The crystal structure of the Z-isomer of 2,4-diamino-5-[2-(2′-methoxyphenyl)-propenyl]-furo[2,3-d]pyrimidine shows an unusual packing arrangement in which the conserved active site Arg70 forms a salt bridge to the side chain of Glu44 from a symmetry-related molecule.
Keywords: human dihydrofolate reductase; 2,4-diaminofuro[2,3-d]pyrimidine antifolate
Abstract
The crystal structure of the ternary complex of human dihydrofolate reductase (hDHFR) with NADPH and the Z isomer of 2,4-diamino-5-[2-(2′-methoxyphenyl)propenyl]-furo[2,3-d]pyrimidine (Z1) shows that the Z isomer binds in the normal antifolate orientation in which the furo oxygen occupies the 8-amino position observed in the binding of 2,4-diaminopteridine antifolates such as methotrexate and with the methoxyphenyl moiety cis to and coplanar with the furo[2,3-d]pyrimidine ring. The hDHFR ternary complex crystallized in the orthorhombic space group P212121 and its structure was refined to 1.7 Å resolution. Although other hDHFR complexes crystallize in this space group, these data provide only the second example of an unusual packing arrangement in which the conserved active-site Arg70 forms a salt bridge to the side chain of Glu44 from a symmetry-related molecule. As a result, the conformations of Phe31 and Gln35 shift with respect to those observed in the structure of mouse DHFR bound to Z1, which crystallizes in the monoclinic space group P21 and shows that Gln35 interacts with Arg70.
1. Introduction
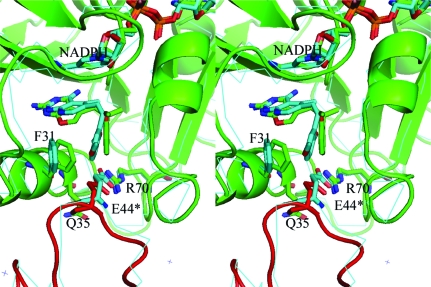
Recent studies have shown that 2,4-diamino-5-substituted furo[2,3-d]pyrimidines are moderately active inhibitors of human dihydrofolate reductase (hDHFR), while also having inhibitory potency against several receptor tyrosine kinases (Gangjee et al., 2005 ▶). These data revealed that a 2:1 mixture of the E and Z isomers of 2,4-diamino-5-[2-(2′-naphthyl)propenyl]furo[2,3-d]pyrimidine (Fig. 1 ▶; Z/E-32) had potent inhibitory activity against human and Toxoplasma gondii DHFR and also possessed potent anti-angeogenic activity. It was shown that an unsaturated C8–C9 bridge was necessary for receptor kinase activity and that the E isomer of a 2′-methoxyphenyl analogue (E1; Fig. 1 ▶) also had significant dual-function inhibitory activity; when separated, there was a ninefold difference in the activity against hDHFR of the E and Z isomers (IC50 values of 16.4 and 1.9 µM, respectively). Structure–activity correlations indicated that the nature of the side-chain aromatic substituent plays an important role in the combined inhibitory activity of this series (Gangjee et al., 2009 ▶). Structure–activity data have also been reported for 2,4-diamino pyrido[2,3-d]pyrimidines substituted with either a quinolyamino (Fig. 1 ▶; LIH) or the Z isomer of a dimethoxyphenyl ethen-1-yl substituent (Fig. 1 ▶; LII), which are potent inhibitors of rat liver DHFR (IC50 values of 7.0 and 12.3 nM, respectively; Piper et al., 1996 ▶; Klon et al., 2002 ▶).
Figure 1.
Schematic representation of the 2,4-diamino-furo-[2,3-d]pyrimidines, pyrrolopyrimidines and pyridopyrimidines under study. The E isomer is defined as having the 2′-methoxyphenyl ring trans to the furopyrimidine ring, while the Z isomer is defined as having these groups cis to each other.
