The Mms21–Smc5 protein complex has been crystallized. The diffraction of the crystals was improved greatly by glutaraldehyde treatment and X-ray diffraction data sets were collected to resolutions of 2.3 and 3.9 Å from native and selenomethionine-derivative protein crystals, respectively.
Keywords: Mms21, Smc5/6 complex, SMC proteins, SUMO, DNA damage repair
Abstract
Smc5/6, a protein complex that belongs to the structural maintenance of chromosome (SMC) family, plays a key role in DNA replication, sister chromatid recombination and DNA damage repair. The complex contains eight subunits, including a SUMO E3 ligase Mms21 (Nse2). The activity of Mms21 is important for regulation of Smc5/6 in the response to DNA damage. Mms21 and the Mms21-binding region of Smc5 were overexpressed and purified individually in Escherichia coli with a C-terminal LEHHHHHH tag. The Mms21–Smc5 protein complex was crystallized. The diffraction of the crystals was improved greatly by glutaraldehyde treatment. X-ray diffraction data sets were collected to resolutions of 2.3 and 3.9 Å from native and selenomethionine-derivative protein crystals, respectively. The crystals belonged to space group C2221, with unit-cell parameters a = 47.465, b = 97.574, c = 249.215 Å for the native crystals.
1. Introduction
The Smc5/6 holocomplex is essential for cell viability and is required for sister chromatid recombination. The complex is important for double-stranded DNA damage repair as well as telomere and rDNA repetitive-site stability maintenance (De Piccoli et al., 2009 ▶; Murray & Carr, 2008 ▶). It contains eight individual subunits: the heterodimer Smc5 and Smc6 and six nonstructural maintenance of chromosome (SMC) elements, i.e. Nse1, Mms21 (Nse2) and Nse3–Nse6. Both Smc5 and Smc6 contain five regions: two globular domains at each end associate together to form a head domain and two long helices intertwine to form a coiled-coil domain, with the hinge domain in the middle. The hinge domain from Smc5 associates with the hinge domain of Smc6 to form the heterodimer Smc5/6; these are the core subunits in the complex (Pebernard et al., 2006 ▶; Duan et al., 2009 ▶; Sergeant et al., 2005 ▶; Taylor et al., 2008 ▶). One of the non-SMC elements, Mms21 (Nse2), associates with the long coiled-coil region of Smc5 and regulates Smc5/6 function (Zhao & Blobel, 2005 ▶; Potts & Yu, 2005 ▶; Andrews et al., 2005 ▶). The other non-SMC elements associate with different regions of the Smc5/6 heterodimer (Palecek et al., 2006 ▶; Sergeant et al., 2005 ▶; Pebernard et al., 2006 ▶).
Mms21, a small ubiquitin-like modifier (SUMO) E3 ligase, contains a SUMO signature motif in the C-terminus. Sumoylation plays many important roles in the cellular response by changing the properties of modified substrates, including regulating DNA transcription, translation and cellular signaling (Ulrich, 2008 ▶, 2009 ▶; Geiss-Friedlander & Melchior, 2007 ▶). Sumoylation also regulates the DNA damage response of the Smc5/6 complex, as mutations in the signature motif of Mms21 cause the cells to be more sensitive to such responses (Zhao & Blobel, 2005 ▶; Potts & Yu, 2005 ▶; Andrews et al., 2005 ▶). However, the mechanism by which Mms21 regulates the function of the Smc5/6 complex is unclear. As a first step toward elucidating the regulatory effect of Mms21 on Smc5/6, we expressed, purified and crystallized a protein complex containing full-length Mms21 and the Mms21-binding region of Smc5. Details of the structural analysis of this complex should provide clues to understanding the molecular mechanism.
2. Materials and methods
2.1. Cloning
Genomic DNA from Saccharomyces cerevisiae was used as a template for the polymerase chain reaction (PCR) to amplify full-length Smc5 and Mms21. The Mms21-binding region of Smc5 was amplified by three steps using full-length Smc5 as the template. Firstly, the fragment encoding residues 302–369 was amplified using forward primer F1, 5′-GGGCCATGGGATAAAAAACCATTTGCAAATAC-3′, and reverse primer R1, 5′-TTCTAGCTTCACGTTTTGATCCTGATCCTAGTTTTTTGGTTCTTC-3′. Secondly, the region encoding residues 733–813 was amplified using forward primer F2, 5′-GAAGAACCAAAAAACTAGGATCAGGATCAAAACGTGAAGCTAGAA-3′, and reverse primer R2, 5′-GGGGAGCTCTTGGCTCTTCAAATCAG-3′. Primer R1 contains an overlapping region with primer F2, which enables them to anneal together. Codons encoding a flexible linker Gly-Ser-Gly-Ser were inserted into the middle of the primers R1 and F2. The two PCR products were mixed together and diluted 100-fold. A third PCR was performed by taking 1 µl of the diluted mixture as a template. Primers F1 and R2 were used to obtain the fragments 302–369 and 733–813. This amplified fragment was cloned into pET24D vector, which adds a C-terminal tag containing residues LEHHHHHH to the expressed recombinant protein. Full-length Mms21 was cloned into the pET24D vector using the standard cloning strategy. The vector also adds a C-terminal LEHHHHHH tag to the expressed recombinant protein. Both constructs were verified by DNA sequencing.
