Abstract
Plasmodium falciparum, the causative agent of malignant malaria, is among the most severe human infectious diseases. The closest known relative of P. falciparum is a chimpanzee parasite, Plasmodium reichenowi, of which one single isolate was previously known. The co-speciation hypothesis suggests that both parasites evolved separately from a common ancestor over the last 5–7 million years, in parallel with the divergence of their hosts, the hominin and chimpanzee lineages. Genetic analysis of eight new isolates of P. reichenowi, from wild and wild-born captive chimpanzees in Cameroon and Côte d'Ivoire, shows that P. reichenowi is a geographically widespread and genetically diverse chimpanzee parasite. The genetic lineage comprising the totality of global P. falciparum is fully included within the much broader genetic diversity of P. reichenowi. This finding is inconsistent with the co-speciation hypothesis. Phylogenetic analysis indicates that all extant P. falciparum populations originated from P. reichenowi, likely by a single host transfer, which may have occurred as early as 2–3 million years ago, or as recently as 10,000 years ago. The evolutionary history of this relationship may be explained by two critical genetic mutations. First, inactivation of the CMAH gene in the human lineage rendered human ancestors unable to generate the sialic acid Neu5Gc from its precursor Neu5Ac, and likely made humans resistant to P. reichenowi. More recently, mutations in the dominant invasion receptor EBA 175 in the P. falciparum lineage provided the parasite with preference for the overabundant Neu5Ac precursor, accounting for its extreme human pathogenicity.
Keywords: chimpanzees, human evolution, Plasmodium falciparum, Plasmodium reichenowi, zoonosis
Malaria counts among the worst scourges of humankind, accounting for some 500 million clinical cases per year and more than one million deaths, mostly children (1). It amounts to an immeasurable health burden and inhibits economic prosperity in numerous tropical countries, most extensively in Africa. Plasmodium falciparum is the most virulent among the four Plasmodium species parasitic to humans, accounting for ≈85% of all malaria cases, and nearly all of the mortality. The extreme pathogenicity of P. falciparum has suggested that it is a recent human parasite, acquired by transfer from a nonhuman host (2). Some early molecular phylogenies seemed to be consistent with this hypothesis, because they showed P. falciparum to be more closely related to Plasmodium gallinaceum, a chicken parasite, than to any of the other human parasite species (3). A considered possibility was that P. falciparum evolved from an avian parasite following a horizontal host transfer, perhaps in association with the Neolithic domestication of the chicken. It was, however, shown by Escalante and Ayala (4) and Escalante et al. (5) that the closest relative of P. falciparum is P. reichenowi, a malaria parasite isolated from a captive chimpanzee that had not been included in earlier studies. These authors showed that P. falciparum and P. reichenowi form an independent clade distinct from other malaria parasites, including the other three human malaria parasites, which appear to have originated in Old World monkeys (4, 5). The close phylogenetic relationship between P. falciparum and P. reichenowi, their distinctness from the other human malaria parasites, and their remoteness from bird or lizard parasites was soon confirmed by other studies (6–8).
Three mutually exclusive hypotheses could account for the relationship between P. falciparum and P. reichenowi. (i) Co-speciation: P. falciparum and P. reichenowi evolved from a common ancestor parasite, independently in their respective hosts, humans and chimpanzees, over the last 5–7 million years; (ii) Human origin: P. reichenowi evolved from an introduction of P. falciparum into chimpanzee hosts; or (iii) Chimpanzee origin: P. falciparum evolved from the introduction of the chimpanzee P. reichenowi into the human lineage (Fig. 1).
Fig. 1.
Alternative hypotheses for the origin of P. falciparum as a human parasite. Green- and blue-shaded areas correspond to parasites infecting chimpanzee and human hosts, denoted as C and H, respectively. The hypothetical ancestors are denoted as F (for P. falciparum), R (for P. reichenowi), and RF (for their common ancestor). (Left) Parasites of humans and chimpanzees diverged as their respective hosts did, starting 5–7 million years ago (5, 52). Alternatively, a human parasite lineage could have given rise to the chimpanzee lineage (Center), or vice versa (Right). The present data overwhelmingly support the latter alternative, identifying chimpanzees as the ancestral host for the agent of malignant human malaria.
These hypotheses can readily be tested by determining the phylogenetic relationships among the two parasite species and by comparing their levels of genetic polymorphism, particularly with respect to silent nucleotides, which are expected to accumulate as a function of the rate of mutation and the time elapsed since the origin of a species. If co-speciation occurred, P. falciparum and P. reichenowi should form distinct, sister clades. If hypotheses ii or iii are correct, the level of neutral polymorphism should be greater in the ancestral parasite than in the derived species. In the absence of the molecular characterization of multiple strains of P. reichenowi, of which only one strain was available, Escalante and Ayala (4) and Escalante et al. (5) favored the co-speciation hypothesis, as soon did other investigators (6–8). The two alternative transfer hypotheses were largely ignored because of the availability of only one known isolate of P. reichenowi (from Cameroon, described in 1917 and 1920, ref. 9). However, Martin et al. (10) suggested a mechanism compatible with hypothesis iii on the basis of differential expression of host sialic acid (Sia) ligands and differences in parasite receptor Sia-binding preferences.
Rich, Ayala, and collaborators soon demonstrated that P. falciparum has very low levels of neutral polymorphism (11–13), a result that was subsequently confirmed by other investigators (14–18). The scarcity of neutral polymorphisms in P. falciparum was interpreted to be the result of a recent world expansion of the species, which was estimated to have happened a few thousand years ago, rather than millions of years ago (the Malaria Eve hypothesis; ref. 13).