Modeling studies of the binding of the E and Z isomers of the 2′-naphthyl furopyrimidine (Fig. 1 ▶; Z/E-32) in the active site of human DHFR (hDHFR) revealed that the Z isomer fitted while the E isomer could not easily be accommodated unless the 2,4-diaminofuropyrimidine ring was ‘flipped’ such that Glu30 interacted with the 2-amino and N3 of the pyrimidine ring and the furo O atom occupied the N4 amino position (Gangjee et al., 2005 ▶). Structural data for the E and Z isomers of 2,4-diamino-5-[2-(2′-methoxyphenyl)propenyl]-furo[2,3-d]pyrimidines (E1 and Z1, respectively; Fig. 1 ▶) complexed with mouse DHFR (mDHFR) revealed that as predicted the binding orientation of E1 was indeed ‘flipped’ such that the 4-amino group bound in the position of N8 of the antifolate methotrexate (MTX; Gangjee et al., 2009 ▶). Other examples of such ‘flipped’ binding have been observed in the previously reported crystal complexes of two pyrrolo[2,3-d]pyrimidines (GPB and TMP-2; Fig. 1 ▶; Gangjee et al., 2000 ▶; Kuyper et al., 1996 ▶), in which the pyrrolo N7 occupies the 4-amino position in the active site. On the other hand, complexes of human and Pneumocystis carinii DHFR with the furo[2,3-d]pyrimidines MOT and MOT-S (Fig. 1 ▶; Gangjee et al., 1998 ▶; Cody, Galitsky, Luft, Pangborn, Gangjee et al., 1997 ▶; Cody et al., 1998 ▶) revealed the normal 2,4-diamino binding orientation.
To verify the binding of the E/Z isomers of 2,4-diamino-5-[2-(2′-methoxyphenyl)propenyl]-furo[2,3-d]pyrimidines with hDHFR, we cocrystallized a ternary complex of Z1 with hDHFR and NADPH and report the final refinement and analysis of this structure and compare it with the Z1 isomer in the mDHFR complex (Gangjee et al., 2009 ▶).
2. Methods
2.1. Crystallization of wild-type hDHFR
Recombinant hDHFR was expressed and purified as described previously (Cody et al., 2009 ▶). The protein was washed in a Centricon-10 with 100 mM K2HPO4 buffer pH 6.9 and concentrated to 8.2 mg ml−1. The protein was incubated with a 10:1 molar excess of NADPH and the inhibitor Z1 for 1 h over ice prior to crystallization using the hanging-drop vapor-diffusion method. Droplets containing 8 µl protein in 100 mM K2HPO4 pH 6.9 with 30% saturated ammonium sulfate were suspended over a reservoir solution consisting of 100 mM K2HPO4 pH 6.9 and 60% saturated ammonium sulfate with 3%(v/v) ethanol. Crystals grew over several days and were orthorhombic, space group P212121, with diffraction to 1.4 Å resolution. Data were collected on beamline 9-2 at the Stanford Synchrotron Radiation Laboratory using the remote-access system (McPhillips et al., 2002 ▶; Cohen et al., 2002 ▶; González et al., 2008 ▶). Data were processed to 1.7 Å resolution owing to a loss of intensity at higher resolution using both DENZO (Otwinowski & Minor, 1997 ▶) and MOSFLM (Collaborative Computational Project, Number 4, 1994 ▶). Diffraction statistics are shown in Table 1 ▶.
Table 1. Crystal and refinement parameters for the Z isomer of 2,4-diamino-5-[2-(2′-methoxyphenyl)propenyl]-furo[2,3-d]pyrimidine as a ternary complex with human DHFR and NADPH.
Values in parentheses are for the highest resolution shell.
