Abstract
Telomerase is an essential cellular ribonucleoprotein (RNP) that solves the end replication problem and maintains chromosome stability by adding telomeric DNA to the termini of linear chromosomes1–3. Genetic mutations that abrogate normal assembly of telomerase RNP cause human disease4. It is thus of fundamental and medical importance to decipher cellular strategies for telomerase biogenesis, which will require new insights into how specific interactions occur in a precise order along the RNP assembly pathway. Here, we demonstrate a single-molecule approach to dissect the individual assembly steps of telomerase. Direct observation of complex formation in real time revealed two sequential steps of protein-induced RNA folding, establishing a hierarchical RNP assembly mechanism: interaction with the telomerase holoenzyme protein p65 induces structural rearrangement of telomerase RNA, which in turn directs binding of the telomerase reverse transcriptase (TERT) to form the functional ternary complex. This hierarchical assembly process is facilitated by an evolutionarily conserved structural motif within the RNA. These results identify the RNA folding pathway during telomerase biogenesis and define the mechanism of action for an essential telomerase holoenzyme protein.
Telomerase RNP functions as a multi-subunit holoenzyme consisting of telomerase RNA, TERT, and additional protein cofactors. Catalytically active telomerase enzyme can be reconstituted from RNA and TERT in rabbit reticulocyte lysate (RRL) wherein general chaperone activities promote RNP assembly5,6. However, the endogenous process of telomerase biogenesis appears to require a more specific assembly pathway7. Supporting this view, cellular accumulation of telomerase RNP is promoted by a number of specific RNA-binding proteins, including dyskerin in vertebrates, Sm proteins in yeasts, and La-motif proteins in ciliates8–11. In this work, we exploited single-molecule fluorescence resonance energy transfer (FRET)12–14 to explore the mechanism for telomerase RNP biogenesis, using the ciliate Tetrahymena thermophila as a model system.
The Tetrahymena telomerase RNA is a 159 nucleotide transcript (Fig. 1a) that provides a template for telomere synthesis and functions in adapting the polymerase to its specialized task of reiterative repeat synthesis15–17. Using a refined DNA-splinted RNA ligation method18 (Supplementary Fig. 1), we strategically placed a FRET donor (Cy3) and acceptor (Cy5) flanking the regions important for interaction with TERT and the holoenzyme protein p65, a La-motif protein that promotes telomerase RNP accumulation in vivo11(Fig. 1a). To facilitate real time observation of telomerase RNP assembly, RNA was surface immobilized through an extension of stem II that does not perturb telomerase activity in vitro or in vivo19. Standard DNA primer extension assays performed with both labelled and unlabeled RNA yielded normal profiles of product synthesis (Fig. 1a, inset), demonstrating that dye labelling did not interfere with RNA function.
Figure 1.
Telomerase proteins p65 and TERT induce distinct conformational changes in telomerase RNA. a, Full-length telomerase RNA labelled with FRET donor (Cy3) and acceptor (Cy5). RNA molecules were immobilized on a streptavidin-coated surface by a biotin engineered onto an extension of stem II. The interaction sites with p65 (light red) and TERT (light green) are highlighted20,24. (inset) Telomerase primer extension assay with dye-labelled (L) and unlabeled (U) RNA or without RNA (−). Numbers to the right indicate the number of nucleotides added to generate each product. b, FRET histograms of RNA molecules in the absence of protein (grey), presence of 10 nM p65 (red) or 10 nM p65 + 32 nM TERT1–516 (green). A peak at FRET = 0 due to ~ 30% of molecules without active Cy5 (Supplementary Fig. 2) was removed from these histograms.