The recent world expansion of P. falciparum could have come about in two ways. First, as a consequence of a severe bottleneck, so that all extant populations of the species would have derived from a few surviving strains. An alternative, equally viable explanation would be that falciparum malaria had been restricted to a local population (somewhere in tropical Africa), from where it would have expanded through much of the African tropics and beyond as a consequence of recent environmental and vector changes (12, 13, 19–21). These changes would include, among others, the introduction of agriculture in tropical Africa during the late Neolithic and consequent deforestation, creating pools of standing water and other conditions favorable for mosquito breeding; the evolution of anthropophilic Anopheles vectors (19, 20); and the gradual warming of the planet that eventually allowed the geographic expansion of the vector–parasite association (11, 13, 22). These three sets of events are timed within the past 10,000 years, which is consistent with the estimated time of the world expansion of P. falciparum (13, 21, 22). The current availability of additional isolates of P. reichenowi makes it now possible to investigate comparatively these hypotheses.
Results
We have sampled wild and wild-born captive chimpanzees from Côte d'Ivoire (n = 10) and Cameroon (n = 84). From the tissues collected from these chimpanzees, we have identified eight new isolates of P. reichenowi by amplifying three gene fragments that have been extensively used in Plasmodium studies: mitochondrial cytB, apicoplast clpC, and nuclear 18S rRNA. Five isolates were derived from the 84 Cameroonian chimpanzees and three from the 10 chimpanzees from Côte d'Ivoire. One of the Ivoirian chimpanzees, Rafiki, was infected with two strains of Plasmodium, characterized by distinct nucleotide sequences in each of the three gene fragments. Because our blood samples were collected on filter paper, we were unable to determine whether the parasites' gametocytes were crescent-shaped, an attribute exclusively characteristic of P. falciparum and P. reichenowi (23). However, the diagnostic repeat region from the circumsporozoite protein (Csp) gene from five sequenced isolates (Bana, Gabon, Max, Nino, and Loukoum) shows that each gene encodes amino acid repeat motifs that specifically distinguish P. falciparum and P. reichenowi from all other malarial parasites (Fig. 2) (24), consistent with their designation as P. reichenowi.
Fig. 2.
Circumsporozoite surface protein (Csp) repeat composition of several Plasmodium species showing that repeat units of Csp genes are diagnostic of species type (24). All characteristic Csp amino acid repeats of P. falciparum, including NVDP and NANP are also found in P. reichenowi. Some P. reichenowi alleles show additional repeat types (NADP and NP).
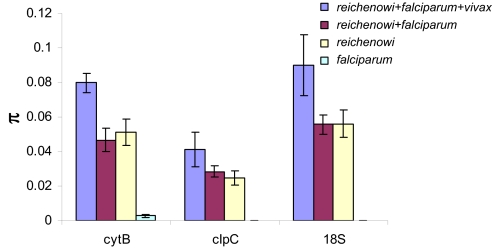
We have examined the nucleotide polymorphisms among the three gene fragments. As reported previously, nucleotide polymorphisms among P. falciparum isolates are extremely scarce (18, 21, 25, 26). Among the P. reichenowi isolates, there is considerably greater nucleotide polymorphism than in P. falciparum, nearly all of which occurs among silent (synonymous) sites in the two amino acid coding regions (cytB and clpC) (Fig. 3). The number of synonymous differences among sequences is expected to increase when evolutionarily independent lineages are combined. However, the mean pairwise number of nucleotide differences (π) is unaffected when sequences from 133 isolates of P. falciparum are added to the nine isolates of P. reichenowi (Fig. 3). This observation, coupled with the related observation of much greater diversity among P. reichenowi isolates than among P. falciparum isolates (even though these are greatly more numerous) is inconsistent with the co-speciation hypothesis and supports the hypothesis that P. falciparum evolved from a chimpanzee parasite.
Fig. 3.
Pairwise nucleotide polymorphism (π) at three genetic loci—cytB, clpC, and 18S rRNA—among Plasmodium species (see SI Text). Diversity in malaria parasites is estimated for (i) P. falciparum isolates alone, (ii) P. reichenowi isolates alone, (iii) P. falciparum + P, reichenowi, and (iv) P. falciparum + P. reichenowi + P. vixax. The addition of P. vivax significantly inflates estimates of nucleotide variation (π), which is consistent with the evolutionary independence of P. vivax. The addition to P. reichenowi of the nucleotide polymorphism found in a very large sample of 133 P. falciparum isolates does not substantially increase the polymorphism found in P. reichenowi alone.
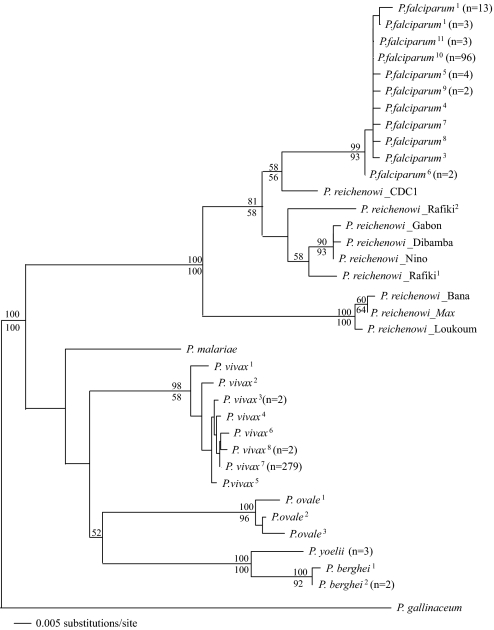
Fig. 4 shows the phylogenetic relationships among the eight new isolates of P. reichenowi, plus the one previously reported isolate (CDC1) together with 133 strains of P. falciparum, based on the cytB gene sequences (see Table S1). We have included in this phylogeny the three other human malaria parasites (P. vivax, P. ovale, and P. malariae), as well as two malaria parasites from African rodents (P. yoelii and P. berghei), and an avian malaria parasite (P. gallinaceum). The cytB gene (Fig. 4), which is thought to be best for recovering the deeper divergences within the genus (7, 8), well illustrates the results from all three gene fragments (for the clpC and 18S rRNA phylogenies, see Fig. S1 and Fig. S2, respectively). It demonstrates that the genetic diversity of P. falciparum is unambiguously nested within the set of the chimpanzee parasites.