2.2. Expression and purification
The plasmids containing either Mms21 or Smc5 were transformed into Rosetta BL21 (DE3) cells (Novagen) individually for expression of native protein. Cells were cultured at 310 K in 4 l LB medium containing 100 µg ml−1 kanamycin to an optical density of approximately 0.5 at 600 nm. The cultures were then cooled to 293 K. Protein expression was induced by treating the cells with 0.4 mM isopropyl β-d-1-thiogalactopyranoside (IPTG) overnight at 293 K. The plasmids were transformed into the methionine-auxotrophic B834 (DE3) strain (Novagen) separately for selenomethione-derivative (SeMet) protein expression. The cells were grown using a defined LeMaster medium containing 125 mg l−1 seleomethionine, other amino acids excluding methionine, salt, vitamins, metal ions and glucose (Sreenath et al., 2005 ▶). Similar to the expression of the native protein, the cells were grown at 310 K to an optical density of approximately 0.5 at 600 nm and induced using IPTG at 293 K. The cells were spun down by centrifugation. The cell pellets were resuspended and lysed by sonication in 80 ml lysis buffer containing 50 mM Tris pH 8.0, 200 mM NaCl and 25 mM imidazole. The lysate was then centrifuged at 15 000 rev min−1 for 30 min at 277 K. The supernatants were mixed with 1 ml Ni–NTA resin (Qiagen), incubated for 30 min at 277 K and applied onto a gravity-flow column (Bio-Rad). The unbound bacterial proteins were removed from the column by washing with lysis buffer. Mms21 and Smc5 proteins were eluted from the column with elution buffer containing 50 mM Tris pH 8.0, 200 mM NaCl and 300 mM imidazole. The eluate from the Ni–NTA column was then applied onto a Superdex 200 10/300 GL gel-filtration column (GE Healthcare), which was pre-equilibrated with buffer containing 50 mM Tris pH 8.0 and 50 mM NaCl. Mms21 and Smc5 were eluted individually with the same buffer. The peak fractions were collected and their concentrations were measured using the Quick Start Bradford Protein Assay (Bio-Rad). The samples were 95% pure as estimated by SDS–PAGE. The complex for crystallization was formed by mixing Mms21 and Smc5 in a 1:1 molar ratio.
2.3. Crystallization
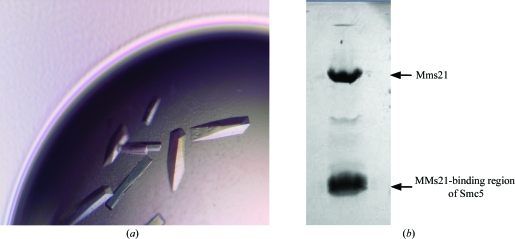
Initial crystallization screening was performed using the hanging-drop vapor-diffusion method at 293 K in 24-well trays with Hampton Research crystallization kits. 2 µl protein solution at 12 mg ml−1 protein concentration was mixed with an equal volume of reservoir solution and equilibrated against 0.5 ml reservoir solution. Several conditions including PEG 3350 and PEG 4000 with either CaCl2 or calcium acetate produced crystals. Final optimization was carried out by testing various gradients of CaCl2. Crystals grew rapidly to dimensions of 0.3 × 0.3 × 1.0 mm overnight. Most contained a hollow space in the middle of the crystal. The crystallization process was slowed by setting up crystallization trays at 277 K. However, the diffraction of crystals grown at 277 K showed no significant improvement compared with those grown at room temperatures. The best crystals formed in conditions contisting of 100 mM Tris–HCl pH 7.5, 18% PEG 3350 and 100 mM CaCl2 (Fig. 1 ▶ a).
Figure 1.
(a) Crystals of the Mms21–Smc5 complex; (b) the two protein bands observed on SDS–PAGE analysis of the crystals.
To examine whether the crystals truly contained the Mms21–Smc5 complex, five crystals were picked out from crystallization droplets and washed carefully with reservoir solution to remove residual proteins from the crystal surfaces. The crystals were then dissolved in water and examined by SDS–PAGE. Two clear bands at molecular weights of about 30 and 20 kDa were consistent with the expected molecular weights of Mms21 and the Mms21-binding region of Smc5, indicating that the crystals indeed consisted of the protein complex (Fig. 1 ▶ b).