The FRET distribution for full-length telomerase RNA showed a single population centred at FRET = 0.29 (Fig. 1b and Supplementary Fig. 2a). Time traces of individual RNA molecules showed a stable FRET level without significant fluctuations except for transient excursions to FRET = 0 due to blinking of the acceptor dye (Supplementary Fig. 3). Addition of purified p65 gave rise to a second population with steady FRET level at 0.46 (Fig. 1b and Supplementary Fig. 3). Estimating the relative abundance of each population as a function of p65 concentration yielded a non-cooperative p65 binding isotherm with K = 1.3 nM (Supplementary Fig. 4), in agreement with ensemble characterization of p65-RNA affinity20. Similar results were obtained with telomerase RNA immobilized by a 5' extension (Supplementary Fig. 5), showing that surface attachment did not perturb telomerase RNP assembly. Control experiments designed to prevent a distance change between donor and acceptor dyes upon p65 binding showed that direct dye-p65 interaction, if present, did not induce any significant change in FRET (Supplementary Fig. 6). Furthermore, binding of p65 did not alter fluorescence intensities from the RNA singly labelled with Cy3 or Cy5 at the same locations (data not shown). Taken together, these results indicate that the p65-induced FRET increase represents a conformational change within the RNA, wherein the ends of stem I and stem IV are brought closer together in the tertiary RNA fold.
To investigate the assembly of p65-RNA-TERT ternary complex, we used a purified TERT polypeptide containing the N-terminal 516 amino acids (TERT1–516). This polypeptide contains all of the primary sites of RNA-TERT interaction, but unlike the full-length TERT, can be expressed in a soluble recombinant form20. Addition of TERT1–516 to the p65-RNA complex further shifted the FRET value to a steady level of 0.65 (Fig. 1b and Supplementary Fig. 3), with a binding affinity of K = 14.4 nM (Supplementary Fig. 4). The binding of TERT1–516 did not alter fluorescence intensities from RNA singly labelled with Cy3 or Cy5 (data not shown), suggesting the TERT1–516-induced FRET change signifies another bona fide structural rearrangement in telomerase RNA. Notably, FRET measurements on catalytically active telomerase RNPs assembled in RRL with p65 and full-length TERT gave a similar FRET distribution (compare Supplementary Fig. 7a with Fig 1b), demonstrating that the high FRET conformation (FRET = 0.65) observed in the p65-RNA-TERT1–516 ternary complex represents the RNA structure in the functional enzyme.
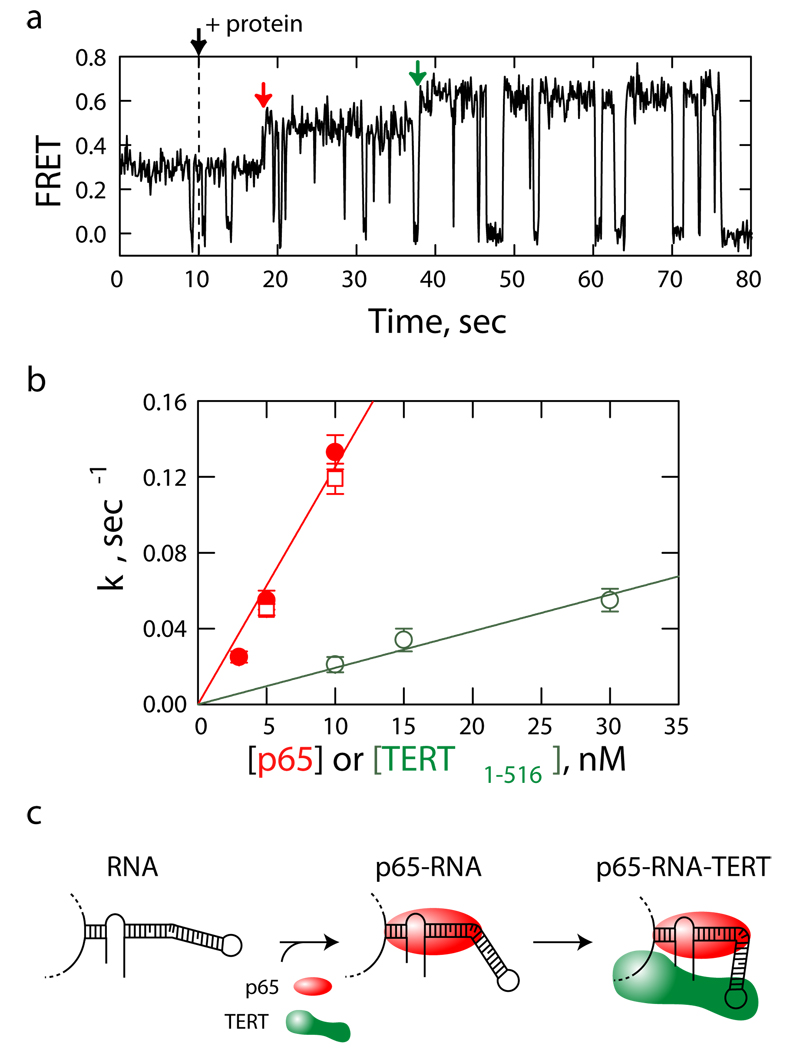
The distinct FRET signatures of the telomerase RNA alone, the p65-RNA complex, and the p65-RNA-TERT1–516 ternary complex allowed us to examine the assembly of individual telomerase RNP particles in real time. To this end, we added protein mixtures containing both p65 and TERT1–516 to telomerase RNA and monitored the FRET signal of individual RNA molecules during the assembly process. The majority of assembly trajectories (74%, n = 173) displayed a prominent kinetic intermediate with FRET = 0.46, equivalent to that measured from p65-RNA complex, before attaining a final state with FRET = 0.65 indicative of the p65-RNA-TERT ternary complex (Fig. 2a). The remaining minor fraction of assembly trajectories exhibited less defined intermediates with irregular FRET fluctuations (Supplementary Fig. 8). The fraction of molecules