Fig. 4.
Cytochrome B gene (cytB) phylogeny of nine P. reichenowi strains (eight newly discovered plus the previously known CDC1 strain) together with a global representation of P. falciparum (133 strains), as well as representative Plasmodium species of rodent and avian parasites. The new P. reichenowi strains are designated by the name of the host chimpanzee from which they were isolated. Three isolates are from Côte d'Ivoire (two Rafiki and Loukoum), the other five are from Cameroon. Multiple strains with identical sequences occur on each of several terminal branches of P. falciparum and other Plasmodium species (n = number of strains). Terminal branches with multiple strains are superscripted; associated GenBank accession numbers for all sequences are included in Table S1. Two distinct nucleotide sequences were identified from Rafiki, and are arbitrarily labeled 1 and 2. The numbers at the nodes are bootstrap support; above the node is support from maximum parsimony whereas the number below indicates maximum likelihood support (no number at a node indicates <50% support for that node).
The competing hypotheses concerning the origins of P. falciparum and P. reichenowi (Fig. 1) can also be tested by estimating the evolutionary “cost” imposed by constraining phylogenetic searches to only those trees consistent with co-speciation. This constraint lengthens the most parsimonious trees for the cytB, clpC, and 18S rRNA by 12, 2, and 3 steps, respectively, and reduces the probability of the observed results by 70-fold, 15.8-fold, and 3.6-fold, by using optimized evolutionary models (see Materials and Methods). The evidence against the reciprocal monophyly of P. falciparum and P. reichenowi (i.e., common ancestral origin; Fig. 1, Left) relative to other phylogenetic alternatives is strong for clpC and decisive for cytB and 18S rRNA (Bayes factors >11, 949, and 910, respectively) (27–29). Evidently, the polymorphism observed in these genes did not accumulate independently in parasite lineages that were restricted to either human or chimpanzee hosts.
Discussion
Our results do not support the co-speciation hypothesis and strongly favor hypothesis iii, namely that P. falciparum evolved from P. reichenowi. Indeed, our data favor five significant conclusions, two that confirm previous results and three new ones: (I) P. falciparum and P. reichenowi combined are monophyletic (4, 5); (II) P. falciparum has very low levels of polymorphism (12–18). Fig. 4 includes 133 strains of P. falciparum, representative of the world distribution of the species. Yet, the total variation is extremely small; for example, the terminal branch labeled “P. falciparum10” includes 96 strains from three different continents, with an average pairwise polymorphism of 0.01 (8). (III) P. reichenowi is a geographically widespread and genetically very polymorphic species, with levels of polymorphism greater than the average divergence between the previously known CDC1 strain of P. reichenowi and all strains of P. falciparum (see Figs. 3 and 4). The overall polymorphism of only nine P. reichenowi isolates is much greater than the polymorphism of all 133 P. falciparum isolates. (IV) The 133 strains representative of the world populations of P. falciparum are all included as one monophyletic branch of the P. reichenowi tree. (V) Based on our data, there has likely been only one host transfer from chimpanzee to humans (P. reichenowi to P. falciparum), shown by the fact that all world strains of P. falciparum are connected to a single branch of the P. reichenowi tree (see Fig. 4 for cytB and Figs. S1 and S2 for ClpC and 18S rRNA).
Ollomo et al. (30) have recently isolated two strains of Plasmodium parasites from two wild-born chimpanzees. The cytB sequences of the two strains are 99.8% identical (out of 866 nucleotides), but they are quite different (91–92%) from the cytB from the previously known P. reichenowi strain (CDC1). Ollomo et al. estimate that the divergence between their newly isolated strains and CDC1 may have occurred as early as 21 ± 9 Mya, if the divergence between P. reichenowi CDC1 and P. falciparum is assumed to be 4–7 million years (Myr) old, consistent with the co-speciation hypothesis and with previous estimates (12, 13, 22). If the divergence between P reichenowi and P. falciparum occurred during the last 2.8 Myr, as proposed by Martin et al. (10), the divergence between P. falciparum and the two new Plasmodium isolates would have occurred within the last 10 Myr. Ollomo et al. (30) have proposed that the two new strains be considered a new species, named Plasmodium gaboni. We have analyzed the cytB sequence of the two new strains in comparison with our newly isolated strains from chimpanzees, and they fit within the clearly distinct clade consisting of three of our strains: Bana, Max, and Loukoum (Fig. 4). Ollomo et al.'s (30) proposal implies that these three strains would belong to the new species, P. gaboni. This would be a reasonable inference, which we are for the moment leaving in abeyance until more definitive estimates of the divergence times or other information becomes available.
