The sulfide:quinone oxidoreductase from A. ferrooxidans ATCC 23270 was overexpressed in E. coli and purified. Crystallization and preliminarily X-ray crystallographic analysis were performed for the recombinant enzyme.
Keywords: sulfide:quinone reductase oxidoreductases
Abstract
The gene product of open reading frame AFE_1293 from Acidithiobacillus ferrooxidans ATCC 23270 is annotated as encoding a sulfide:quinone oxidoreductase, an enzyme that catalyses electron transfer from sulfide to quinone. Following overexpression in Escherichia coli, the enzyme was purified and crystallized using the hanging-drop vapour-diffusion method. The native crystals belonged to the tetragonal space group P42212, with unit-cell parameters a = b = 131.7, c = 208.8 Å, and diffracted to 2.3 Å resolution. Preliminary crystallographic analysis indicated the presence of a dimer in the asymmetric unit, with an extreme value of the Matthews coefficient (V M) of 4.53 Å3 Da−1 and a solvent content of 72.9%.
1. Introduction
Acidithiobacillus ferrooxidans is a motile Gram-negative acidophilic chemolithotrophic bacterium that utilizes FeII, H2S, S0, reduced inorganic sulfur compounds and molecular hydrogen as energy sources (Temple & Delchamps, 1953 ▶). The oxidation of sulfur produces sulfuric acid, which is responsible for some of the interesting characteristics of A. ferrooxidans. It is generally found in acidic environments such as mining dumps and acid mine drainages. This organism plays one of the key roles in microbial communities by taking part in the bacterial chemical processes of bioleaching under mesophilic conditions (Olson et al., 2003 ▶).
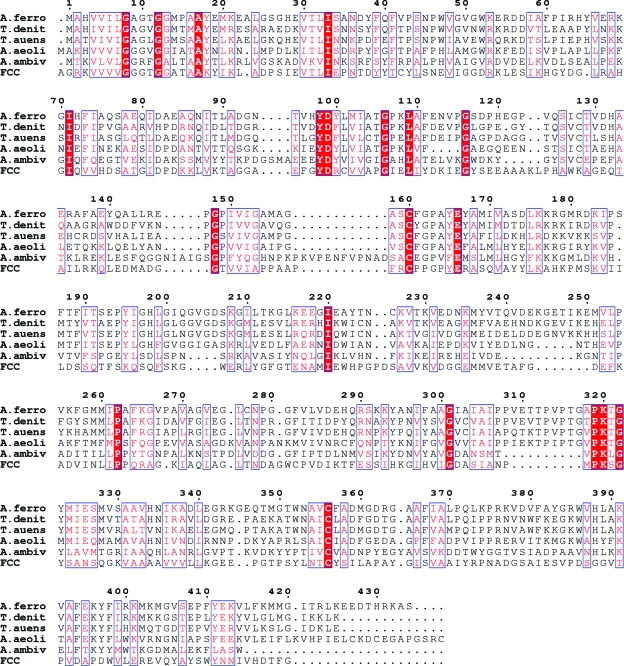
Sulfur can exist in various oxidation states from −2 to +6, complicating studies of the enzymatic steps involved. Based on comparative genomic analysis, sulfur oxidation in A. ferrooxidans is catalyzed by the sulfide:quinone oxidoreductase (SQR) system and the initial step of the SQR system is the oxidation of sulfide to elemental sulfur by SQR. The sulfur may be further oxidized via S2O3 2− and SO3 2− to the final oxidation state +6 in SO4 2− by other enzymes of the SQR system (Valdes et al., 2008 ▶). The mechanism of SQR catalysis includes electron transfer from sulfide anions to quinone, resulting in the reduction of the latter. SQRs are ancient flavoproteins that are present in many organisms from archaea to humans. Several SQRs have been characterized by biochemical methods (Griesbeck et al., 2002 ▶; Theissen & Martin, 2008 ▶; Arieli et al., 1994 ▶; Shibata & Kobayashi, 2006 ▶; Wakai et al., 2007 ▶; Shibata et al., 2007 ▶; Vande Weghe & Ow, 1999 ▶). Recently, two structures of SQR from the hyperthermoacidophilic archaeon Acidianus ambivalens (PDB code 3h8l; Brito et al., 2009 ▶) and from the hyperthermophilic bacterium Aquifex aeolicus (PDB codes 3h27, 3h28 and 3h29; Marcia et al., 2009 ▶) have been reported. The structure of a related protein, flavocytochrome c sulfide