assembled through the well-defined two-step (0.29 ➔ 0.46 ➔ 0.65) pathway did not vary substantially with altered ratios of p65 to TERT1–516 concentrations (at the p65:TERT1–516 ratio of 1:1, 1:3, and 1:10, the fraction was 73% (n = 37), 76% (n = 101) and 69% (n = 35), respectively). The rates of conversion from FRET = 0.29 to 0.46 and from FRET = 0.46 to 0.65 increased linearly with p65 and TERT1–516 concentrations, respectively (Fig. 2b). These results suggest a hierarchical telomerase assembly process directed by sequential steps of protein-induced RNA folding: assembly is initiated by p65 binding, stabilizing a RNA structural intermediate, which in turn promotes functional co-assembly of TERT with telomerase RNA (Fig. 2c).
Figure 2.
Real-time assembly of individual telomerase RNP complexes. a, A single molecule FRET trajectory showing hierarchical RNP assembly upon addition of a protein mixture of p65 (10 nM) and TERT1–516 (10 nM), characterized by a p65-induced FRET change (red arrow) followed by a second FRET transition upon complete assembly of the ternary p65-RNA-TERT1–516 complex (green arrow). Excursions to FRET = 0 correspond to blinking of Cy5 (Supplementary Fig. 3) b, The conversion rate (k) from FRET = 0.29 to 0.46 (red) or from FRET = 0.46 to 0.65 (green) as a function of p65 or TERT1–516 concentration, respectively. Open red squares indicate conversion rates measured in the presence of p65 only and solid red circles indicate those measured in the presence of both p65 and TERT1–516. Error bars indicate the s.e.m. (n ≥ 32 for all conditions). Linear fits of the data (red and green lines) yield binding rate constants of 1.3 × 107 and 2 × 106 M−1sec−1 for p65 and TERT1–516, respectively. c, The interaction of p65 (red) induces compaction in stem I–IV of the RNA, followed by co-assembly with TERT and concomitant additional RNA folding in the ternary complex. The conserved GA bulge introduces a kink in stem IV in the absence of protein25,26. The additional bending in stem IV in the presence of proteins is drawn to indicate RNA compaction as measured by FRET, and is not meant to represent specific structural orientation. Note that tertiary RNA contacts not shown in the model may be formed within the telomerase holoenzyme.
To structurally characterize the p65-induced assembly intermediate, we generated a series of truncated constructs comprised of telomerase RNA stems I and IV, harbouring the primary p65 interaction sites20 (Fig. 3a–e). When FRET donor and acceptor were incorporated in stem I and distal stem IV (approximating the labelling sites in the full-length RNA construct), we observed a significant increase in FRET upon p65 binding (Fig. 3a), consistent with results described above (Fig. 1b). Experiments with FRET donor and acceptor relocated to flank the ACAA linker (Fig. 3b) or the di-nucleotide GA bulge (Fig. 3c) indicated that the primary p65-induced structural rearrangement occurs between proximal and distal stem IV linked by the GA bulge rather than between stem I and stem IV. Furthermore, substitution (Fig. 3d, GA➔UU) or deletion (ΔGA, Fig. 3e) of the GA bulge abolished any stable p65-induced FRET change. At the concentration of p65 used here (30 nM), the majority of RNA molecules were bound to p65 (Supplementary Fig. 9). Thus, the p65-induced RNA conformational change occurs within stem IV and requires the central stem IV GA bulge, which is conserved across all Tetrahymena species21.