The distinguished anthropologist Frank B. Livingstone conjectured that P. falciparum may have been acquired by a transfer to humans of a chimpanzee parasite (31). The plausibility of Livingstone's hypothesis was based on the supposition that, as humans developed increasingly larger agricultural societies, they encroached upon the dwindling forest habitats of species such as the chimpanzee, and so there may have been repeated opportunities for horizontal transfer. Today, human encroachment into the last forest habitats has further extended, leading to a higher risk of transfer of new pathogens, including new malaria parasites. Our results confirm Livingstone's conjecture and, moreover, suggest that the world's extant populations of P. falciparum derive from a single transfer of P. reichenowi from chimpanzees to humans.
How and when did the host transfer occur? A hypothesis proposed in the past was that the ancestors of P. falciparum would have been transferred from another host to humans as our Neolithic ancestors transitioned from hunter-gatherers to agriculturalists some 10,000 years ago (22, 31). This proposal was based on anthropological information about the history of our species, but also on the estimated age of hemoglobin mutants that render humans resistant to malaria infection. The Malaria Eve hypothesis, based on P. falciparum's very low levels of neutral polymorphisms, is consistent with this hypothesis (13, 21), which is also supported by the recent evolution of anthropophilic vectors and by climatic considerations (19, 20, 22). Coluzzi and colleagues have shown that the rapid incipient speciation of the principal African vectors of P. falciparum was driven by postagricultural human conditions, which were “a key influence on the origin of the modern P. falciparum from an ancestral, less pathogenic, taxon” (20).
Other investigators subsequently have sought to determine the time of the most recent common ancestor of extant P. falciparum populations. A time-period consistent with a postagricultural introduction remains most probable (18, 25). Indirect corroboration of this hypothesis lies within the human genome. Specific genetic mechanisms that have given humans resistance to malaria, such as G6PD deficiency and other hemoglobinopathies, appear to have arisen within a time-frame consistent with a postagricultural origin (16, 17).
Our results confirm that the extant populations of P. falciparum have originated in the recent past, because of their very low genetic diversity, but our results do not tell us when the transfer to humans may have occurred. This transfer must have happened much earlier, allowing for the differentiation of P. falciparum from P. reichenowi, which may have originally consisted of nothing more than a change in binding specificity. At present, the divergence between the two species is not large, although much larger than the divergence among the extant P. falciparum. It seems likely that considerable time, on the order of many tens or hundreds of thousands of years, may have elapsed from the time of the host transfer to the time when genetic changes in the parasite and/or the human lineages made possible the rapid expansion of P. falciparum.
Martin et al. (10) have suggested that the classic failure of experimental cross-infection of P. falciparum into chimpanzees and of P. reichenowi into humans can be explained by a human-specific mutation event. Whereas the P. falciparum merozoite can use multiple pathways to invade erythrocytes (32), the dominant invasion receptor appears to be the erythrocyte-binding-like (EBA)-175. The underlying polypeptide sequence of its primary target molecule in erythrocytes is the Sia-capped N-terminal domain of the major erythrocyte glycoprotein glycophorin-A (GYPA), which itself is undergoing rapid evolution (33), presumably due to selection pressure from the parasite.
Several million years after divergence from the chimpanzee lineage, a major biochemical change in Sia biology occurred in the human ancestral lineage (34, 35). The most common Old World primate Sias are N-glycolylneuraminic acid (Neu5Gc) and its metabolic precursor, N-acetylneuraminic acid (Neu5Ac), which differ by a single oxygen atom. A mutation in the CMAH gene makes humans unable to produce Neu5Gc from Neu5Ac, which accumulates in great excess on human erythrocytes. Chimpanzees carry both Sias, with Neu5Gc being dominant (10, 36). P. reichenowi EBA-175 has a marked preference for Neu5Gc, which was suggested to represent the ancestral condition, whereas P. falciparum EBA-175 has a preference for Neu5Ac, which accumulates in humans as a consequence of the CMAH mutation (10, 36). Martin et al. (10) thus conjectured that the loss of Neu5Gc in the human lineage (by way of the CMAH mutation) would have provided our emerging Homo ancestors, perhaps as early as 2–3 million years ago, with temporary relief from P. reichenowi malaria (8, 10). Indeed, it is possible that the CMAH mutation was driven to fixation by continued selection pressure from the then extant form of P. reichenowi. The parasitic malignancy of P. falciparum would have come about later, by selective evolution of its EBA-175, which preferentially recognizes the Neu5Ac-rich human erythrocytes (10). It seems also likely that there would have been an intermediate stage, wherein EBA-175 of the P. reichenowi ancestor would have relaxed its specificity to accommodate binding of Neu5Ac. The final EBA-175 mutations potentially responsible for the malignancy and rapid expansion of P. falciparum may have occurred relatively recently, perhaps ≈5,000–10,000 years ago, which would account for the scarcity of neutral DNA polymorphisms in P. falciparum, consistent with a recent worldwide population expansion of this parasite (11, 12, 18, 22, 25).
Our investigations suggest that P. falciparum has only once established itself among human hosts. The zoonotic origin of P. falciparum elevates interest in the possible ongoing transmission of other malaria parasites of primate origin into the human population (37). The repeated emergence of human malaria parasites from zoonotic reservoirs raises the question of whether ongoing transmission of P. reichenowi from chimpanzees to humans may be possible (or vice versa). The fact that this transmission has not happened repeatedly may reflect the difficulty in changing the sialic acid binding specificity of the parasite-binding proteins. In this regard, it is interesting that a major barrier limiting cross-transmission of avian influenza into humans (and vice versa) is due to differences in sialic acid linkage binding specificity. In this case, it is the relative preference of the human and avian virus hemoglutinins for binding α-2–6- and α-2–3-linked sialic acids, respectively, on epithelial cells in target tissues (38–41). This is also another instance wherein a human-specific change in sialic acid biology is relevant to infectious disease transmission, as chimpanzees and other great apes do not express human upper airway epithelial α-2–6-linked sialic acid targets for human influenza viruses (42, 43).