| PDB code | 3gyf |
| Space group | P212121 |
| Unit-cell parameters (Å) | |
| a | 39.91 |
| b | 57.48 |
| c | 74.97 |
| Beamline | SSRL 9-2 |
| Resolution (Å) | 45.64–1.70 (1.79–1.70) |
| Wavelength (Å) | 0.979 |
| Rmerge† (%) | 0.06 (0.22) |
| Completeness (%) | 97.2 (93.9) |
| Observed reflections | 196156 (25778) |
| Unique reflections | 19108 (2656) |
| I/σ(I) | 22.2 (9.0) |
| Multiplicity | 10.3 (9.7) |
| Refinement and model quality | |
| Resolution range (Å) | 31.40–1.70 (1.70–1.79) |
| No. of reflections | 18089 (1310) |
| R factor‡ | 21.3 (26.0) |
| Rfree factor§ | 26.4 (29.3) |
| Total protein atoms | 1626 |
| Total water atoms | 126 |
| Average B factor (Å2) | 24.4 |
| R.m.s. deviation from ideal | |
| Bond lengths (Å) | 0.023 |
| Bond angles (°) | 2.03 |
| E.s.d. from Luzzati plot (Å) | 0.22 |
| Ramachandran plot | |
| Most favored regions (%) | 95.7 |
| Additional allowed regions (%) | 2.7 |
| Generously allowed regions (%) | 1.6 |
| Disallowed regions (%) | 0.0 |
R
merge = 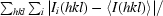
 , where 〈I(hkl)〉 is the mean intensity of a set of equivalent reflections.
, where 〈I(hkl)〉 is the mean intensity of a set of equivalent reflections.
R factor = 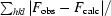
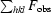 , where F
obs and F
calc are observed and calculated structure-factor amplitudes.
, where F
obs and F
calc are observed and calculated structure-factor amplitudes.
The R free factor was calculated as for the R factor but for a random 5% subset of all reflections.
2.2. Structure determination
The structure was solved by the molecular-replacement method using the coordinates of human DHFR (PDB code 1u72; Cody et al., 2005 ▶) in the program MOLREP (Collaborative Computational Project, Number 4, 1994 ▶). Inspection of the resulting difference electron-density maps was made using the program Coot (Emsley & Cowtan, 2004 ▶) running on a Mac G5 workstation and revealed density for a ternary complex (Fig. 2 ▶). To monitor the refinement, a random subset of all reflections was set aside for the calculation of R free (5%). The template for the inhibitor Z1 was created based on the structure of MOT (Cody et al., 1997 ▶) using the builder function in SYBYL (Tripos, St Louis, Missouri, USA) and the parameter file for the inhibitor was prepared using the Dundee PRODGR2 server website (http://davapc1.bioch.dundee.ac.uk/programs/prodrg; Schüttelkopf & van Aalten, 2004 ▶). The final cycles of refinement were carried out using the program REFMAC5 from the CCP4 suite of programs (Collaborative Computational Project, Number 4, 1994 ▶). The Ramachandran conformational parameters from the last cycle of refinement generated by PROCHECK (Laskowski et al., 1993 ▶) showed that 95.7% of the residues in the Z1–hDHFR complex have the most favored conformation and none are in the disallowed regions. Coordinates for this structure have been deposited in the Protein Data Bank (PDB code 3gyf).
Figure 2.
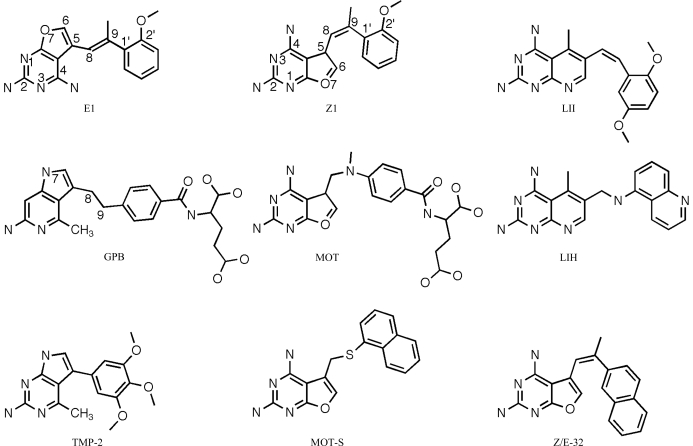
Stereoview of the 2F o − F c difference electron-density map (1.2σ) for the ternary complex of hDHFR with Z1 and NADPH, highlighting the interactions of Glu44 from a symmetry-related molecule (green) with Arg70 in the active site of hDHFR. The red X is a water.