Figure 3.
The p65-induced RNA structural change occurs within stem IV and requires the conserved GA bulge. a–e, Schematic illustrations of stem I–IV constructs (right panels) and their corresponding FRET histograms (left panels) in the absence (grey) or presence (red) of p65. Stem I–IV constructs were labelled with FRET donor (green spheres) and acceptor (red spheres) and surface-immobilized via a 5' RNA extension (thick black line) annealed to a biotinylated complementary DNA oligonucleotide (pink line).
To probe whether the p65-induced RNA conformation is an essential intermediate during hierarchical telomerase RNP biogenesis, we compared the assembly observations described above to reactions that lacked p65 or lacked the GA bulge in telomerase RNA. Telomerase RNA in the presence of TERT1–516 alone did not yield the stable high FRET state (FRET = 0.65) observed for the p65-RNA-TERT1–516 ternary complex (compare Fig. 4a with Fig. 1b). Instead, the FRET trajectories showed characteristic fluctuations with amplitudes and rates that were independent of TERT1–516 concentration (1–100 nM) (Fig. 4b and Supplementary Fig. 10), indicating that the FRET dynamics were not due to repetitive binding and dissociation of protein. The fraction of molecules showing such fluctuations increased with the concentration of TERT1–516 (data not shown), with an affinity comparable to the previously determined value for telomerase RNA and TERT1–516(20). These results demonstrate that the RNA fold in the RNA-TERT1–516 complex is structurally dissimilar to that in the p65-RNA-TERT1–516 ternary complex. Addition of p65 to preformed RNA-TERT1–516 complexes recovered the stable high-FRET conformation (data not shown), suggesting that p65 can rescue misassembled RNA-TERT complexes by directing productive interactions between TERT and telomerase RNA. Furthermore, p65 substantially stimulated complex formation and catalytic activity of telomerase RNPs reconstituted in RRL (Supplementary Fig. 7). Thus the role of p65 cannot be fully recapitulated by heterologous factors within RRL.
Figure 4.
The p65 protein and conserved GA bulge in stem IV of telomerase RNA are required for telomerase assembly. a, FRET histograms of full-length RNA in the absence (grey) or presence (green) of 10 nM TERT1–516. b, A single-molecule FRET trajectory of telomerase RNA in the presence of 10 nM TERT1–516. c, FRET histograms of full-length RNA lacking GA bulge (ΔGA) in the absence of protein (grey), presence of 10 nM p65 (red), or presence of 10 nM p65 + 30 nM TERT1–516 (green). d, A singlemolecule FRET trajectory of ΔGA RNA in the presence of 10 nM p65.
When p65 was added to full-length telomerase RNA lacking the GA bulge, no stable FRET change was observed (Fig. 4c). Instead, the FRET traces only transiently visited a higher FRET state (Fig. 4d), with a rate independent of p65 concentration (data not shown), suggesting that disruption of the GA bulge renders the stem IV region too rigid to be folded by p65 in a stable manner. Subsequent addition of TERT1–516 did not substantially change the FRET dynamics or induce the FRET = 0.65 conformation indicative of the functional p65-RNA-TERT complex (compare Fig. 4c with Fig. 1b). Furthermore, deletion or UU substitution of the conserved GA bulge in vivo prevented incorporation of telomerase RNA into a stable RNP, resulting in the failure of these RNA variants to accumulate relative to endogenous telomerase RNA (Supplementary Fig. 11). Taken together, these results show that the p65-induced RNA conformational change is critical for formation of the native p65-RNA-TERT complex.