2.4. Diffraction improvement
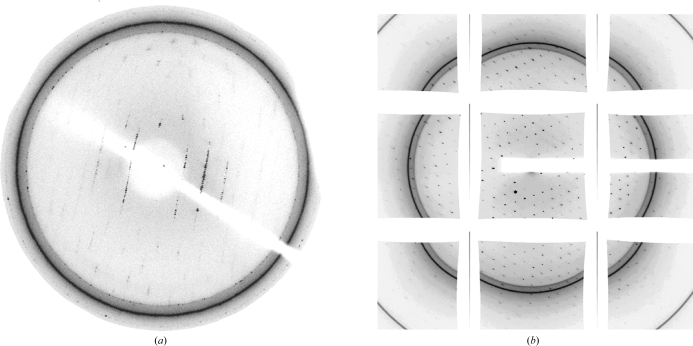
A single crystal was picked up and soaked for 10 s in reservoir solution supplemented with 20% glycerol as a cryoprotectant. The crystal then was mounted in a nylon-fiber loop and flash-cooled to 100 K using a nitrogen-gas stream. The native crystal diffracted to about 5 Å resolution at the home X-ray source (Rigaku) equipped with a MAR 345 image-plate detector (Fig. 2 ▶ a) and the SeMet crystal diffracted to 7 Å resolution. To improve the diffraction, 5 µl of the cross-linking reagent glutaraldehyde was added to the 500 µl reservoir solution (Cohen-Hadar et al., 2006 ▶). Crystals were equilibrated at 293 K for 5 min, 10 min, 30 min, 2 h, 3 h and 4 h before being mounted on the loop. The best cross-linking time was 2 h and the crystal diffraction resolution was greatly improved. Diffraction data were collected at a synchrotron to 2.3 Å resolution (Fig. 2 ▶ b) for native crystals and to 3.9 Å resolution for SeMet crystals. The crystal diffraction was lost completely if the crystals were treated for longer than 3 h.
Figure 2.
(a) A representative image collected from a native crystal that diffracted to about 5.0 Å resolution at the home X-ray source. The mosaicity is high, especially in the higher resolution areas. (b) A representative image collected from a native crystal treated with the cross-linking reagent glutaraldehyde with improved resolution. The image was collected at a synchrotron.
2.5. Data collection and processing
All diffraction data sets were collected at the Advanced Photon Source (APS). The data set from the native crystal was collected on beamline 19-ID (SER-CAT) and that from the SeMet crystal was collected on beamline 24-ID (NE-CAT) with a wavelength of 0.97916 Å. The data sets were indexed and processed using HKL-2000 (Otwinowski & Minor, 1997 ▶). Diffraction data statistics for the native and SeMet crystals are shown in Table 1 ▶. Preliminary X-ray diffraction analysis of the native crystal data showed that the crystal belonged to space group C2221, with unit-cell parameters a = 47.465, b = 97.574, c = 249.215 Å. The Matthews coefficient V M (Matthews, 1968 ▶) for space group C2221 using the molecular weight of the complex (50 kDa) was 2.89 Å3 Da−1, which corresponds to a solvent content of 57.4% and indicates the presence of one complex molecule in each asymmetric unit.
Table 1. Diffraction data statistics for Mms21–Smc5 complex crystals.
Values in parentheses are for the last resolution shell.
| Native | SeMet | |
|---|---|---|
| X-ray source | APS 19-BM | APS 24-ID |
| Wavelength (Å) | 0.97899 | 0.97616 |
| Resolution (Å) | 50–2.3 | 50–3.9 |
| Space group | C2221 | C2221 |
| Unit-cell parameters (Å) | a = 47.465, b = 91.574, c = 249.215 | a = 47.138, b = 91.566, c = 250.583 |
| No. of observations | 105443 | 29652 |
| Unique reflections | 21519 | 4942 |
| Redundancy | 4.9 (1.9) | 6.0 (3.9) |
| Completeness (%) | 88.0 (77.3) | 93.4 (85.6) |
| I/σ(I) | 19.1 (1.6) | 32.8 (5.6) |
| Rmerge | 0.09 (0.41) | 0.07 (0.19) |
The structure of the Mms21–Smc5 complex was determined by single-wavelength anomalous diffraction (SAD) phasing using the SeMet data set at 3.9 Å resolution. There are seven selenium sites, which were located using SOLVE (Terwilliger & Berendzen, 1996 ▶). The same result was confirmed by HKL2MAP (Pape & Schneider, 2004 ▶).
