Expression, purification, crystallization, and preliminary X-ray diffraction analysis of the DNA-binding effector domain from an atypical OmpR response regulator homolog, ChxR, encoded by Chlamydia trachomatis are reported.
Keywords: ChxR, DNA-binding domains, OmpR/PhoB subfamily, Chlamydia trachomatis
Abstract
Two-component signal transduction systems in bacteria are a primary mechanism for responding to environmental stimuli and adjusting gene expression accordingly. Generally in these systems a sensor kinase phosphorylates a response regulator that regulates transcription. Response regulators contain two domains: a receiver domain and an effector domain. The receiver domain is typically phosphorylated and as a result facilitates the DNA-binding and transcriptional activity of the effector domain. The OmpR/PhoB subfamily is the largest of the response-regulator subfamilies and is primarily defined by the winged helix–turn–helix DNA-binding motif within the effector domain. The overall structure of effector domains is highly conserved and contains three defined elements that are critical for transcriptional regulation: a DNA major-groove binding helix, a DNA minor-groove binding wing and a transcriptional activation loop. These functional elements are often diverse in sequence and conformation and reflect the functional differences observed between individual subfamily members. ChxR from Chlamydia trachomatis is an atypical OmpR/PhoB response regulator homolog that has transcriptional activity in the absence of phosphorylation. To facilitate the precise identification of the functional elements of the ChxR effector domain, this protein was cloned, expressed, purified and crystallized. Crystals were obtained from two separate mother liquors, producing two morphologically distinct crystals. The space group of both crystals was P43212 (or its enantiomorph P41212) with isomorphous unit-cell parameters; the crystals diffracted to 2.2–2.5 Å resolution.
1. Introduction
Bacteria use two-component signal transduction systems as a primary means of responding to stimuli within their environment and adjusting gene expression accordingly (Hoch, 2000 ▶). These systems regulate a wide variety of processes, including bacterial development, virulence and physiology (Stock et al., 2000 ▶; Jones, 2005 ▶). Two-component signal transduction systems are predominantly comprised of a membrane-bound histidine (sensor) kinase and an associated response regulator. Frequently, the sensor kinase undergoes an autophosphorylation event in response to environmental stimuli. The phosphate group is then transferred to a highly conserved active site within a cognate response regulator. Phosphorylation of the response regulator controls the activity of the response regulator, which is frequently transcriptional regulation.
OmpR/PhoB is one of the largest subfamilies of response regulators (Baikalov et al., 1996 ▶). The function of the OmpR/PhoB subfamily of response regulators appears to be controlled by interactions between the two domains: the receiver domain and the effector domain. The receiver domain typically becomes phosphorylated at a highly conserved active site that promotes homodimerization through a conserved receiver-domain interface. As a result of the receiver domain becoming phosphorylated, the affinity of the effector domain for DNA becomes enhanced (Kenney, 2002 ▶). It is the winged helix–turn–helix motif in the effector domain that largely defines the OmpR/PhoB subfamily of response regulators (Brennan, 1993 ▶).
Chlamydia trachomatis is a medically important obligate intracellular bacterium that encodes an OmpR/PhoB-subfamily homolog termed ChxR. It has previously been demonstrated that ChxR is an atypical OmpR/PhoB response regulator that does not require phosphorylation for transcriptional activity (Koo et al., 2006 ▶). Despite this fundamental difference from OmpR/PhoB response regulators, we hypothesize that the DNA-binding effector domain of ChxR (ChxREff) is in a structural conformation similar to those of other OmpR/PhoB response regulators. This is supported by structural analysis of other atypical response regulators (Hong et al., 2007 ▶). In addition to demonstrating overall structural similarity, determination of the three-dimensional structure of ChxREff is expected to reveal the residues within and the orientation of the functional regions (β6–β7 DNA minor-groove binding wing, α3 DNA major-groove binding helix and α2–α3 transactivation loop). Here, we report the production, purification, crystallization and preliminary X-ray analysis of ChxREff.
2. Methods and materials
2.1. Expression and purification
The coding region for ChxREff (residues 115–227) was PCR-amplified using genomic DNA from C. trachomatis serovar L2/434/Bu. The primers used for amplification were 5′-GGAATTCCATATGCATTCTGTTCCGGAAAGTT-3′ (forward, containing a 5′ NdeI site) and 5′-CCGCTCGAGCTAAGAAAGCTTTGTATCTTGTTG-3′ (reverse, containing an XhoI site) (Integrated DNA Technologies). The PCR product was digested with NdeI/XhoI and ligated into a similarly digested pET29b vector (Novagen). The resultant plasmid encodes ChxREff with a C-terminal histidine tag. The chxR Eff plasmid was transformed into Escherichia coli BL21 (DE3) (Invitrogen). The protein was expressed in cells grown in LB medium containing 50 µg ml−1 kanamycin at 310 K with shaking at 250 rev min−1. Protein expression was induced at an optical density of 0.7 by the addition of 1 mM isopropyl β-d-1-thiogalactopyranoside. The cells were harvested by centrifugation (4000g, 20 min, 277 K) after overnight incubation at 288 K and 250 rev min−1.