In summary, our experiments directly demonstrate a hierarchical assembly mechanism for telomerase RNP in which the protein subunits mould a specific RNA tertiary structure in a stepwise fashion. First, the telomerase holoenzyme protein p65 induces a marked structural change within the stem IV region of the RNA, a domain important for processive telomere synthesis15–17. This stable structural rearrangement strictly requires the central stem IV GA bulge, accounting for the phylogenetic conservation of this sequence motif. Second, the RNA conformation in the p65-RNA complex is further altered by binding of TERT, resulting in a compact RNA tertiary fold within the functional telomerase RNP. By monitoring the assembly of individual RNP complexes in real time, we demonstrated that the p65-induced RNA conformation is a fundamental structural intermediate during hierarchical telomerase assembly. These results provide a likely mechanism for telomerase RNP assembly in vivo, wherein a holoenzyme protein repositions distant RNA binding sites for TERT, promoting correct co-assembly of the catalytic subunit TERT with telomerase RNA (Fig. 1a and Fig. 2c). Consistent with this notion, removal of p65(11) or mutation of the GA bulge prevents incorporation of telomerase RNA into a stable RNP in vivo (Supplementary Fig. 11). An analogous hierarchical biogenesis pathway has been proposed for the ribosome22,23, which contains a RNA rather than protein catalytic centre. Hierarchical steps of protein-mediated RNA folding might thus represent a general assembly strategy for cellular RNPs, providing multiple layers of control to circumvent formation of long-lived misassembled intermediates.
Methods
NA and protein preparation
Full-length FRET-labelled RNAs were prepared by a refined DNA-splinted RNA ligation method detailed in Supplementary methods and Supplementary Fig. 1. Preparation of truncated RNA constructs is described in Supplementary methods. Histidine tagged p65 and TERT1–516 proteins were expressed and purified from E. coli as described20.
Single-molecule FRET
Single-molecule FRET measurements were performed as described in Supplementary Methods. The FRET value is defined as IA / (IA + ID), where ID and IA are the fluorescence intensities measured in the donor and acceptor channel, respectively.
RRL reconstitution of telomerase RNP and in vivo RNA accumulation assays
Protein expression in RRL was performed by standard methods (Supplementary methods) using the pCITE plasmid system as described16. In vivo telomerase RNA accumulation assays are described in Supplementary methods.
Supplementary Material
Acknowledgements
The authors thank M. Bates for LabView software for data acquisition. This work was supported in part by NIH (GM 068518) and Packard Foundation (to X.Z.), and NIH (GM 54198, to K.C.). X.Z. is a Howard Hughes Medical Institute investigator. MDS is a NIH Ruth L. Kirschstein NSRA Fellow.
Footnotes
Supplementary Information accompanies the paper on www.nature.com/nature.
Competing interests statement The authors declare that they have no competing financial interests.
References
- 1.Greider CW, Blackburn EH. Tracking telomerase. Cell. 2004;116:S83–S86. doi: 10.1016/s0092-8674(04)00053-4. [DOI] [PubMed] [Google Scholar]
- 2.Cech TR. Beginning to understand the end of the chromosome. Cell. 2004;116:273–279. doi: 10.1016/s0092-8674(04)00038-8. [DOI] [PubMed] [Google Scholar]
- 3.Collins K. The biogenesis and regulation of telomerase holoenzymes. Nat. Rev. Mol. Cell Biol. 2006;7:484–494. doi: 10.1038/nrm1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wong JM, Collins K. Telomere maintenance and disease. Lancet. 2003;362:983–988. doi: 10.1016/S0140-6736(03)14369-3. [DOI] [PubMed] [Google Scholar]
- 5.Weinrich SL, et al. Reconstitution of human telomerase with the template RNA component hTR and the catalytic protein subunit hTRT. Nat. Genet. 1997;17:498–502. doi: 10.1038/ng1297-498. [DOI] [PubMed] [Google Scholar]
- 6.Holt SE, et al. Functional requirement of p23 and Hsp90 in telomerase complexes. Genes Dev. 1999;13:817–826. doi: 10.1101/gad.13.7.817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Harrington L. Biochemical aspects of telomerase function. Cancer Lett. 2003;194:139–154. doi: 10.1016/s0304-3835(02)00701-2. [DOI] [PubMed] [Google Scholar]
