Abstract
Some breast cancers have been shown to contain a small fraction of cells characterized by CD44+/CD24−/low cell-surface antigen profile that have high tumor-initiating potential. In addition, breast cancer cells propagated in vitro as mammospheres (MSs) have also been shown to be enriched for cells capable of self-renewal. In this study, we have defined a gene expression signature common to both CD44+/CD24−/low and MS-forming cells. To examine its clinical significance, we determined whether tumor cells surviving after conventional treatments were enriched for cells bearing this CD44+/CD24−/low-MS signature. The CD44+/CD24−/low-MS signature was found mainly in human breast tumors of the recently identified “claudin-low” molecular subtype, which is characterized by expression of many epithelial-mesenchymal-transition (EMT)-associated genes. Both CD44+/CD24−/low-MS and claudin-low signatures were more pronounced in tumor tissue remaining after either endocrine therapy (letrozole) or chemotherapy (docetaxel), consistent with the selective survival of tumor-initiating cells posttreatment. We confirmed an increased expression of mesenchymal markers, including vimentin (VIM) in cytokeratin-positive epithelial cells metalloproteinase 2 (MMP2), in two separate sets of postletrozole vs. pretreatment specimens. Taken together, these data provide supporting evidence that the residual breast tumor cell populations surviving after conventional treatment may be enriched for subpopulations of cells with both tumor-initiating and mesenchymal features. Targeting proteins involved in EMT may provide a therapeutic strategy for eliminating surviving cells to prevent recurrence and improve long-term survival in breast cancer patients.
Keywords: CD44+/CD24−/low markers, gene expression signature, tumor-initiating cancer cells, mesenchymal features, treatment resistance
Despite recent improvements in breast cancer mortality, many patients relapse after an initial response to conventional endocrine therapy and chemotherapies. Several alternative but not necessarily mutually exclusive hypotheses have been proposed to explain this treatment failure and recurrence. In particular, it has been suggested that a small subpopulation of cells within tumors, often designated as “tumor-initiating cells” or “cancer stem cells,” may be resistant to therapy and hence may reinitiate tumor growth after treatment (1). The population containing these cells can be isolated by FACS using specific cell surface markers. For example, in cells from metastatic pleural effusions, the CD44+/CD24−/low/lineage− subpopulation was shown to be enriched for tumor-initiating cells (2). Recently, we have reported that this CD44+/CD24−/lowsubpopulation of cells appears to be more relatively resistant to chemotherapy in paired primary human breast cancer biopsies (3). This increase was associated with an enhanced mammosphere (MS)-forming ability, an in vitro surrogate assay of self-renewal capacity (3).
Global gene expression analyses have identified at least four major subtypes of human breast cancers (luminal A/B, basal-like, and ERBB2-enriched) (4, 5). More recently Herschkowitz et al. (6) described another subtype, involving relatively uncommon breast cancers, termed “claudin-low,” which are characterized by comparatively high expression of mesenchymal markers such as vimentin and low expression of epithelial markers, notably claudins and E-cadherin. During the course of normal development, the transdifferentiation of epithelial cells to a more mesenchymal state (7–13) known as epithelial-to-mesenchymal transition (EMT), is an essential process; this process has also been implicated in cancer progression (14). Indeed, an increased representation after therapy of cancer cells expressing mesenchymal markers, notably those described in claudin-low tumors, may account for treatment resistance (15–17). It is not known directly at this time whether the claudin-low tumors show elevated resistance to therapy.
Defining the regulatory pathways in subpopulations of residual tumor cells that survive conventional therapy could ultimately provide important new therapeutic targets. With this objective in mind, we first derived a gene signature from CD44+/CD24−/low/lineage− cells and formed cancer MS cells, both isolated from human breast cancers. We then compared this CD44+/CD24−/low-MS signature to the previously defined intrinsic subtypes of breast cancer to determine whether there was any subtype that was highly enriched for this subpopulation. To examine the signature's clinical and therapeutic significance, we evaluated it in breast tumors before and after therapy (letrozole or docetaxel), doing so with the hypothesis that the tumor cells surviving after treatment would have an increased expression of the CD44+/CD24−/low-MS signature. Finally, relevant key markers from this signature were confirmed in additional paired before and after treatment patient specimens.
Results
Gene Transcription Patterns in Breast Cancer Cells with Tumor-Initiating Potential.
To define a gene expression signature, we used two methods to obtain breast cancer subpopulations that have been suggested previously to be enriched for putative tumor-initiating cells: CD44+/CD24−/low markers and formed cancer MSs (2). These analyses involved 36 tumors representing all subtypes of breast cancer (18 luminal A/B, 13 basal-like, and 5 ERBB2-enriched) (Fig. S1). We performed comparative gene expression profiling in the populations that have been shown by others to be enriched for tumor-initiating cells (CD44+/CD24−/low or formed cancer MSs) vs. non-tumor-initiating cells (all “other” flow-sorted fractions or bulk tumor, respectively), and then analyzed to look for significant overlap between the gene expression patterns of cells isolated by the two alternative enrichment methods.