dehydrogenase (FCC) from Chromatium vinosum, was published almost 15 years ago (PDB code 1fcd; Chen et al., 1994 ▶). It has been found that Cys160 and Cys356 (in the A. ferrooxidans numbering) are the active-site residues (Fig. 1 ▶). Interestingly, all reported structures had the flavin adenine dinucleotide (FAD) cofactor molecules covalently bound to the protein through a linkage of the C8M group of FAD to a Cys128 residue that is different from the active-site cysteines. Several other SQRs, including that presented here, have noncovalently bound FAD (Griesbeck et al., 2002 ▶) that can sometimes be lost during purification steps. However, the exact mechanism of the electron transfer between the sulfides and quinone is not entirely clear. Further structural and mutagenic studies are needed.
Figure 1.
Sequence alignment of SQR from A. ferrooxidans (A.ferro) with SQRs from Thiobacillus denitrificans (T.denit), Tolumonas auensis (T.auens), Aquifex aeolicus (A.aeoli) and Acidianus ambivalens (A.ambiv) and FCC (flavocytochrome c sulfide dehydrogenase) from Chromatium vinosum. Conserved residues are shown on a red background, whereas residues with similar chemical properties are shown in red. The sequence identities between A.ferro and T.denit, T.auens, A.aeoli, A.ambiv and FCC are 57, 51, 40, 27 and 25%, respectively. Sequence alignment was performed using the program ClustalW (Thompson et al., 1994 ▶) and the figure was generated with the program ESPript (Gouet et al., 1999 ▶).
In this study, we report the molecular cloning, protein purification and preliminary X-ray crystallographic analysis of the AFE_1293 gene product from A. ferrooxidans, which has been annotated as a sulfide:quinone oxidoreductase (SQR). It has a bright yellow colour arising from the nonconvalently bound cofactor FAD (data on FAD binding are not presented here).
2. Materials and methods
2.1. Cloning, expression and purification
Genomic template DNA from A. ferrooxidans ATCC 23270 was prepared using the EZ-10 spin-column genomic DNA-isolation kit from Bio Basic Inc. according to the manufacturer’s instructions for bacterial DNA extraction. The SQR open reading frame (ORF) was amplified by the polymerase chain reaction (PCR) using primers that were designed to add six continuous histidine codons to the 3′ primer. The sequence of the forward primer was 5′-CGCGCGGATCCAGGAGGAATTTAAAATGAGAGGATCGGCACATGTGGTAATTTTGGGTG-3′, containing a BamHI site (GGATCC), a ribosome-binding site (AGGAGGA), codons for the amino-acid sequence MRGS (start codon) and codons for amino acids 2–8 of SQR. The sequence of the reverse primer was 5′-CTGCAGGTCGACTCAGTGATGGTGATGGTGATGGGAGGCCTTACGATGGGTATCTTCTT-3′, containing a SalI site (GTCGAC), a stop anticodon (TTA) and anticodons for the hexa-His tag and the last eight amino acids of SQR. The resulting PCR product was gel purified, doubly digested and ligated into a pLM1 (Sodeoka et al., 1993 ▶) expression vector, resulting in the pLM1::SQR plasmid.