- 8.Mitchell JR, Wood E, Collins K. A telomerase component is defective in the human disease dyskeratosis congenita. Nature. 1999;402:551–555. doi: 10.1038/990141. [DOI] [PubMed] [Google Scholar]
- 9.Seto AG, Zaug AJ, Sobel SG, Wolin SL, Cech TR. Saccharomyces cerevisiae telomerase is an Sm small nuclear ribonucleoprotein particle. Nature. 1999;401:177–180. doi: 10.1038/43694. [DOI] [PubMed] [Google Scholar]
- 10.Aigner S, Postberg J, Lipps HJ, Cech TR. The Euplotes La motif protein p43 has properties of a telomerase-specific subunit. Biochemistry. 2003;42:5736–5747. doi: 10.1021/bi034121y. [DOI] [PubMed] [Google Scholar]
- 11.Witkin KL, Collins K. Holoenzyme proteins required for the physiological assembly and activity of telomerase. Genes Dev. 2004;18:1107–1118. doi: 10.1101/gad.1201704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stryer L, Haugland RP. Energy transfer: a spectroscopic ruler. Proc. Nat. Acad. Sci. USA. 1967;58:719–726. doi: 10.1073/pnas.58.2.719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ha T, et al. Probing the interaction between two single molecules: fluorescence resonance energy transfer between a single donor and a single acceptor. Proc. Natl. Acad. Sci. USA. 1996;93:6264–6268. doi: 10.1073/pnas.93.13.6264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhuang X. Single-molecule RNA science. Annu. Rev. Biophys. Biomol. Struct. 2005;34:399–414. doi: 10.1146/annurev.biophys.34.040204.144641. [DOI] [PubMed] [Google Scholar]
- 15.Sperger JM, Cech TR. A stem-loop of Tetrahymena telomerase RNA distant from the template potentiates RNA folding and telomerase activity. Biochemistry. 2001;40:7005–7016. doi: 10.1021/bi0103359. [DOI] [PubMed] [Google Scholar]
- 16.Lai CK, Miller MC, Collins K. Roles for RNA in telomerase nucleotide and repeat addition processivity. Mol. Cell. 2003;11:1673–1683. doi: 10.1016/s1097-2765(03)00232-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mason DX, Goneska E, Greider CW. Stem-loop IV of tetrahymena telomerase RNA stimulates processivity in trans. Mol. Cell Biol. 2003;23:5606–5613. doi: 10.1128/MCB.23.16.5606-5613.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Moore MJ, Query CC. Joining of RNAs by splinted ligation. Methods Enzymol. 2000;317:109–123. doi: 10.1016/s0076-6879(00)17009-0. [DOI] [PubMed] [Google Scholar]
- 19.Cunningham DD, Collins K. Biological and biochemical functions of RNA in the tetrahymena telomerase holoenzyme. Mol. Cell Biol. 2005;25:4442–4454. doi: 10.1128/MCB.25.11.4442-4454.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Prathapam R, Witkin KL, O'Connor CM, Collins K. A telomerase holoenzyme protein enhances telomerase RNA assembly with telomerase reverse transcriptase. Nat. Struct. Mol. Biol. 2005;12:252–257. doi: 10.1038/nsmb900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ye AJ, Romero DP. Phylogenetic relationships amongst tetrahymenine ciliates inferred by a comparison of telomerase RNAs. Int. J. Syst. Evol. Microbiol. 2002;52:2297–2302. doi: 10.1099/00207713-52-6-2297. [DOI] [PubMed] [Google Scholar]
- 22.Adilakshmi T, Ramaswamy P, Woodson SA. Protein-independent folding pathway of the 16S rRNA 5' domain. J. Mol. Biol. 2005;351:508–519. doi: 10.1016/j.jmb.2005.06.020. [DOI] [PubMed] [Google Scholar]
- 23.Talkington MW, Siuzdak G, Williamson JR. An assembly landscape for the 30S ribosomal subunit. Nature. 2005;438:628–632. doi: 10.1038/nature04261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.O'Connor CM, Lai CK, Collins K. Two purified domains of telomerase reverse transcriptase reconstitute sequence-specific interactions with RNA. J. Biol. Chem. 2005;280:17533–17539. doi: 10.1074/jbc.M501211200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Richards RJ, et al. Structural study of elements of Tetrahymena telomerase RNA stem-loop IV domain important for function. RNA. 2006 doi: 10.1261/rna.112306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen Y, et al. Structure of stem-loop IV of Tetrahymena telomerase RNA. EMBO J. 2006 doi: 10.1038/sj.emboj.7601195. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.