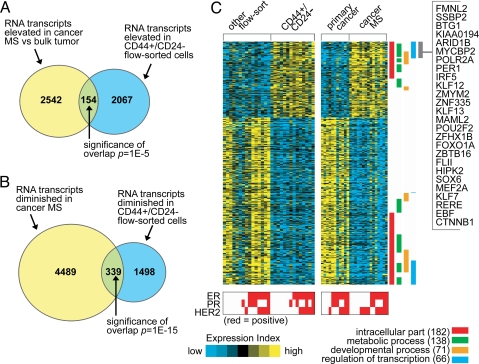
In the first comparison (consisting of 14 CD44+/CD24−/low vs. 15 other profiles, representing 20 patients including nine patient pairs), 2,221 RNA transcripts (1,424 named genes) were elevated (P < 0.01 unpaired, two-sided t test; fold change >1.5; FDR ≈0.2) in the flow-sorted CD44+/CD24−/low vs. other cells. In the second comparison (consisting of 15 MS vs. 11 primary cancer profiles, representing 16 patients including 10 patient pairs), 2,696 transcripts (1,890 named genes) were elevated (P < 0.01 unpaired, two-sided t test; fold change >1.5; FDR ≈0.25) in the MSs vs. primary cancers. The numbers of elevated genes appearing in both of the enrichment methods exceeded the chance overlap expected (P = 1E − 5, one-sided Fisher's exact test) (Fig. 1 A, Fig. S2 A, and SI Text). Between the transcripts with decreased expression, 339 transcripts (263 genes) significantly overlapped (P = 1E − 15, one-sided Fisher's exact test) (Fig. 1B, Fig. S2 B, and SI Text). Therefore, we defined here a “CD44+/CD24−/low-MS gene signature” which comprised the relative “up” and “down” patterns of the 493 transcripts (154 overexpressed and 339 underexpressed) present in the significant overlap between both comparisons (heat map in Fig. 1C, complete gene list in Dataset S1).
Fig. 1.
A gene transcription signature of breast cancer cells with putative tumor-initiating potential. (A) Venn diagram of the intersection between genes elevated in cancer mammospheres (MSs, rich in CD44+/CD24−/low cells) compared with primary cancers and genes elevated in CD44+/CD24−/low cells obtained from FACS assay (P < 0.01, fold change >1.5 for each comparison). P value for the overlap between the two gene sets by one-sided Fisher's exact test. (B) As with A but for genes lower in cancer MSs and genes lower in CD44+/CD24−/low flow-sorted cells. (C) Heat map of genes in the CD44+/CD24−/low-MS signature (from parts A and B). Each row represents a transcript; each column, a sample (yellow: high expression). Associations of the genes with selected Gene Ontology (GO) annotation terms are indicated. Genes in the indicated region are listed by name.
The CD44+/CD24−/low-MS Signature Is Enriched in Human Breast Tumors of the Claudin-Low Molecular Subtype.
Most human breast cancers can be robustly classified into one of the intrinsic subtypes: luminal A and B, HER2-enriched, basal-like, and claudin-low (4–6). The claudin-low subtype identified by Herschkowitz et al. (6) is characterized by the high expression of mesenchymal genes and the low expression of cell–cell contact genes, such as claudins 3, 4, and 7, and E-cadherin. We compared the CD44+/CD24−/low-MS signature to each subtype to determine whether any group showed enrichment. For each tumor in the Herschkowitz dataset, we derived an “R value” in relation to our CD44+/CD24−/low-MS signature, with the R value being a measure for how much each individual Herschkowitz tumor manifested our signature (see Methods).
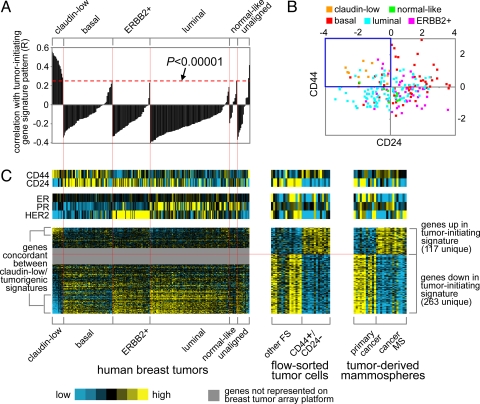
Only the previously defined claudin-low group showed a clear association with the CD44+/CD24−/low-MS signature (Fig. 2A). We obtained the same results using a number of other analytical approaches, including one-sided Fisher's exact tests and GSEA (see SI Text and Fig. S3). Furthermore, relative to the other tumors, claudin-low tumors showed low mRNA expression of CD24 and high mRNA expression of CD44 (Fig. 2B), consistent with the notion that these tumors are enriched for tumor-initiating cells bearing these markers. The above correlation patterns were further evident when viewing a heat map of the genes in the CD44+/CD24−/low-MS signature within the published Herschkowitz dataset (Fig. 2C).