Materials and Methods
Sample Collection and Preparation.
In Côte d'Ivoire, tissue and blood samples were collected from ten chimpanzees that had died due to anthrax (44), respiratory disease (45), or other reasons in the research area of Taï National Park between 1998 and 2002 (46). DNA extractions from tissue were performed by using DNAeasy tissue kits (Qiagen). DNA was stored in several aliquots.
In Cameroon, samples were collected from captive animals in three wildlife sanctuaries. Animals were primarily wild-born and brought to the sanctuaries after confiscation by authorities or abandonment by owners. Blood samples were collected during routine health examinations, quarantine, or recaptures by sanctuary staff after escapes. Whole blood was collected via venipuncture into a syringe or EDTA vaccutainer and 1 mL was spotted onto Whatman #3 filter paper or Whatman/S&S 903 filter paper. Spots were allowed to dry and were placed in an envelope and sealed in a cliplock bag with silica gel. Dried samples were frozen at −80 °C once in the laboratory in the US. Dry Blood Spot filter papers were processed with Epicenter Master Complete DNA and RNA purification kits (Epicenter Technologies) to obtain genomic DNA following the manufacturer protocols. Total DNA was first dissolved in 30 μL molecular grade H2O.
Gene Fragment Analysis.
As noted by Martinsen et al. (47), reliable determination of species phylogeny is best accomplished by analyses of multiple gene fragments. Accordingly, we amplified gene fragments from the three principal genomes of Plasmodium parasites, mitochondrial (cytB), apicoplast (clpC), and nuclear (18S rDNA). The 50-μL PCRs used 1 μL stock DNA as template, 1× PCR buffer containing 2.5 mM MgCl2, 0.2 mM dNTPs, 0.25 μM of each primer, and 1.25 unit TaqDNA polymerase. PCR for cytB gene used outer primers DW2 (TAATGCCTAGACGTATTCCTGATTATCCAG) and DW3 (TGCTGTATCATACCCTAAAG) (6), and inter primers DW1 (TCAACAATGACTTTATTTGG) and cytb1 (GGATCACTTACAGTATATCCTCC). Amplification for the clpC gene used outer primers TFM1421+ (AAAACTGAATTAGCAAAAATATTA) and TFM1423RC (CGAGCTCCATATAAAGGAT) (48), and inter primers CLPCF1 (TCTAAACAATTATTTGGTTCTG) and CLPCRC1 (TTGGACAACCTAAATTACTTG). PCR for 18S gene used outer primers 18SF1 (TGTAATTGGAATGGTGGGA) and 18SRC1 (TGATCGTCTTCACTCCCTTAA), and inter primers 18SF2 (TTCCAGCTCCAATAGCGTATA) and 18SRC2 (GACATCTGAATACGAATGTCCC).
A touchdown PCR program was used with the annealing temperature starting at 54 °C and decreasing by 3 °C in the first 3 cycles, then 3 °C in the second 3 cycles, followed by 30 cycles at 48 °C. In each cycle, denaturing (at 94 °C) and annealing (at 54–48 °C) were 20 s, and extensions (at 68 °C) were 60 s in duration. An initial 2-min denaturation and a final 6-min extension step were also included. PCR products were separated by electrophoresis in a 1% agarose gel, excised from the gel, and purified by using a QIAquick Gel Extraction Kit (Qiagen). Purified PCR products were cloned by using a TOPO TA cloning for sequencing kit as suggested by the manufacturer, Invitrogen. For each sample, 8–48 clones were picked up and Sanger-sequenced in complementary directions by using T7 and T3 primers. The resulting nucleotide sequences were aligned by using CLUSTALW (49), with corrections made by eye to account for inappropriate alignments (e.g., frame shifts from improperly aligned codon sequences).
Phylogenetic Comparisons.
We conducted a phylogenetic analysis of each gene fragment independently. These nucleotide sequences were compared with those of other human parasites (P. falciparum, P. vivax, P. malariae, and P. ovale), of African rodents (P. berghei and P. yoelii), and one isolate of bird malaria associated with chickens (P. gallinaceum). The sampling of these genetic loci varies greatly among the taxa, with the data for cytB being the most extensive in the public database. For present purposes, we have included only unique haplotypes in our phylogenetic analysis, but the occurrence of multiple representatives of that haplotype are indicated in Table S1 as well as in the resulting phylogenies.
Phylogenetic reconstructions were performed by using maximum parsimony (MP) and maximum likelihood (ML) criteria. MP and ML trees were estimated by using PAUP*4.0b4 (50). Gene-specific evolutionary models were determined empirically for ML analyses by using ModelTest (51). Not surprisingly, the genes from the different genomes adhere to markedly different models of molecular evolution. We used the hierarchical likelihood ratio tests to choose the most suitable model parameters for each genetic locus. The best fit models for cytB, clpC, and 18S rRNA are the F81+G, GTR+G, and HKY+I+G models, respectively.
Each gene tree is shown as a phylogram with branch lengths correspondent to the degree of divergence, with the scale shown at the bottom (Fig. 4, Figs. S1 and S2). The topology of these trees is derived by the Neighbor-Joining method using gene-specific maximum likelihood parameters. Bootstrap analyses using 1,000 replicates of the MP and ML methods were also conducted. The bootstrap values for nodes supported by ≥50% are shown in the tree for both MP and ML iterative analyses. (Bootstrap support values are not shown if <50%.) The resulting consensus dendrograms from MP and ML bootstrap analyses vary slightly in the degree to which they reliably resolve polytomies, particularly among the nonhuman primate parasites. However, in every instance there was extremely high support (93–100%) for the monophyly of the P. reichenowi–P. falciparum clade.