3. Discussion
The success in obtaining an Mms21–Smc5 complex crystal that diffracted to high resolution was the result of several efforts. Firstly, we obtained the correct constructs of the Smc5 fragments. The coiled-coil region of Smc5 contains two helices, each of which extends over about 300 residues. As such, many different constructs of Smc5 were tried with various regions for the individual helices and different-length linkers between the two helices. Most of them either led to no crystal formation or poor crystals that we were not able to optimize. The best construct, which was found through enzyme digestion (unpublished results) and which we used to obtain the large crystal, contained residues 302–369 and 733–813 linked by four amino acids, Gly-Ser-Gly-Ser. Secondly, although the complex of Mms21 and Smc5 could be co-purified by gel filtration, the best crystals were formed by mixing the two proteins in a 1:1 ratio. Finally, the crystal resolution was improved significantly by the cross-linking method. The mosaicity decreased significantly according to the appearance of the images. While most of the native crystals diffracted to 5 Å resolution at the home X-ray source, the cross-linking treatment improved their diffraction resolution greatly.
Acknowledgments
We thank Xiaoqiang Wang and Andrew Marsh for critical review and proofreading of the manuscript. We are grateful to Drs Craig Ogata at NE-CAT (24-ID) of the Advanced Photon Source (APS) and Stephan Ginnell at SBC-CAT (19-BM) of the APS for help with data collection. We also thank Drs John Arnez and Mark Mashuta for maintaining our X-ray facility. This work is supported by the NIH Center of Biomedical Research Excellence in Molecular Targets grant (5P20 RR 018733) to the James Graham Brown Cancer Center at the University of Louisville and R01GM079516 to HY.
References
- Andrews, E. A., Palecek, J., Sergeant, J., Taylor, E., Lehmann, A. R. & Watts, F. Z. (2005). Mol. Cell. Biol.25, 185–196. [DOI] [PMC free article] [PubMed]
- Cohen-Hadar, N., Wine, Y., Nachliel, E., Huppert, D., Gutman, M., Frolow, F. & Freeman, A. (2006). Biotechnol. Bioeng.94, 1005–1011. [DOI] [PubMed]
- De Piccoli, G., Torres-Rosell, J. & Aragon, L. (2009). Chromosome Res.17, 251–263. [DOI] [PubMed]
- Duan, X., Yang, Y., Chen, Y. H., Arenz, J., Rangi, G. K., Zhao, X. & Ye, H. (2009). J. Biol. Chem.284, 8507–8515. [DOI] [PMC free article] [PubMed]
- Geiss-Friedlander, R. & Melchior, F. (2007). Nature Rev. Mol. Cell Biol.8, 947–956. [DOI] [PubMed]
- Matthews, B. W. (1968). J. Mol. Biol.33, 491–497. [DOI] [PubMed]
- Murray, J. M. & Carr, A. M. (2008). Nature Rev. Mol. Cell Biol.9, 177–182. [DOI] [PubMed]
- Otwinowski, Z. & Minor, W. (1997). Methods Enzymol.276, 307–326. [DOI] [PubMed]
- Palecek, J., Vidot, S., Feng, M., Doherty, A. J. & Lehmann, A. R. (2006). J. Biol. Chem.281, 36952–36959. [DOI] [PubMed]
- Pape, T. & Schneider, T. R. (2004). J. Appl. Cryst.37, 843–844.
- Pebernard, S., Wohlschlegel, J., McDonald, W. H., Yates, J. R. III & Boddy, M. N. (2006). Mol. Cell. Biol.26, 1617–1630. [DOI] [PMC free article] [PubMed]
- Potts, P. R. & Yu, H. (2005). Mol. Cell. Biol.25, 7021–7032. [DOI] [PMC free article] [PubMed]
- Sergeant, J., Taylor, E., Palecek, J., Fousteri, M., Andrews, E. A., Sweeney, S., Shinagawa, H., Watts, F. Z. & Lehmann, A. R. (2005). Mol. Cell. Biol.25, 172–184. [DOI] [PMC free article] [PubMed]
- Sreenath, H. K., Bingman, C. A., Buchan, B. W., Seder, K. D., Burns, B. T., Geetha, H. V., Jeon, W. B., Vojtik, F. C., Aceti, D. J., Frederick, R. O., Phillips, G. N. Jr & Fox, B. G. (2005). Protein Expr. Purif.40, 256–267. [DOI] [PubMed]
- Taylor, E. M., Copsey, A. C., Hudson, J. J., Vidot, S. & Lehmann, A. R. (2008). Mol. Cell. Biol.28, 1197–1206. [DOI] [PMC free article] [PubMed]
- Terwilliger, T. C. & Berendzen, J. (1996). Acta Cryst. D52, 749–757. [DOI] [PubMed]
- Ulrich, H. D. (2008). Mol. Cell, 32, 301–305. [DOI] [PubMed]
- Ulrich, H. D. (2009). Methods Mol. Biol.497, 3–16. [DOI] [PubMed]
- Zhao, X. & Blobel, G. (2005). Proc. Natl Acad. Sci. USA, 102, 4777–4782. [DOI] [PMC free article] [PubMed]