The positive plasmid was confirmed by DNA-sequencing analysis (ABI23270) and transformed into Escherichia coli strain BL21 (DE3) competent cells for expression purposes. An overnight culture from a single colony was used to inoculate 2 l Terrific Broth (TB) supplemented with 100 mg ml−1 ampicillin. Shaking of the culture at 310 K was continued until its OD600nm reached 0.8. Subsequently, the temperature of the culture was shifted to 298 K and protein overexpression was induced with 0.3 mM isopropyl β-d-1-thiogalactopyranoside (IPTG) for 22 h. Production of the selenomethionine-substituted protein followed a published procedure (Van Duyne et al., 1993 ▶). Cells were harvested by centrifugation and resuspended in 40 ml buffer A (50 mM MOPS pH 7.0, 300 mM NaCl, 5 mM β-mercaptoethanol, 200 µM PMSF). The cells were ruptured in an EmulsiFlex high-pressure cell homogenizer (Avestin) at ∼103 MPa and ultracentrifuged at 40 000 rev min−1 for 90 min at 277 K. The supernatant was filtered through a 0.45 µm filter (Sartorius) and loaded onto a 5 ml Hi-Trap column (GE Healthcare) pre-equilibrated with buffer A. The column was washed with wash buffer (50 mM MOPS pH 7.0, 300 mM NaCl, 5 mM β-mercaptoethanol, 200 µM PMSF, 150 mM imidazole) and the His-tagged protein was eluted in the same buffer with the inclusion of 300 mM imidazole and was precipitated by the addition of ammonium sulfate to 70% saturation. Dissolved precipitate was loaded onto a Superdex 200 gel-filtration column (GE Healthcare) and eluted in a suitable buffer for crystallization (5 mM Tris–HCl pH 7.0, 300 mM NaCl, 1 mM DTT). The resulting solution was concentrated to 5 mg ml−1 using a YM-30 Centricon tube (Amicon). All steps of protein purification subsequent to the initial thawing of cells were performed at 277 K and the result of each step was monitored by 12% SDS–PAGE (Laemmli, 1970 ▶).
2.2. Crystallization, data collection and preliminary crystallographic analysis
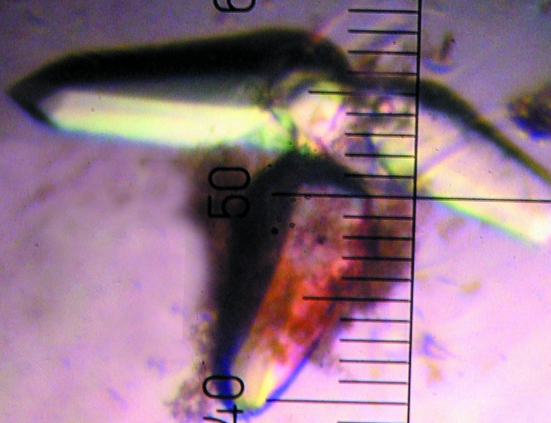
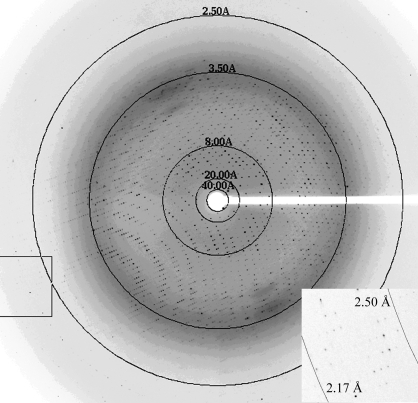
The search for crystallization conditions for SQR from A. ferrooxidans was performed by the sitting-drop vapour-diffusion method in Intelli-Plates (Hampton Research) using a crystallization robot (Hydra96 Plus One, Robbins Scientific) and the commercially available Crystal Screens I and II and Index Screen (Hampton Research). Equal volumes (0.3 µl) of protein and reservoir solutions were mixed and equilibrated against the reservoir solutions at room temperature. Several crystallization conditions were found in the initial screens. After optimization of the best screening conditions, X-ray diffraction-quality tetragonal crystals grew in hanging drops from 30% PEG 600, 0.1 M bis-tris pH 5.5, 0.1 M ammonium sulfate in 1–2 d (Fig. 2 ▶). For data collection, the crystals were flash-cooled in liquid nitrogen. Native data were collected on beamline 9-2 at the Stanford Synchrotron Radiation Laboratory (SSRL) using a MAR 325 image-plate detector (Fig. 3 ▶). The raw data were processed using the HKL2000 program suite (Otwinowski & Minor, 1997 ▶). The crystal structure was solved by molecular replacement using MOLREP (Vagin & Teplyakov, 1997 ▶; Collaborative Computational Project, Number 4, 1994 ▶) and Phaser (McCoy et al., 2005 ▶). A truncated molecule A (residues 2–375) of the SQR trimer from Aquifex aeolicus (PDB code 3h27; 40% sequence identity) was used as a search model.