Fig. 2.
The CD44+/CD24−/low-MS gene signature is enriched in human breast tumors of the claudin-low molecular subtype. (A) Correlation (R value, see Methods) between the CD44+/CD24−/low-MS signature pattern and each tumor in the gene expression profile dataset by Herschkowitz et al. (6) (claudin-low, basal, ERBB2+, luminal, normal-like; averages centered on the mean centroid of the groups). R values above red dotted line are significant at P < 0.00001. (B) Scatter plot of CD24 versus CD44 mRNA expression for the Herschkowitz tumors. (C) Heat map of the corresponding patterns of the CD44+/CD24−/low-MS signature genes within the Herschkowitz tumors. The order of the Herschkowitz tumors is the same for A and C; the order of the genes is the same across datasets.
CD44+/CD24−/low-MS and Claudin-Low Gene Signatures Are Enriched in Human Breast Tumors After Treatment with Endocrine Therapy or Chemotherapy.
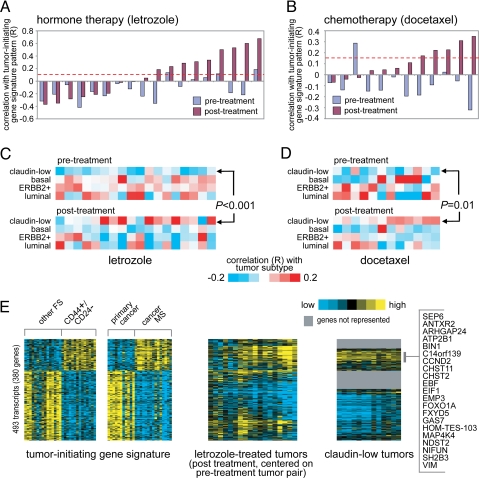
We have reported that breast cancers after chemotherapy are enriched for CD44+/CD24−/low-MS cells by flow cytometric analysis (3). In that earlier study, postchemotherapy specimens showed an increase in cells bearing CD44+/CD24−/low and an increase in MS-forming efficiency (3). If tumor-initiating cells are resistant to chemotherapy and endocrine therapy, then we would expect to see strong enrichment of the CD44+/CD24−/low-MS signature in both the endocrine- and chemotherapy-treated residual tumors. We tested this notion in two datasets. First, we profiled 36 ER-positive human breast tumors from patients receiving neoadjuvant endocrine therapy; these represented 18 patient pairs before and after treatment (3 months) with the aromatase inhibitor letrozole. We computed an R value in relation to the CD44+/CD24−/low-MS signature pattern, to measure the degree of signature enrichment (see SI Text and Fig. S4). The letrozole posttreatment tumors had higher R values than the pretreatment tumors (P < 0.01, paired t test) (Fig. 3A).
Fig. 3.
The CD44+/CD24−/low-MS gene signature is enriched in a subset of human breast tumors after treatment with hormone therapy or chemotherapy. (A) Correlation between the CD44+/CD24−/low-MS signature pattern and each of 36 human breast tumor profiles, representing 18 pairs before and after treatment with letrozole. R values above red dotted line are significant at P < 0.01. (B) Correlation between the CD44+/CD24−/low-MS signature pattern and each of 24 tumor profiles, representing 12 pairs before and after treatment with docetaxel. (C) Heat map of the correlations between each letrozole-treated tumor profile (part A) and the average expression for each of the four major Herschkowitz molecular tumor profile subtypes (claudin-low, basal, ERBB2+, luminal; averages centered on the mean centroid of the groups). P value comparing pretreatment and posttreatment R values for the claudin-low group by paired t test. (D) As with part C, but for the docetaxel-treated tumors (B). (E) Heat maps of the corresponding patterns of the CD44+/CD24−/low-MS signature genes within the letrozole posttreatment tumors (each tumor centered on corresponding pretreatment pair) and the Herschkowitz claudin-low tumors (centered on mean centroid of tumor groups). Gene ordering is the same across datasets.
Next, we examined our published dataset of predocetaxel and postdocetaxel treatment, representing 12 patient pairs (18, 19). Again, we found that postdocetaxel tumors had higher correlations with the CD44+/CD24−/low-MS signature (P = 0.01, paired t test) (Fig. 3B). Thus, the CD44+/CD24−/low-MS signature was increased after either chemotherapy or endocrine therapy. Similarly, the claudin-low patterns were increased in posttreated compared with pretreated tumors in both the letrozole and docetaxel cohorts (P < 0.001 and P = 0.01, respectively, comparing R values by paired t test) (Fig. 3 C and D). We also examined the CD44+/CD24−/low-MS signature as heat maps in both the letrozole-treated and claudin-low samples (Fig. 3E). We found that the overlapping patterns between the letrozole treatment signature and our CD44+/CD24−/low-MS signature were significant by both one-sided Fisher's exact tests and GSEA (SI Text). Likewise, the overlap between the letrozole-treated tumors and the CD44+/CD24−/low-MS signature was largely shared by the claudin-low tumors in the heat map representation (Fig. 3E). Notably, all of the claudin-low tumors in the Herschkowitz dataset had been sampled from patients before therapy, yet this claudin-low profile was more evident after therapy.
