. Angew. Chem.Int. Engl. 2008;47:8473–8477. doi: 10.1002/anie.200802525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fan R, Vermesh O, Srivastava A, Yen BKH, Qin L, Ahmad H, Kwong GA, Liu CC, Gould J, Hood L, Heath JR. Nat. Biotechnol. 2008;26:1373–1378. doi: 10.1038/nbt.1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ramachandiran V, Grigoriev V, Lan L, Ravkov E, Mertens SA, Altman JD. J. Immunol. Methods. 2007;319:13–20. doi: 10.1016/j.jim.2006.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cameron TO, Cochran JR, Yassine-Diab B, Sekaly RP, Stern LJ. J. Immunol. 2001;166:741–745. doi: 10.4049/jimmunol.166.2.741. [DOI] [PubMed] [Google Scholar]
- 28.Reznik GO, Vajda S, Cantor CR, Sano T. Bioconj. Chem. 2001;12:1000–1004. doi: 10.1021/bc015507t. [DOI] [PubMed] [Google Scholar]
- 29.Green NM. Methods Enzymol. 1970;18:418–424. [Google Scholar]
- 30.Johnson LA, Heemskerk B, Powell DJ, Cohen CJ, Morgan RA, Dudley ME, Robbins PF, Rosenberg SA. J. Immunol. 2006;177:6548–6559. doi: 10.4049/jimmunol.177.9.6548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nishimura MI, Avichezer D, Custer MC, Lee CS, Chen C, Parkhurst MR, Diamond RA, Robbins PF, Schwartzentruber DJ, Rosenberg SA. Cancer Res. 1999;59:6230–6238. [PubMed] [Google Scholar]
- 32.Wargo JA, Robbins PF, Li Y, Zhao Y, El-Gamil M, Caragacianu D, Zheng Z, Hong JA, Downey S, Schrump DS, Rosenberg SA, Morgan RA. Cancer Immunol. Immunother. 2009;58:383–394. doi: 10.1007/s00262-008-0562-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Moon JJ, Chu HH, Pepper M, McSorley SJ, Jameson SC, Kedl RM, Jenkins MK. Immunity. 2007;27:203–213. doi: 10.1016/j.immuni.2007.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.For a typical homogeneous sorting experiment, 106 T cells are applied to an array containing 144 spots. Each spot can bind to ~1486 ± 62 T cells. The recovery, R, is calculated as R = 100% x (144 × 1486)/106 ≈ 20%. The recovery is valid even with heterogeneous samples (e.g. 1:1 mixture of two antigen-specific T cell populations) when the entire array is used to sort a single target population (R = 100% x (144 spots x 661)/(5×105) ≈ 20%). For multiplexed experiments, R is proportional to the fraction of the 144 spots dedicated for cell capture. For example, sorting 2 populations from a 1:1 mixture, R = 100% x (72 spots x 661)/(5×105) ≈ 10%
- 35.Schumacher TNNatRev. Immunol. 2002;2:512–519. doi: 10.1038/nri841. [DOI] [PubMed] [Google Scholar]
- 36.Morgan RA, Dudley ME, Wunderlich JR, Hughes MS, Yang JC, Sherry RM, Royal RE, Topalian SL, Kammula US, Restifo NP, et al. Science. 2006;314:126–129. doi: 10.1126/science.1129003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rosenberg SA. Immunity. 1999;10:281–287. doi: 10.1016/s1074-7613(00)80028-x. [DOI] [PubMed] [Google Scholar]
- 38.Rosenberg SA. Nature. 2001;411:380–384. doi: 10.1038/35077246. [DOI] [PubMed] [Google Scholar]
- 39.Dudley ME, et al. Science. 2002;298:850–854. doi: 10.1126/science.1076514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hunder NN, Wallen H, Cao J, Hendricks DW, Reilly JZ, Rodmyre R, Jungbluth A, Gnjatic S, Thompson JA, Yee CN. Engl. J. Med. 2008;358:2698–2703. doi: 10.1056/NEJMoa0800251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Toebes M, Coccoris M, Bins A, Rodenko B, Gomez R, Nieuwkoop NJ, van de Kasteele, W.; Rimmelzwaan GF, Haanen J, Ovaa H, Schumacher TN. Nat. Med. 2006;12:246–251. doi: 10.1038/nm1360. [DOI] [PubMed] [Google Scholar]
- 42.Grotenbreg GM, Roan NR, Guillen E, Meijers R, Wang J, Bell GW, Starnbach MN, Ploegh HL. Proc. Natl. Acad. Sci. USA. 2008;105:3831–3836. doi: 10.1073/pnas.0711504105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bakker AH, Hoppes R, Linnemann C, Toebes M, Rodenko B, Berkers CR, Hadrup SR, van Esch WJ, Heemskerk MH, Ovaa H, Schumacher TN. Proc. Natl. Acad. Sci. USA. 2008;105:3825–3830. doi: 10.1073/pnas.0709717105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sano T, Cantor CR. Proc. Natl. Acad. Sci. USA. 1990;87:142–146. doi: 10.1073/pnas.87.1.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Szymczak AL, Workman CJ, Wang Y, Vignali KM, Dilioglou S, Vanin EF, Vignali DAA. Nat. Biotechnol. 2004;22:589–594. doi: 10.1038/nbt957. [DOI] [PubMed] [Google Scholar]
- 46.Garboczi DN, Hung DT, Wiley DC. Proc. Natl. Acad. Sci. USA. 1992;89:3429–3433. doi: 10.1073/pnas.89.8.3429. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.