Figure 2.
Tetragonal crystals of SQR from A. ferrooxidans.
Figure 3.
A 0.5° oscillation image of diffraction from the SQR crystal. Details of the boxed region are shown in the inset.
3. Results
A complete X-ray diffraction data set was collected from a single crystal at 100 K without additional cryoprotection. The crystals belonged to space group P42212, with unit-cell parameters a = b = 131.7, c = 208.8 Å. The crystallographic statistics of the native data are summarized in Table 1 ▶. The data set was 100% complete to 2.3 Å resolution. The full-length SQR molecule contains 434 amino-acid residues and has a molecular mass of approximately 50 kDa. Calculation of the Matthews coefficient V M (Matthews, 1968 ▶) suggested the presence of four or three molecules in the asymmetric unit, with V M values of 2.26 Å3 Da−1 (45.7% solvent) or 3.02 Å3 Da−1 (59.2% solvent), respectively. However, the molecular-replacement programs MOLREP and Phaser were only able to find two molecules in the asymmetric unit. Attempts to find a third or fourth molecule in the asymmetric unit failed. The correlation score calculated by MOLREP for the dimer had a value of 0.200. Similarly, the log-likelihood gain Phaser) for the dimer had a value of 354. The solution containing a dimer in the asymmetric unit has been accepted. As a result, the Matthews coefficient V M is 4.53 Å3 Da−1 and the solvent content is 72.9%. These values are outside the usual values for the Matthews coefficient of between 1.62 and 3.53 Å3 Da−1. However, the high solvent content does not prevent the SQR crystals from diffracting to 2.3 Å resolution. Structure refinement and detailed structural analysis of SQR are in progress.
Table 1. Crystal parameters and data-collection statistics.
Values in parentheses are for the highest resolution shell.
| Space group | P42212 |
| Unit-cell parameters (Å) | a = b = 131.73, c = 208.86 |
| No. of molecules in the unit cell (Z) | 16 |
| Solvent content (%) | 72.9 |
| Temperature (K) | 100 |
| Detector | MAR Q325 |
| Wavelength (Å) | 0.97946 |
| Resolution (Å) | 50.00–2.30 (2.38–2.30) |
| No. of unique reflections | 82166 (8046) |
| Multiplicity | 8.0 (7.6) |
| 〈I/σ(I)〉 | 19.5 (1.94) |
| Completeness (%) | 100 (100) |
| Rmerge† | 0.097 (0.986) |
R
merge = 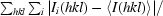
 , where I
i(hkl) is the ith intensity measurement and 〈I(hkl)〉 is the mean of all measurements of I(hkl).
, where I
i(hkl) is the ith intensity measurement and 〈I(hkl)〉 is the mean of all measurements of I(hkl).
Acknowledgments
This work was partially supported by the National Basic Research Program of China (2004CB619201) and the National Natural Science Group Foundation of the People’s Republic of China (No. 50621063). Research in the laboratories of MNGJ and JHW was supported by the Canadian Institutes of Health Research (CIHR FRN-37770 and CIHR MOP-15292), and MNGJ also received support from the Alberta Heritage Foundation for Medical Research (AHFMR). We thank the SSRL staff for their help in data collection.