Mesenchymal-Associated Genes Are Increased in Postletrozole Breast Cancer Specimens.
The association between the CD44+/CD24−/low-MS signature and the claudin-low subtype suggests that residual tumor cells may display mesenchymal features. We therefore examined a number of mesenchymal markers described in a literature review (20) and found that, as predicted, a number of these markers were elevated (FN1, SNAI2, VIM, FOXC2, MMP2, and MMP3) or diminished (CDH1 and DSP) at the RNA level in the cancer MSs vs. primary bulk tumor (Fig. 4A). The overlap between the mesenchymal up genes (20) and the cancer MS up genes was statistically significant (P < 0.001, one-sided Fisher's exact test). These patterns were not as clearly evident in the CD44+/CD24−/low vs. other flow sort profile dataset, with only some mesenchymal genes elevated (VIM) or decreased (CDH1 and DSP) significantly (Fig. 4A).
Fig. 4.
Specific markers of mesenchymal cells are overexpressed in both CD44+/CD24−/low-MS cells and letrozole-treated patient tumors. (A) For the flow sort and cancer MS profile datasets, heat map of previously associated mesenchymal genes in a review by Lee et al. (31) (genes in bold, significant with P < 0.01; the entire set of mesenchymal-associated proteins from the review that we could map to specific genes in our dataset are represented). (B) Protein expression of vimentin and pan-cytokeratin (CK) in a patient tumor after letrozole treatment. (C) Immunofluorescence analysis of vimentin (red) and pan-cytokeratin (green) after chemotherapy. Yellow arrows mark double-positive cells, whereas red and green arrows mark vimentin and CK positive epithelial cells, respectively. (D) MMP2mRNA levels in tumors before and after letrozole. (P values in B and C by paired t test).
We next analyzed the expression of a number of these identified mesenchymal genes (VIM, E-cadherin, and MMP2) in additional paired letrozole-treated breast cancer specimens. We selected these markers based on the availability of commercial antibodies and established methods for detecting them. Protein validation of additional genes like SNAI2 was not performed, because procedures for detecting these by immunohistochemistry of these gene products had not been established. We performed immunoflorescence analysis of the mesenchymal gene vimentin (red) with the epithelial pan-cytokeratin marker (green). A subset of tumor epithelial cells postletrozole were both cytokeratin and vimentin positive (Fig. 4B), suggesting that these may represent epithelial cancer cells that have acquired some mesenchymal features.
To quantify this apparent increase in mesenchymal properties in epithelial cells, we performed automated quantitative analysis (AQUA) of vimentin expression in the epithelial compartments of the tumor in 26 paired patient samples that were delineated by its/their cytokeratin expression. The percentage of vimentin+/CK+ double-positive cells in biopsies taken before vs. after letrozole treatment showed a statistically significant increase (P = 0.034, paired t test) (Fig. 4C). The expected decrease in expression of E-cadherin was not observed in postletrozole treated specimens, possibly because of the difficulty in quantifying decreases in biomarker expression by immunohistochemistry (IHC) in tumors with low baseline values. In addition, we quantified RNA levels by quantitative reverse transcription PCR for MMP2 in tumors before and after letrozole therapy from 60 patients, and showed an increased expression both at 10–14 days and at 3 months (P < 0.0001 and P < 0.00001, respectively, paired t test) (Fig. 4D).
Discussion
The present work has led to the identification of a unique molecular signature of therapy-resistant CD44+/CD24−/low-MS cells. We derived our signature by using two published enrichment methods for putative tumor-initiating cells (2): flow cytometric sorting for selected markers (CD44+/CD24−/low) and a biological functional assay for self-renewal (formed cancer MSs). Our previous study showed an increase in the CD44+/24−/low cell population by flow cytometric analysis in primary cancers surviving neoadjuvant chemotherapy, which was associated with an increase in mammosphere-forming ability and an increase in xenograft outgrowths in immunocompromised mice; the latter measurement, however, was not statistically significant (3). In the present study, we demonstrate a far stronger manifestation of the CD44+/CD24−/low-MS signature after both neoadjuvant chemotherapy, and endocrine therapy. Taken together, these data provide evidence that residual after conventional treatment breast tumors are enriched for cells exhibiting both tumor-initiating and mesenchymal features. The gene expression patterns of the CD44+/CD24−/low-MS cells showed significant correlation with the subtype of breast cancers classified as claudin-low, which have been found to exhibit up-regulation of mesenchymal genes (6). One possible explanation of this correlation is that conventional therapies yield tumors that are enriched for “tumor-initiating” cells, which may be more resistant to these treatments (3); accordingly, the tumor cells surviving treatment also show more mesenchymal-like features that are characteristic of the cells in claudin-low tumors.