The E. coli cells were resuspended in 50 mM Tris–HCl pH 7, 400 mM NaCl, disrupted by sonication and subjected to centrifugation (14 000g, 30 min, 277 K) to remove cell debris. The lysate was further clarified by passing the supernatant through a 0.22 µm filter before protein purification. The cell extract was applied onto a Talon Metal (Co2+) Affinity Column (Clontech) according to the manufacturer’s instructions. The column was washed with 5 mM imidazole, 50 mM Tris–HCl pH 7, 400 mM NaCl. ChxREff was eluted with the above buffer supplemented with 250 mM imidazole. Peak fractions were pooled and further purified by gel-filtration chromatography.
The pooled ChxREff protein (containing a C-terminal six-histidine tag) mixture was applied onto a Sephacryl S-300 16/60 gel-filtration column (GE Healthcare) pre-equilibrated in 50 mM Tris–HCl pH 7, 400 mM NaCl. Fractions containing ChxREff were pooled and concentrated to 8.4 mg ml−1 using an Amicon (Millipore) 3000 molecular-weight cutoff centrifugal filtration device. The purified protein was determined to be >95% pure by Coomassie staining after SDS–PAGE (Fig. 1 ▶).
Figure 1.
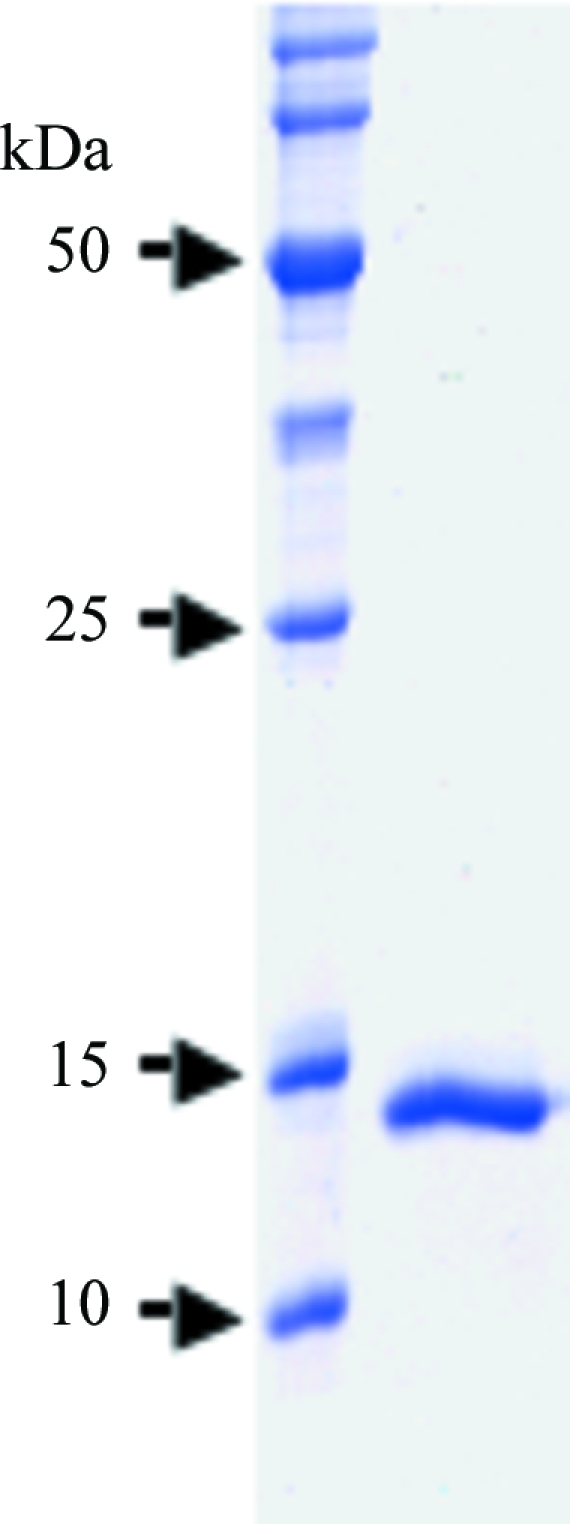
SDS–PAGE gel showing protein purity. 100 µg purified ChxREff was subjected to SDS–PAGE and visualized by Coomassie staining.