Testing Reciprocal Monophyly.
Two means were used to evaluate whether different ancestors could likely have given rise to P. falciparum and P. reichenowi, based on the distribution of variation now evident in the cytB, clpC, and 18S genes. First, trees were searched under the criteria of maximum parsimony and maximum likelihood, with and without the imposition of a topological constraint consistent with the co-speciation hypothesis. The lengths and likelihoods of optimal trees identified in each case were compared. The cost of enforcing reciprocal monophyly, wherein the ancestor of P. reichenowi did not also give rise to P. falciparum, was judged to be considerable.
More formal statistical consideration of this cost was evaluated by means of Bayesian statistical sampling of the posterior distribution of trees identified under the null and alternative (co-speciation) hypotheses. For cytB, the HKY + G model was taken as a prior, which resembles the model identified as optimal by ModelTest (see above) but which also allows different rates of substitution for transitions and transversions. For the clpC and 18S genes, models identified by ModelTest (see above) were taken as priors. Tree topology and model parameters were allowed to vary in 10 million generations of an MCMC algorithm seeking optimal trees constrained to be consistent with the co-speciation hypothesis, or freed from any such constraint. After a “burn-in” period of 10 thousand generations assuming exponential demographic growth and a strict molecular clock, stable estimates of substitution parameters, tree topology, and tree likelihood were obtained. The ratio of the likelihood of each model, given the data, provided a means to evaluate the statistical cost of imposing reciprocal monophyly on each dataset after integrating over other plausible values for model parameters. The “weight of evidence”, representing the ratio of likelihoods, was represented by Bayes factors (28) calculated by using the methods implemented by BEAST v. 1.4.8 (27, 29).
Supplementary Material
Acknowledgments.
We thank the Cameroon Ministry of Forestry and Wildlife, Ministry of Scientific Research and Innovation, Cameroon Wildlife Aid Fund, and Limbe Wildlife Centre for permits and permission to sample animals at Limbe Wildlife Centre, Mvog Betsi Zoo, and Mfou National Park. We thank Dr. John Kiyang, Dr. Felix Lankster, Babila Talfon, Jean-Michel Takuo, Felix Nkom, Mary-Chantal Bindele, and Mamo Benadette Adzenyuy for Cameroonian field support. We thank the Ministry of the Environment and Forests as well as the Ministry of Research, the directorship of the Taï National Park, and the Swiss Research Center in Abidjan in Côte d'Ivoire. For Ivorian field support, we thank Ilka Herbinger, Rebecca Stumpf, Yasmin Moebius, Cristina Gomes, Tobias Deschner, and Emmanuel Normand. Additional support was provided by the Robert Koch-Institut, the Max-Planck-Society, the Global Viral Forecasting Initiative, Google.org, and The Skoll Foundation. We are extremely grateful to Dr. François Renaud (Centre National de la Recherche Scientifique-International Relief and Development, Montpellier, France) for making available to us the DNA sequences of Plasmodium gaboni. Comments or assistance were provided by Norman Johnson, Dr. Mike Dean, Russell Hanson, Charles Wolfe, Dr. David Sintasath, Lisa Krain, Bill Switzer, and Dr. Jared Diamond. We are grateful to Drs. François Renaud, Nora Besansky, and Ajit Varki for helpful comments about the manuscript. Research and experimentation was funded by National Institutes of Health Grants NIH-RO1GM70077 and NIH-RO1GM60759 (to S.M.R.), with additional support from Cummings School of Veterinary Medicine at Tufts University. Support for collection of specimens in Cameroon was provided by the National Institutes of Health Director's Pioneer Award DP1-OD00370 (to N.D.W.), the National Institutes of Health Fogarty International Center International Research Scientist Development Award 5 K01 TW000003–05 (to N.D.W.), and the National Geographic Society Committee for Research and Exploration.
Footnotes
The authors declare no conflict of interest.
Data deposition: The genetic sequences reported in this paper have been deposited in the GenBank database. Accession numbers are listed in Tables S1–S3.
See Commentary on page 14739.
This article contains supporting information online at www.pnas.org/cgi/content/full/0907740106/DCSupplemental.