Alternatively, it is possible that stromal reactions to therapy result in an increase in tumor-associated cells expressing mesenchymal genes. This appears to be less likely, because the samples analyzed here were obtained from women who received a variety of treatments, including both chemotherapy and endocrine treatments. Whereas the claudin-low tumors described by Herschkowitz et al. analyzed tumors prepared from patients before therapy, our own data indicate that cells having a similar profile are more evident after therapy. The mechanistic basis for these observations has yet to be determined. Because the claudin-low tumors have only been just described, it is not known whether this subtype is more or less resistant to conventional therapy. However, a recent paper described similarities in gene expression patterns between metaplastic cancers and the “claudin low” subtype; metaplastic cancers are known to be more resistant to conventional chemotherapy (18).
As mentioned, studies have identified increased mesenchymal features in basal-like breast cancers (19). These studies in cell lines have suggested that the switch to a mesenchymal phenotype might be related to the aggressiveness and metastatic spread of tumors. In this regard, one of the genes elevated in the CD44+/CD24−/low-MS signature, FOXC2, has been suggested to play a key role in orchestrating this mesenchymal phenotype (20). Recently, the same authors noted that induction of EMT in immortalized human mammary epithelial cells resulted in the acquisition of mesenchymal traits and expression of stem-cell markers (21). Our current observations suggest that the residual tumor cells surviving after therapy may also display mesenchymal features in human clinical specimens. The association of the mesenchymal phenotype with treatment resistance has been inferred by several articles describing the association between resistance to endocrine therapy (tamoxifen) (22) and chemotherapy (oxaliplatin, paclitaxel, and others) and a mesenchymal phenotype (15–17). Here, we provide clinical evidence that markers of the mesenchymal phenotype, assessed by both protein and gene expression, are higher in tumors after chemotherapy and endocrine therapy.
These data support a growing body of evidence for a mesenchymal-like phenotype in some breast cancers that may be responsible for invasion, metastasis, and possibly, treatment resistance (15–17). In addition, recent evidence indicates that CD44, whose positive expression selects for tumor-initiating cells, may be important in cancer cell proliferation, migration, and metastasis (23). Our results demonstrate that these residual tumors after various treatments also display increased expression of some mesenchymal markers. The future challenge will be to use our CD44+/24−/low cell-MS gene signature to select the most relevant targets to combine with conventional therapy to overcome the intrinsic resistance to therapy of these cells.
Materials and Methods
Patients and Clinical Samples.
We made the following comparisons on breast cancer samples obtained from women undergoing clinical trials: i) flow-sorted samples: CD44+/CD24−/low vs. others; ii) cancer MSs vs. primary tumors; iii) comparison of CD44+/CD24−/low-MS signature with the published Herschkowitz dataset describing the new group of claudin-low vs. basal, luminal, ERBB2+, and normal-like tumors; iv) letrozole and docetaxel: before and after treatment to demonstrate enrichment of the CD44+/CD24−/low-MS signature.
First, to generate the CD44+/CD24−/low-MS signature, tumor biopsies were obtained before any treatment from patients with locally advanced breast cancer enrolled into two phase II clinical trials (approved by the institutional review board) from January 2000 to December 2006 (Figs. S1–S4). These two studies were conducted in the Breast Center at Baylor College of Medicine, Houston, TX. Biopsies were obtained with informed consent for biomarker correlative studies. All of the core biopsies used in these analyses were examined by H&E staining and determined to contain greater than or equal to 50% epithelial cancer cells before analysis. From these two studies, we obtained microarray gene expression analyses of flow-sorted samples (CD44+/CD24−/low vs. others) and cancer MSs vs. primary tumor (Fig. S1), for establishing our CD44+/CD24−/low-MS gene signature.
Second, to examine the clinical and therapeutic significance of the microarray results, we evaluated the gene expression profile in additional breast tumors before and after therapy. For these analyses, there were three patient cohorts, “letrozole,” “docetaxel,” and “additional letrozole,” as follows. In the first, the letrozole cohort, RNAs were provided from biopsies from postmenopausal patients with ER-positive breast cancers treated with neoadjuvant aromatase inhibitor letrozole for 12 weeks. This study was conducted at Western General Hospital, Edinburgh, U.K. In the second, the docetaxel cohort, we used our published analysis of differentially expressed genes before and after docetaxel (24, 25). The third, the additional letrozole cohort, also from Edinburgh, was used to validate certain results by immunohistochemistry on formalin-fixed sections of a further series of paired letrozole-treated samples (before and after 12 weeks of letrozole). There were 23 pairs of samples with sufficient tissue for IHC, where we analyzed costaining of the epithelial marker pan-cytokeratin (CK) together with the mesenchymal marker vimentin (VIM), and loss of E-cadherin staining. In an earlier analysis, we performed QRT-PCR for MMP2 gene expression in paired biopsies from 60 patients before and after letrozole treatment (26).
Analytical Flow Cytometry and MSs.