2.2. Crystallization
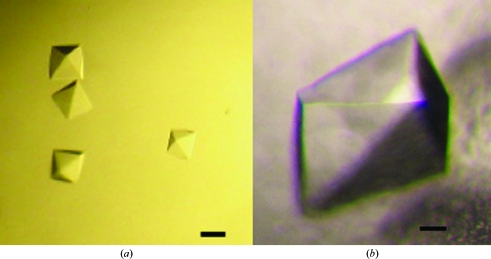
Crystallization was carried out at room temperature by the hanging-drop vapor-diffusion method using 1 µl 8.4 mg ml−1 ChxREff mixed with 1 µl of commercially available sparse-matrix screens (Hampton Research). The protein crystallized readily in a variety of conditions with multiple morphologies after incubation for approximately three months. Two of the crystallization conditions were optimized: (A) 0.2 M ammonium sulfate, 0.1 M bis-tris–HCl pH 6.5, 25%(w/v) PEG 3350 (Fig. 2 ▶ a) and (B) 1 M succinic acid, 0.1 M HEPES pH 7.0, 1%(w/v) PEG MME 2000 (Fig. 2 ▶ b).
Figure 2.
ChxREff crystals (a) grown with PEG 3350 as precipitant and (b) grown with succinic acid as precipitant. The black bars denote 200 µm.
2.3. Confirmation of ChxREff crystallization
To determine that the ChxREff crystals were indeed composed of the desired protein, crystals from crystallization condition A were washed extensively in mother liquor and analyzed by SDS–PAGE. The predominant band corresponded to the approximate molecular weight of ChxREff (∼15 kDa). The sample was further analyzed by combined MS+MS/MS (Mass Spectrometry Core facility at the University of Texas Medical Branch). All expected proteolytic fragments of ChxREff were present and the predicted sequences matched identically. Only predicted peptide sequences that matched ChxREff were detected, indicating that the crystals were predominantly comprised of ChxREff.
2.4. Data collection and processing
ChxREff crystals were soaked for about 20 s in mother liquor supplemented with 10% ethylene glycol and flash-cooled by immersion in liquid nitrogen. Data collection was carried out at 100 K on Stanford Synchrotron Radiation Laboratory (SSRL) beamline 9-2. For crystals grown in condition A, the exposure time for each 0.5° oscillation image was 5 s using a MAR 325 detector and a wavelength of 0.1 nm. For crystals grown in condition B, 0.7° oscillation images were collected for 5 s using the same detector and wavelength as described above. The resulting data sets were processed and scaled using MOSFLM and SCALA from the CCP4 suite (Collaborative Computational Project, Number 4, 1994 ▶). The data-collection statistics are summarized in Table 1 ▶.
Table 1. ChxREff data collection and processing.
Values in parentheses are for the last shell.
| Crystal form | A | B |
|---|---|---|
| Resolution (Å) | 43.5–2.5 (2.56–2.5) | 59.9–2.2 (2.32–2.2) |
| Space group | P41212 or P43212 | P41212 or P43212 |
| Unit-cell parameters (Å) | a = b = 109.3, c = 95.7 | a = b = 110.0, c = 93.9 |
| Unique observations | 20605 | 29023 |
| Total observations | 164342 | 224352 |
| Completeness (%) | 98.1 (99.3) | 97.8 (93.5) |
| Rmerge† | 0.08 (0.40) | 0.05 (0.41) |
| I/σ(I) | 22.2 (4.2) | 22.9 (3.1) |
| Redundancy | 8.1 (8.2) | 7.7 (6.0) |
R
merge = 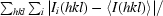
 .
.
3. Results and discussion
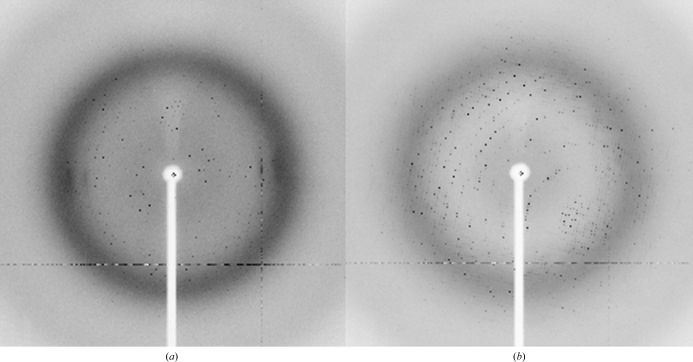
The effector domain of ChxR was cloned from C. trachomatis, overexpressed in E. coli and purified by cobalt affinity (owing to the addition of a C-terminal histidine tag) and gel-filtration chromatography. The purified ChxREff with the C-terminal histidine tag still attached (molecular weight of ∼15 kDa) was determined to be >95% pure by Coomassie-stained SDS–PAGE (Fig. 1 ▶). Sparse-matrix screening of this protein sample using commercially available crystal screens produced many crystallization hits, two of which (Fig. 2 ▶) produced crystals of diffraction quality (Fig. 3 ▶).