References
- 1.Snow RW, Guerra CA, Noor AM, Myint HY, Hay SI. The global distribution of clinical episodes of Plasmodium falciparum malaria. Nature. 2005;434:214–217. doi: 10.1038/nature03342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boyd MF. Malariology: A comprehensive survey of all aspects of this group of diseases from a global standpoint. Philadelphia: Saunders; 1949. pp. 1–47. [Google Scholar]
- 3.Waters AP, Higgins DG, McCutchan TF. Plasmodium falciparum appears to have arisen as a result of lateral transfer between avian and human hosts. Proc Natl Acad Sci USA. 1991;88:3140–3144. doi: 10.1073/pnas.88.8.3140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Escalante AA, Ayala FJ. Phylogeny of the malarial genus Plasmodium, derived from rRNA gene sequences. Proc Natl Acad Sci USA. 1994;91:11373–11377. doi: 10.1073/pnas.91.24.11373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Escalante AA, Barrio E, Ayala FJ. Evolutionary origin of human and primate malarias: Evidence from the circumsporozoite protein gene. Mol Biol Evol. 1995;12:616–626. doi: 10.1093/oxfordjournals.molbev.a040241. [DOI] [PubMed] [Google Scholar]
- 6.Escalante AA, Freeland DE, Collins WE, Lal AA. The evolution of primate malaria parasites based on the gene encoding cytochrome b from the linear mitochondrial genome. Proc Natl Acad Sci USA. 1998;95:8124–8129. doi: 10.1073/pnas.95.14.8124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hagner SC, Misof B, Maier WA, Kampen H. Bayesian analysis of new and old malaria parasite DNA sequence data demonstrates the need for more phylogenetic signal to clarify the descent of Plasmodium falciparum. Parasitol Res. 2007;101:493–503. doi: 10.1007/s00436-007-0499-6. [DOI] [PubMed] [Google Scholar]
- 8.Perkins SL, Schall JJ. A molecular phylogeny of malarial parasites recovered from cytochrome b gene sequences. J Parasitol. 2002;88:972–978. doi: 10.1645/0022-3395(2002)088[0972:AMPOMP]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- 9.Reichenow E. The presence of human malaria parasites in the African apes (Translated from German) Cent F Bakt I Abt Orig. 1920;85:207. [Google Scholar]
- 10.Martin MJ, Rayner JC, Gagneux P, Barnwell JW, Varki A. Evolution of human-chimpanzee differences in malaria susceptibility: Relationship to human genetic loss of N-glycolylneuraminic acid. Proc Natl Acad Sci USA. 2005;102:12819–12824. doi: 10.1073/pnas.0503819102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ayala FJ, Rich SM. Genetic variation and the recent worldwide expansion of Plasmodium falciparum. Gene. 2000;261:161–170. doi: 10.1016/s0378-1119(00)00478-9. [DOI] [PubMed] [Google Scholar]
- 12.Rich SM, Ayala FJ. Population structure and recent evolution of Plasmodium falciparum. Proc Natl Acad Sci USA. 2000;97:6994–7001. doi: 10.1073/pnas.97.13.6994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rich SM, Licht MC, Hudson RR, Ayala FJ. Malaria's Eve: Evidence of a recent population bottleneck throughout the world populations of Plasmodium falciparum. Proc Natl Acad Sci USA. 1998;95:4425–4430. doi: 10.1073/pnas.95.8.4425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Escalante AA, Cornejo OE, Rojas A, Udhayakumar V, Lal AA. Assessing the effect of natural selection in malaria parasites. Trends Parasitol. 2004;20:388–395. doi: 10.1016/j.pt.2004.06.002. [DOI] [PubMed] [Google Scholar]
- 15.Hartl DL, et al. The paradoxical population genetics of Plasmodium falciparum. Trends Parasitol. 2002;18:266–272. doi: 10.1016/s1471-4922(02)02268-7. [DOI] [PubMed] [Google Scholar]
- 16.Tishkoff SA, et al. Haplotype diversity and linkage disequilibrium at human G6PD: Recent origin of alleles that confer malarial resistance. Science. 2001;293:455–462. doi: 10.1126/science.1061573. [DOI] [PubMed] [Google Scholar]
- 17.Tishkoff SA, Verrelli BC. In: Infectious Disease and Host-Pathogen Evolution. Dronamraju KR, editor. Cambridge, UK: Cambridge Univ Press; 2004. pp. 113–140. [Google Scholar]
- 18.Volkman SK, et al. Recent origin of Plasmodium falciparum from a single progenitor. Science. 2001;293:482–484. doi: 10.1126/science.1059878. [DOI] [PubMed] [Google Scholar]
- 19.Ayala FJ, Coluzzi M. Chromosome speciation: Humans, Drosophila, and mosquitoes. Proc Natl Acad Sci USA. 2005;102(Suppl 1):6535–6542. doi: 10.1073/pnas.0501847102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Coluzzi M. The clay feet of the malaria giant and its African roots: Hypotheses and inferences about origin, spread and control of Plasmodium falciparum. Parassitologia. 1999;41:277–283. [PubMed] [Google Scholar]
- 21.Rich SM, Ferreira MU, Ayala FJ. The origin of antigenic diversity in Plasmodium falciparum. Parasitol Today. 2000;16:390–396. doi: 10.1016/s0169-4758(00)01741-5. [DOI] [PubMed] [Google Scholar]
- 22.Ayala FJ, Escalante AA, Rich SM. Evolution of Plasmodium and the recent origin of the world populations of Plasmodium falciparum. Parassitologia. 1999;41:55–68. [PubMed] [Google Scholar]
- 23.Gilles HM, Warrel DA. Bruce-Chwatt's Essential Malariology. 3rd Ed. London: Edward Arnold; 1993. [Google Scholar]
- 24.McCutchan TF, et al. Comparison of circumsporozoite proteins from avian and mammalian malarias: Biological and phylogenetic implications. Proc Natl Acad Sci USA. 1996;93:11889–11894. doi: 10.1073/pnas.93.21.11889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Joy DA, et al. Early origin and recent expansion of Plasmodium falciparum. Science. 2003;300:318–321. doi: 10.1126/science.1081449. [DOI] [PubMed] [Google Scholar]