Core biopsies were taken before and after treatment, and placed immediately in cold RPMI-1640 supplemented with 10% heat-inactivated newborn calf serum (HINCS, Invitrogen). The methods for flow cytometry and mammosphere growth have been described in detail elsewhere (3). In summary, the samples were digested in collagenase and ≈106 single cells were resuspended, incubated for 15 min with anti-CD44 (APC), anti-CD24, and anti-lineage mixture antibodies (PE-conjugated anti-CD2, CD3, CD10, CD16, CD18, CD31, and CD 140B) (PharMingen) using the manufacturer's suggested concentrations, and then analyzed using Dako MoFlo flow cytometry. Side and forward scatter were used to eliminate debris and cell doublets, and the Lin cells were further analyzed for expression of the CD44 and CD24 markers. A portion of the unsorted cells was used in MS assays by plating these cells onto nonadherent (polyhema-coated) plastic, counting with a hemocytometer, and seeding 20,000 cells into six-well ultralow attachment plates.
Gene Expression Profiling.
RNA was purified from sorted CD44+/CD24−/low cells vs. all others (CD24+ and CD44−/CD24−), cancer-derived MSs vs. primary breast cancer, and before and after treatment with 3 months of letrozole for use in gene expression profiling, as described in refs. 24 and 25. Briefly, total RNA was isolated using TRIzol (Invitrogen) and passed over a Qiagen RNeasy column (Qiagen). Double-stranded cDNA was synthesized by a chimeric oligonucleotide with an oligo(dT) and a T7 RNA polymerase promoter. Reverse transcription was then carried out, followed by biotin labeling and an ≈250-fold linear amplification by in vitro transcription (Enzo Biochem). From each sample, ≈5 μg of labeled cRNA was hybridized onto an Affymetrix U133 Plus2 GeneChip.
IHC, AQUA, and Real-Time PCR (qPCR) Analysis.
Standard methods for immunohistochemical analysis have been described in detail elsewhere (27). All slides were scored by two pathologists (M.C.G. and D.R.) blinded to the treatment status (before vs. after treatment). For immunofluorescence, 4-μm paraffin-embedded formalin-fixed tumor biopsies were incubated with antibodies against vimentin (dilution, 1:250; AB-1 Clone V9, Oncogene Research Products) and pan-keratin (dilution, 1:100; polyclonal, Signet) for 1 h at room temperature, followed by secondary antibodies conjugated to Alexafluor 594 and 488, respectively, for 30 min. Slides were mounted in Vectashield Mounting Media containing DAPI (Vector Laboratories) for nuclear staining.
For quantitative analysis of vimentin, we performed AQUA analysis as described in ref. 28. In brief, a tumor-specific mask was generated by thresholding the image of a marker (cytokeratin) that differentiates tumor from surrounding stroma and/or leukocytes. This creates a binary mask (each pixel is either on or off). In this study, total signal nuclear, cytoplasmic, and the ratio of nuclear to cytoplasmic signal were analyzed. Scores were adjusted according to amount of area covered by the subcellular compartments within the masked area. Histospots containing <5% tumor, by mask area (automated), were excluded from further analysis. AQUA scores were normalized on a 0 to 100 scale for each cohort by dividing by the maximum AQUA score. As part of this study, we conducted qPCR for six differentially expressed genes, including MMP2, and these data were used as part of the validation of mesenchymal genes (Fig. 4).
Data Analysis.
We normalized gene expression arrays using DNA-Chip Analyzer software (dChip). The array data have been deposited in the public Gene Expression Omnibus (GEO) database (GSE7513, GSE7515, and GSE10281). For each of the profile datasets analyzed, we centered gene expression values on the centroid mean of the comparison groups of interest (flow sort: CD44+/CD24−/low and others; MS: cancer MS and primary tumor; Herschkowitz tumors: claudin-low, basal, luminal, ERBB2+, letrozole and docetaxel: before and after treatment). In the case of the Herschkowitz dataset, where multiple array probe sets referred to the same gene, we selected the probe set with the greatest variation to represent the gene. (In the Herschkowitz et al. analysis, luminal A and B tumors were assigned to one “luminal” group based on cluster analysis, and we used the subtype labels assigned by the Herschkowitz group for our own analysis.) Two-sided t tests using log-transformed data determined significant differences in mean gene mRNA levels between groups of samples. We computed fold changes between groups as in ref. 29 and estimated the false discovery rate (FDR) using 100 permutations of the profile group labels. Expression values were visualized as color maps using the Java TreeView software. The mapping of transcripts or genes between independent array datasets was made on the Affymetrix probe set identifier, in the case where both datasets were on the Affymetrix U133 Plus 2 or U133A platforms, or on the Entrez Gene identifier, in the case where the datasets were from different platforms. We determined the significance of overlap between gene sets using one-sided Fisher's exact tests, with the reference gene population being either the 54675 U133 Plus 2 probe sets or the 14112 genes in both U133 Plus 2 and the Herschkowitz Agilent platforms, depending on from which array platforms the two gene sets were obtained. We also carried out gene set enrichment analysis (GSEA) as an alternative enrichment test (see SI Text) (30).