3. Results and discussion
Compared with other reports of human DHFR complexes (Cody & Schwalbe, 2006 ▶), there are no significant changes in the overall structure of the ternary complex of hDHFR with NADPH and Z1. As illustrated in Table 2 ▶, the conformational parameters for Z1 bound to hDHFR are similar to those reported for Z1 bound to mDHFR (Gangjee et al., 2009 ▶). In both structures, the furo[2,3-d]pyrimidine ring in the Z1 isomer binds such that the 2-amino group and N1 interact via hydrogen bonds to Glu30, similar to the binding observed for the classical furo[2,3-d]pyrimidine analogue MOT (Cody, Galitsky, Luft, Pangborn, Gangjee et al., 1997 ▶; Cody et al., 1998 ▶) and the nonclassical furo[2,3-d]pyrimidine analogue MOT-S (Fig. 1 ▶; Gangjee et al., 1998 ▶). The 4-amino group of Z1 makes a number of hydrogen-bonding interactions with the backbone functional groups of Ile7, Val115, Tyr121 and the nicotinamide of NADPH. These contacts are similar to those observed for methotrexate (Cody et al., 2005 ▶) and contribute towards the tight binding of antifolates. The Z isomer Z1 places the 2-methoxyphenyl ring cis to the furo[2,3-d]pyrimidine ring. In this orientation, the 9-methyl group makes van der Waals contact with the methyl group of Thr56 (3.5 Å), while the closest contact for the 2′-methoxy methyl group is with the Cβ atom of Ser59 (4.1 Å).
Table 2. Torsion angles (°) for selected analogues bound to DHFR and shown in Fig. 1 ▶ .
| Torsion angle | Z1 (human) | Z1 (mouse) | E1 (mouse) | MOT | GPB | LII |
|---|---|---|---|---|---|---|
| C6—C5—C8—C9 | 27.0 | 8.3 | −16.2 | 22.2 | −39.9 | 51.3 |
| C5—C8—C9—C1′ | −8.3 | −9.7 | −171.5 | 51.9 | −91.1 | 4.1 |
| C8—C9—C1′—C2′ | −88.2 | −88.1 | −82.6 | −152.0 | −96.2 | 48.5 |
| C5—C8—C9—CH3 | 168.0 | 176.4 | 10.8 | −102.5 | — | — |
| CH3—C9—C1′—C2′ | 96.8 | 86.1 | 95.2 | −3.1 | — | — |
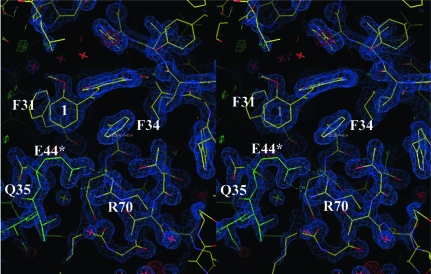
The binding of classical folates such as methotrexate or folate to DHFR shows that the α-carboxylate forms a salt bridge with the conserved Arg70 in the active site (Cody et al., 2005 ▶; Cody & Schwalbe, 2006 ▶). However, the packing arrangement observed for the hDHFR ternary complex with Z1 is such that Glu44 of a symmetry-related molecule forms a salt bridge with the conserved Arg70 in the active site (Fig. 2 ▶). The conformations of Phe31 and Gln35 shift relative to those observed in the structure of the mDHFR–Z1 ternary complex to accommodate the insertion of the carboxylate of Glu44 from the symmetry-related molecule (Fig. 3 ▶; Gangjee et al., 2009 ▶).
Figure 3.
Stereo comparison of the crystal structures of hDHFR in complex with NADPH and Z1 (green) and of mDHFR in complex with NADPH and Z1 (yellow; Gangjee et al., 2009 ▶). The side chains of residues Glu30, Phe31, Gln35, Glu44* and Arg70 are shown. The interactions of Glu44 (dotted van der Waals surface) from a symmetry-related molecule (red) is shown, illustrating how Glu44 fits into the active-site pocket and interacts with the conserved Arg70 of hDHFR in the orthorhombic P212121 lattice. This figure was produced using PyMOL (DeLano, 2002 ▶).