- 26.Su XZ, Mu J, Joy DA. The “Malaria's Eve” hypothesis and the debate concerning the origin of the human malaria parasite Plasmodium falciparum. Microbes Infect. 2003;5:891–896. doi: 10.1016/s1286-4579(03)00173-4. [DOI] [PubMed] [Google Scholar]
- 27.Drummond AJ, Rambaut A. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol Biol. 2007;7:214. doi: 10.1186/1471-2148-7-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kass RE, Raftery AE. Bayes factors. J Amer Stat Assoc. 1995;90:773–795. [Google Scholar]
- 29.Suchard MA, Weiss RE, Sinsheimer JS. Bayesian selection of continuous-time Markov chain evolutionary models. Mol Biol Evol. 2001;18:1001–1013. doi: 10.1093/oxfordjournals.molbev.a003872. [DOI] [PubMed] [Google Scholar]
- 30.Ollomo B, et al. A new malaria agent in African hominids. PLoS Pathog. 2009;5:e1000446. doi: 10.1371/journal.ppat.1000446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Livingstone FB. The distribution of the sickle cell gene in Liberia. Am J Hum Genet. 1958;10:33–41. [PMC free article] [PubMed] [Google Scholar]
- 32.Baum J, Pinder M, Conway DJ. Erythrocyte invasion phenotypes of Plasmodium falciparum in The Gambia. Infect Immun. 2003;71:1856–1863. doi: 10.1128/IAI.71.4.1856-1863.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang HY, Tang H, Shen CK, Wu CI. Rapidly evolving genes in human. I. The glycophorins and their possible role in evading malaria parasites. Mol Biol Evol. 2003;20:1795–1804. doi: 10.1093/molbev/msg185. [DOI] [PubMed] [Google Scholar]
- 34.Chou HH, et al. Inactivation of CMP-N-acetylneuraminic acid hydroxylase occurred prior to brain expansion during human evolution. Proc Natl Acad Sci USA. 2002;99:11736–11741. doi: 10.1073/pnas.182257399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hayakawa T, Aki I, Varki A, Satta Y, Takahata N. Fixation of the human-specific CMP-N-acetylneuraminic acid hydroxylase pseudogene and implications of haplotype diversity for human evolution. Genetics. 2006;172:1139–1146. doi: 10.1534/genetics.105.046995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Varki A, Nelson DL. Genomic comparisons of humans and chimpanzees. Annu Rev Antrhopol. 2007;36:191–209. [Google Scholar]
- 37.Cox-Singh J, et al. Plasmodium knowlesi malaria in humans is widely distributed and potentially life threatening. Clin Infect Dis. 2008;46:165–171. doi: 10.1086/524888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shinya K, et al. Avian flu: Influenza virus receptors in the human airway. Nature. 2006;440:435–436. doi: 10.1038/440435a. [DOI] [PubMed] [Google Scholar]
- 39.Stevens J, et al. Structure and receptor specificity of the hemagglutinin from an H5N1 influenza virus. Science. 2006;312:404–410. doi: 10.1126/science.1124513. [DOI] [PubMed] [Google Scholar]
- 40.Tumpey TM, et al. A two-amino acid change in the hemagglutinin of the 1918 influenza virus abolishes transmission. Science. 2007;315:655–659. doi: 10.1126/science.1136212. [DOI] [PubMed] [Google Scholar]
- 41.Yen HL, et al. Inefficient transmission of H5N1 influenza viruses in a ferret contact model. J Virol. 2007;81:6890–6898. doi: 10.1128/JVI.00170-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Couceiro JN, Paulson JC, Baum LG. Influenza virus strains selectively recognize sialyloligosaccharides on human respiratory epithelium: The role of the host cell in selection of hemagglutinin receptor specificity. Virus Res. 1993;29:155–165. doi: 10.1016/0168-1702(93)90056-s. [DOI] [PubMed] [Google Scholar]
- 43.Gagneux P, et al. Human-specific regulation of alpha 2–6-linked sialic acids. J Biol Chem. 2003;278:48245–48250. doi: 10.1074/jbc.M309813200. [DOI] [PubMed] [Google Scholar]
- 44.Leendertz FH, et al. Anthrax kills wild chimpanzees in a tropical rainforest. Nature. 2004;430:451–452. doi: 10.1038/nature02722. [DOI] [PubMed] [Google Scholar]
- 45.Kondgen S, et al. Pandemic human viruses cause decline of endangered great apes. Curr Biol. 2008;18:260–264. doi: 10.1016/j.cub.2008.01.012. [DOI] [PubMed] [Google Scholar]
- 46.Leendertz FH, et al. Pathogens as drivers of population declines: The importance of systematic monitoring in great apes and other threatened mammals. Biol Cons. 2006;131:325–337. [Google Scholar]
- 47.Martinsen ES, Perkins SL, Schall JJ. A three-genome phylogeny of malaria parasites (Plasmodium and closely related genera): Evolution of life-history traits and host switches. Mol Phylogenet Evol. 2008;47:261–273. doi: 10.1016/j.ympev.2007.11.012. [DOI] [PubMed] [Google Scholar]
- 48.Rathore D, Wahl AM, Sullivan M, McCutchan TF. A phylogenetic comparison of gene trees constructed from plastid, mitochondrial and genomic DNA of Plasmodium species. Mol Biochem Parasitol. 2001;114:89–94. doi: 10.1016/s0166-6851(01)00241-9. [DOI] [PubMed] [Google Scholar]
- 49.Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL_X windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. (2nd Ed) 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Swofford D. Molecular Systematics. Sunderland, MA: Sinauer Associates; 2003. [Google Scholar]
- 51.Posada D, Crandall KA. MODELTEST: Testing the model of DNA substitution. Bioinformatics. 1998;14:817–818. doi: 10.1093/bioinformatics/14.9.817. [DOI] [PubMed] [Google Scholar]
- 52.Escalante AA, Ayala FJ. Evolutionary origin of Plasmodium and other Apicomplexa based on rRNA genes. Proc Natl Acad Sci USA. 1995;92:5793–5797. doi: 10.1073/pnas.92.13.5793. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.