Figure 3.
Diffraction images collected on SSRL beamline 9-2. (a) Oscillation image of a crystal from mother liquor A. The resolution limit is 2.5 Å. (b) Oscillation image of a crystal from mother liquor B. The resolution limit is 2.2 Å.
Despite the differences in the mother liquors, both crystal morphologies belonged to the tetragonal space group P41212 or P43212, had isomorphous unit-cell parameters and diffracted to comparable resolution (Table 1 ▶). OmpR/PhoB effector domains of known structure have been reported to be monomeric in the absence of the receiver domain (Wang et al., 2007 ▶). Therefore, calculation of probable Matthews coefficients suggests that there are most likely to be four monomers in the asymmetric unit, with a V M value of 2.55 Å3 Da−1 and a probable solvent content of 51.7% (Matthews, 1968 ▶). However, three monomers (V M = 3.40 Å3 Da−1; 64.0% solvent content) and five monomers (V M = 2.04 Å3 Da−1; 39.7% solvent content) are also possible in this cell system. The data-collection statistics are summarized in Table 1 ▶.
YycF from Bacillus subtilis is the closest homolog to ChxREff, with a sequence identity of 30% and a similarity of 48%. The YycF DNA-binding domain (PDB code 2d1v; Okajima et al., 2008 ▶) will be used as a model for molecular-replacement experiments, as will all effector domains of response regulators of known structure. ChxREff crystals will also be soaked with heavy metals for multiple isomorphous replacement phasing trials.
Acknowledgments
Diffraction data were collected at the Stanford Synchrotron Radiation Laboratory, a national user facility operated by Stanford University on behalf of the US Department of Energy, Office of Basic Energy Sciences. The SSRL Structural Molecular Biology Program is supported by the Department of Energy, Office of Biological and Environmental Research and by the National Institutes of Health, National Center for Research Resources, Biomedical Technology Program and the National Institute of General Medical Sciences. We would like to thank the staff at the Stanford Synchrotron Radiation Laboratory for their support and assistance. This study was funded by NIH/NCRR COBRE 5P20-RR17780.
References
- Baikalov, I., Schroder, I., Kaczor-Grzeskowiak, M., Grzeskowiak, K., Gunsalus, R. P. & Dickerson, R. E. (1996). Biochemistry, 35, 11053–11061. [DOI] [PubMed]
- Brennan, R. G. (1993). Cell, 74, 773–776. [DOI] [PubMed]
- Collaborative Computational Project, Number 4 (1994). Acta Cryst. D50, 760–763.
- Hoch, J. A. (2000). Curr. Opin. Microbiol.3, 165–170. [DOI] [PubMed]
- Hong, E., Lee, H. M., Ko, H., Kim, D. U., Jeon, B. Y., Jung, J., Shin, J., Lee, S. A., Kim, Y., Jeon, Y. H., Cheong, C., Cho, H. S. & Lee, W. (2007). J. Biol. Chem.282, 20667–20675. [DOI] [PubMed]
- Jones, B. D. (2005). J. Microbiol.43, 110–117.
- Kenney, L. J. (2002). Curr. Opin. Microbiol.5, 135–141. [DOI] [PubMed]
- Koo, I. C., Walthers, D., Hefty, P. S., Kenney, L. J. & Stephens, R. S. (2006). Proc. Natl Acad. Sci. USA, 103, 750–755. [DOI] [PMC free article] [PubMed]
- Matthews, B. W. (1968). J. Mol. Biol.33, 491–497. [DOI] [PubMed]
- Okajima, T., Doi, A., Okada, A., Gotoh, Y., Tanizawa, K. & Utsumi, R. (2008). FEBS Lett.582, 3434–3438. [DOI] [PubMed]
- Stock, A. M., Robinson, V. L. & Goudreau, P. N. (2000). Annu. Rev. Biochem.69, 183–215. [DOI] [PubMed]
- Wang, S., Engohang-Ndong, J. & Smith, I. (2007). Biochemistry, 46, 14751–14761. [DOI] [PMC free article] [PubMed]