Gene Signature R Values.
To score each tumor within a set for similarity to our gene signature, we derived an R value for each tumor in relation to the CD44+/CD24−/low-MS signature, similar to what we have done in previous studies (29). We defined the R value as the Pearson's correlation between the CD44+/CD24−/low-MS gene signature pattern (using “1” and “−1”, for up and down, respectively) and the primary tumor's expression values (which is essentially a t statistic comparing the average of the up genes to the average of the down genes within each tumor). In this way, tumors with high R values would tend to have both high expression of many of the genes high in CD44+/CD24−/low-MS cells and low expression of many of the genes that are low in CD44+/CD24−/low-MS cells (and vice versa for tumors with low R values). For the human tumor datasets, the gene expression values were first centered as described above before computing the R value.
We then determined whether the gene expression pattern of the previously defined claudin-low human subtype was more pronounced in the tumors after hormone therapy or chemotherapy (Fig. 3 C and D) in the following way. For each gene common to our array platform and the Herschkowitz platform, we computed the mean centroid of the four major tumor subtypes: claudin-low, basal-like, ERBB2+, and luminal, and centered each group average on the centroid. We then took the Pearson's correlation (R value, using all 14,112 genes common to both datasets) between the Herschkowitz centered averages and the expression values of each pretreated or posttreated tumor from the letrozole and docetaxel cohorts.
Supplementary Material
Acknowledgments.
We thank Dr. Gary Chamness for his editing of this manuscript. This work was supported in part by the Breast Cancer Research Foundation (J.C.C., C.M.P.), the Helis Foundation (J.C.C., J.M.R., M.T.L.), the National Cancer Institute Breast Cancer SPORE Grants P50 CA50183 (to J.C.C. and M.T.L.) and CA58223–09A1 (to C.M.P.), National Cancer Institute Grant R01 CA112305–01 (to J.C.C.), Breakthrough Research Unit, Edinburgh (J.M.D., A.A.L., L.R., D.F.), Cancer Research United Kingdom (J.M.D., A.A.L., D.F.), National Institutes of Health Grant P30 CA125123 (to J.C.C. and C.J.C.), grant-in-aid from Glaxo Smith Kline (to J.C.C.), and U.S. Army Medical Research and Material Command Grants DAMD17–01-0132 and W81XWH-04–1-0468 (to J.C.C).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0905718106/DCSupplemental.
References
- 1.Dick JE. Stem cell concepts renew cancer research. Blood. 2008;112:4793–4807. doi: 10.1182/blood-2008-08-077941. [DOI] [PubMed] [Google Scholar]
- 2.Al-Hajj M, Wicha MS, Benito-Hernandez A, Morrison SJ, Clarke MF. Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci USA. 2003;100:3983–3988. doi: 10.1073/pnas.0530291100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li X, et al. Intrinsic resistance of tumorigenic breast cancer cells to chemotherapy. J Natl Cancer Inst. 2008;100:672–679. doi: 10.1093/jnci/djn123. [DOI] [PubMed] [Google Scholar]
- 4.Sorlie T, et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci USA. 2001;98:10869–10874. doi: 10.1073/pnas.191367098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Perou CM, et al. Molecular portraits of human breast tumours. Nature. 2000;406:747–752. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- 6.Herschkowitz JI, et al. Identification of conserved gene expression features between murine mammary carcinoma models and human breast tumors. Genome Biol. 2007;8:R76. doi: 10.1186/gb-2007-8-5-r76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Andarawewa KL, et al. Ionizing radiation predisposes nonmalignant human mammary epithelial cells to undergo transforming growth factor beta induced epithelial to mesenchymal transition. Cancer Res. 2007;67:8662–8670. doi: 10.1158/0008-5472.CAN-07-1294. [DOI] [PubMed] [Google Scholar]
- 8.Buijs JT, et al. Bone morphogenetic protein 7 in the development and treatment of bone metastases from breast cancer. Cancer Res. 2007;67:8742–8751. doi: 10.1158/0008-5472.CAN-06-2490. [DOI] [PubMed] [Google Scholar]
- 9.Damonte P, Gregg JP, Borowsky AD, Keister BA, Cardiff RD. EMT tumorigenesis in the mouse mammary gland. Lab Invest. 2007;87:1218–1226. doi: 10.1038/labinvest.3700683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Guarino M, Rubino B, Ballabio G. The role of epithelial-mesenchymal transition in cancer pathology. Pathology. 2007;39:305–318. doi: 10.1080/00313020701329914. [DOI] [PubMed] [Google Scholar]