References
- Arieli, B., Shahak, Y., Taglicht, D., Hauska, G. & Padan, E. (1994). J. Biol. Chem.269, 5705–5711. [PubMed]
- Brito, J. A., Sousa, F. L., Stelter, M., Bandeiras, T. M., Vonrhein, C., Teixeira, M., Pereira, M. M. & Archer, M. (2009). Biochemistry, 48, 5613–5622. [DOI] [PubMed]
- Chen, Z. W., Koh, M., Van Driessche, G., Van Beeumen, J. J., Bartsch, R. G., Meyer, T. E., Cusanovich, M. A. & Mathews, F. S. (1994). Science, 266, 430–432. [DOI] [PubMed]
- Collaborative Computational Project, Number 4 (1994). Acta Cryst. D50, 760–763.
- Gouet, P., Courcelle, E., Stuart, D. I. & Métoz, F. (1999). Bioinformatics, 15, 305–308. [DOI] [PubMed]
- Griesbeck, C., Schutz, M., Schodl, T., Bathe, S., Nausch, L., Mederer, N., Vielreicher, M. & Hauska, G. (2002). Biochemistry, 41, 11552–11565. [DOI] [PubMed]
- Laemmli, U. K. (1970). Nature (London), 227, 680–685. [DOI] [PubMed]
- Marcia, M., Ermler, U., Peng, G. & Michel, H. (2009). Proc. Natl Acad. Sci. USA, 106, 9625–9630. [DOI] [PMC free article] [PubMed]
- Matthews, B. W. (1968). J. Mol. Biol.33, 491–497. [DOI] [PubMed]
- McCoy, A. J., Grosse-Kunstleve, R. W., Storoni, L. C. & Read, R. J. (2005). Acta Cryst. D61, 458–464. [DOI] [PubMed]
- Olson, G. J., Brierley, J. A. & Brierley, C. L. (2003). Appl. Microbiol. Biotechnol.63, 249–257. [DOI] [PubMed]
- Otwinowski, Z. & Minor, W. (1997). Methods Enzymol.276, 307–326. [DOI] [PubMed]
- Shibata, H. & Kobayashi, S. (2006). Can. J. Microbiol.52, 724–730. [DOI] [PubMed]
- Shibata, H., Suzuki, K. & Kobayashi, S. (2007). Can. J. Microbiol.53, 1091–1100. [DOI] [PubMed]
- Sodeoka, M., Larson, C. J., Chen, L., LeClair, K. P. & Verdine, G. L. (1993). Bioorg. Med. Chem. Lett.3, 1089–1094.
- Temple, K. L. & Delchamps, E. W. (1953). Appl. Microbiol.1, 255–258. [DOI] [PMC free article] [PubMed]
- Theissen, U. & Martin, W. (2008). FEBS J.275, 1131–1139. [DOI] [PubMed]
- Thompson, J. D., Higgins, D. G. & Gibson, T. J. (1994). Nucleic Acids Res.22, 4673–4680. [DOI] [PMC free article] [PubMed]
- Vagin, A. & Teplyakov, A. (1997). J. Appl. Cryst.30, 1022–1025.
- Valdes, J., Pedroso, I., Quatrini, R., Dodson, R. J., Tettelin, H., Blake, R. II, Eisen, J. A. & Holmes, D. S. (2008). BMC Genomics, 9, 597. [DOI] [PMC free article] [PubMed]
- Vande Weghe, J. G. & Ow, D. W. (1999). J. Biol. Chem.274, 13250–13257. [DOI] [PubMed]
- Van Duyne, G. D., Standaert, R. F., Karplus, P. A., Schreiber, S. L. & Clardy, J. (1993). J. Mol. Biol.229, 105–124. [DOI] [PubMed]
- Wakai, S., Tsujita, M., Kikumoto, M., Manchur, M. A., Kanao, T. & Kamimura, K. (2007). Biosci. Biotechnol. Biochem.71, 2735–2742. [DOI] [PubMed]