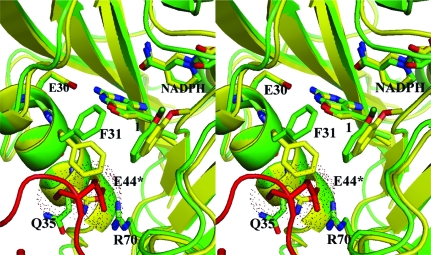
A search of the Protein Data Bank (10 March 2009) reveals that of the 155 entries for DHFR, 25 are for human DHFR. Of the five that crystallize in orthorhombic space groups, only three belong to space group P212121. Of these, two have similar unit-cell parameters to those reported here and are of complexes with LII and LIH (Fig. 1 ▶) that also show the same active-site binding from a symmetry-related Glu44 (Klon et al., 2002 ▶). As illustrated in Fig. 4 ▶, there is little change in the packing environment of hDHFR in the orthorhombic lattices for the Z1 and LII (Klon et al., 2002 ▶) ternary complexes, despite the differences in unit-cell parameters (a = 39.91, b = 57.48, c = 74.97 Å and a = 41.30, b = 54.99, c = 82.68 Å for Z1 and LII, respectively). The other orthorhombic P212121 lattice reported for a hDHF–inhibitor complex has significantly different unit-cell parameters that result in packing orientations of DHFR that do not place the loop containing Glu44 from a symmetry-related molecule in proximity to the active-site pocket (PDB code 1ohk; Cody, Galitsky, Luft, Pangborn, Rosowsky et al., 1997 ▶).
Figure 4.
Stereo comparison of the packing environment between hDHFR–Z1–NADPH (green, with red symmetry-related molecule) and hDHFR–LII (cyan, thin lines), showing the interaction of Glu44* (red, Z1; cyan, LII) from a symmetry-related molecule that binds in the active site of hDHFR. Also shown are residues Phe31, Glu44*, Gln35, Arg70 and NADPH. This figure was produced using PyMOL (DeLano, 2002 ▶).
The unusual packing interactions for these orthorhombic crystal complexes of hDHFR do not appear to be dependent upon crystallization conditions, as those reported by Klon et al. (2002 ▶) (24–33% PEG 4000, 0.2 M LiSO4 and 0.1 M Tris–HCl pH 7.9–8.4, 5% glycerol) differ significantly from those reported here (30% ammonium sulfate, 3% ethanol, 0.1 M K2HPO4 pH 6.9), which usually produce crystals with an H3 lattice (Cody & Schwalbe, 2006 ▶). These observations would suggest that these two crystal forms are iso-energetic as they can arise from the same crystallization solutions. Also, there was little change in the packing interactions between the DHFR molecules despite the differences in the resolution of the data reported (1.05 versus 1.70 Å; Fig. 4 ▶).
The orientation of the 2′-methoxyphenyl ring in the hDHFR–Z1 complex occupies the same conformational space in the active site as observed in mDHFR complexes of the E and Z isomers of the 9-methyl and ethyl analogues (Gangjee et al., 2009 ▶). Modeling data show that if either E1 or Z1 were to bind with the alternate furo[2,3-d]pyrimidine orientation then the 2′-methoxyphenyl ring would make unfavorable contacts to residues Thr56, Ile60 and Leu67 in the DHFR active site. Biochemical data reveal that the Z isomer is ninefold more potent than the E isomer. The results of these structural analyses suggest that features of the DHFR active site can preferentially influence the binding orientation of the E and Z isomers of these substituted furo[2,3-d]pyrimidines. To further explore the factors that influence diaminopyrimidine-ring flipping in the binding of furo[2,3-d]pyrimidine analogues to DHFR, structural studies are currently under way for a series of mixed E/Z isomers of C9-substituted furo[2,3-d]pyrimidines.
Supplementary Material
PDB reference: hDHFR–NADPH–Z1 complex, 3gyf, r3gyfsf
Acknowledgments
This work was supported in part by grants from the National Institutes of Health: GM51670 (VC), AI069966(AG) and CA09885 (AG).