- 11.Kokkinos MI, et al. Vimentin and epithelial-mesenchymal transition in human breast cancer–observations in vitro and in vivo. Cells Tissues Organs. 2007;185:191–203. doi: 10.1159/000101320. [DOI] [PubMed] [Google Scholar]
- 12.Lo HW, et al. Epidermal growth factor receptor cooperates with signal transducer and activator of transcription 3 to induce epithelial-mesenchymal transition in cancer cells via up-regulation of TWIST gene expression. Cancer Res. 2007;67:9066–9076. doi: 10.1158/0008-5472.CAN-07-0575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tiezzi DG, Fernandez SV, Russo J. Epithelial mesenchymal transition during the neoplastic transformation of human breast epithelial cells by estrogen. Int J Oncol. 2007;31:823–827. [PubMed] [Google Scholar]
- 14.Trimboli AJ, et al. Direct evidence for epithelial-mesenchymal transitions in breast cancer. Cancer Res. 2008;68:937–945. doi: 10.1158/0008-5472.CAN-07-2148. [DOI] [PubMed] [Google Scholar]
- 15.Yang AD, et al. Chronic oxaliplatin resistance induces epithelial-to-mesenchymal transition in colorectal cancer cell lines. Clin Cancer Res. 2006;12:4147–4153. doi: 10.1158/1078-0432.CCR-06-0038. [DOI] [PubMed] [Google Scholar]
- 16.Kajiyama H, et al. Twist expression predicts poor clinical outcome of patients with clear cell carcinoma of the ovary. Oncology. 2006;71:394–401. doi: 10.1159/000107108. [DOI] [PubMed] [Google Scholar]
- 17.Kajiyama H, et al. Chemoresistance to paclitaxel induces epithelial-mesenchymal transition and enhances metastatic potential for epithelial ovarian carcinoma cells. Int J Oncol. 2007;31:277–283. [PubMed] [Google Scholar]
- 18.Hennessy BT, et al. Characterization of a naturally occurring breast cancer subset enriched in epithelial-to-mesenchymal transition and stem cell characteristics. Cancer Res. 2009;69:4116–4124. doi: 10.1158/0008-5472.CAN-08-3441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sarrio D, et al. Epithelial-mesenchymal transition in breast cancer relates to the basal-like phenotype. Cancer Res. 2008;68:989–997. doi: 10.1158/0008-5472.CAN-07-2017. [DOI] [PubMed] [Google Scholar]
- 20.Mani SA, et al. Mesenchyme Forkhead 1 (FOXC2) plays a key role in metastasis and is associated with aggressive basal-like breast cancers. Proc Natl Acad Sci USA. 2007;104:10069–10074. doi: 10.1073/pnas.0703900104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mani SA, et al. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell. 2008;133:704–715. doi: 10.1016/j.cell.2008.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hiscox S, et al. Tamoxifen resistance in MCF7 cells promotes EMT-like behaviour and involves modulation of beta-catenin phosphorylation. Int J Cancer. 2006;118:290–301. doi: 10.1002/ijc.21355. [DOI] [PubMed] [Google Scholar]
- 23.Gotte M, Yip GW. Heparanase, hyaluronan, and CD44 in cancers: A breast carcinoma perspective. Cancer Res. 2006;66:10233–10237. doi: 10.1158/0008-5472.CAN-06-1464. [DOI] [PubMed] [Google Scholar]
- 24.Chang JC, et al. Patterns of resistance and incomplete response to docetaxel by gene expression profiling in breast cancer patients. J Clin Oncol. 2005;23:1169–1177. doi: 10.1200/JCO.2005.03.156. [DOI] [PubMed] [Google Scholar]
- 25.Chang JC, et al. Gene expression profiling for the prediction of therapeutic response to docetaxel in patients with breast cancer. Lancet. 2003;362:362–369. doi: 10.1016/S0140-6736(03)14023-8. [DOI] [PubMed] [Google Scholar]
- 26.Miller WR, et al. Changes in breast cancer transcriptional profiles after treatment with the aromatase inhibitor, letrozole. Pharmacogenet Genomics. 2007;17:813–826. doi: 10.1097/FPC.0b013e32820b853a. [DOI] [PubMed] [Google Scholar]
- 27.Allred DC, Harvey JM, Berardo M, Clark GM. Prognostic and predictive factors in breast cancer by immunohistochemical analysis. Mod Pathol. 1998;11:155–168. [PubMed] [Google Scholar]
- 28.Camp RL, Chung GG, Rimm DL. Automated subcellular localization and quantification of protein expression in tissue microarrays. Nat Med. 2002;8:1323–1327. doi: 10.1038/nm791. [DOI] [PubMed] [Google Scholar]
- 29.Creighton CJ, et al. Insulin-like growth factor-I activates gene transcription programs strongly associated with poor breast cancer prognosis. J Clin Oncol. 2008;26:4078–4085. doi: 10.1200/JCO.2007.13.4429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Subramanian A, et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee JM, Dedhar S, Kalluri R, Thompson EW. The epithelial-mesenchymal transition: New insights in signaling, development, and disease. J Cell Biol. 2006;172:973–981. doi: 10.1083/jcb.200601018. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.