References
- Cody, V., Galitsky, N., Luft, J. R., Pangborn, W., Blakley, R. L. & Gangjee, A. (1998). Anticancer Drug Des.13, 307–315. [PubMed]
- Cody, V., Galitsky, N., Luft, J. R., Pangborn, W., Gangjee, A., Devraj, R., Queener, S. F. & Blakley, R. L. (1997). Acta Cryst. D53, 638–649. [DOI] [PubMed]
- Cody, V., Galisky, N., Luft, J. R., Pangborn, W., Rosowsky, A. & Blakley, R. L. (1997). Biochemistry, 36, 13897–13903. [DOI] [PubMed]
- Cody, V., Luft, J. R. & Pangborn, W. (2005). Acta Cryst. D61, 147–155. [DOI] [PubMed]
- Cody, V., Pace, J., Makin, J., Piraino, J., Queener, S. F. & Rosowsky, A. (2009). Biochemistry, 48, 1702–1711. [DOI] [PMC free article] [PubMed]
- Cody, V. & Schwalbe, C. H. (2006). Crystallogr. Rev.12, 301–333.
- Cohen, A. E., Ellis, P. J., Miller, M. D., Deacon, A. M. & Phizackerley, R. P. (2002). J. Appl. Cryst.35, 720–726. [DOI] [PMC free article] [PubMed]
- Collaborative Computational Project, Number 4 (1994). Acta Cryst. D50, 760–763.
- DeLano, W. L. (2002). The PyMOL Molecular Graphics System. DeLano Scientific LLC, San Carlos, California, USA.
- Emsley, P. & Cowtan, K. (2004). Acta Cryst. D60, 2126–2132. [DOI] [PubMed]
- Gangjee, A., Guo, X., Queener, S. F., Cody, V., Galitsky, N., Luft, J. R. & Pangborn, W. (1998). J. Med. Chem.41, 1263–1271. [DOI] [PubMed]
- Gangjee, A., Li, W., Lin, L., Zeng, Y., Ihnat, M., Warnke, L. A., Green, D. W., Cody, V., Pace, J. & Queener, S. F. (2009). Submitted. [DOI] [PMC free article] [PubMed]
- Gangjee, A., Yu, J., McGuire, J. J., Cody, V., Galitsky, N., Kisliuk, R. L. & Queener, S. F. (2000). J. Med. Chem.43, 3837–3851. [DOI] [PubMed]
- Gangjee, A., Zeng, Y., Ihnat, M., Warnke, L. A., Green, D. W., Kisliuk, R. L. & Lin, F.-T. (2005). Bioorg. Med. Chem.13, 5475–5491. [DOI] [PubMed]
- González, A., Moorhead, P., McPhillips, S. E., Song, J., Sharp, K., Taylor, J. R., Adams, P. D., Sauter, N. K. & Soltis, S. M. (2008). J. Appl. Cryst.41, 176–184.
- Klon, A. E., Heroux, A., Ross, L. J., Pathak, V., Johnson, C. A., Piper, J. R. & Borhani, D. W. (2002). J. Mol. Biol.320, 677–693. [DOI] [PubMed]
- Kuyper, L. F., Garvey, J. M., Baccanari, D. P., Champness, J. N., Stammers, D. K. & Beddell, C. R. (1996). Bioorg. Med. Chem.4, 593–602. [DOI] [PubMed]
- Laskowski, R. A., MacArthur, M. W., Moss, D. S. & Thornton, J. M. (1993). J. Appl. Cryst.26, 283–291.
- McPhillips, T. M., McPhillips, S. E., Chiu, H.-J., Cohen, A. E., Deacon, A. M., Ellis, P. J., Garman, E., Gonzalez, A., Sauter, N. K., Phizackerley, R. P., Soltis, S. M. & Kuhn, P. (2002). J. Synchrotron Rad.9, 401–406. [DOI] [PubMed]
- Otwinowski, Z. & Minor, W. (1997). Methods Enzymol.276, 306–327. [DOI] [PubMed]
- Piper, J. R., Johnson, C. A., Krauth, C. A., Carter, R. L., Hosmer, C. A., Queener, S. F., Borotz, S. E. & Pfefferkorn, E. R. (1996). J. Med. Chem.39, 1271–1280. [DOI] [PubMed]
- Schüttelkopf, A. W. & van Aalten, D. M. F. (2004). Acta Cryst. D60, 1355–1363. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PDB reference: hDHFR–NADPH–Z1 complex, 3gyf, r3gyfsf
PDB reference: hDHFR–NADPH–Z1 complex, 3gyf, r3gyfsf