Abstract
Proteome analysis has emerged as a powerful technology to decipher biological processes. One of the main goals is to discover biomarkers for diseases from tissues and body fluids. However, the complexity and wide dynamic range of protein expression present an enormous challenge to separation technologies and mass spectrometry (MS). In this review, we examine the limitations of proteomics, and aim towards the definition of the current key prerequisites. We focus on capillary electrophoresis coupled to mass spectrometry (CE-MS), because this technique continues to show great promise. We discuss CE-MS from an application point of view, and evaluate its merits and vices for biomarker discovery and clinical applications. Finally, we present several examples on the use of CE-MS to determine urinary biomarkers and implications for disease diagnosis, prognosis, and therapy evaluation.
Keywords: Capillary electrophoresis, mass spectrometry, biomarker, urine, clinical proteomics, peptides
I. INTRODUCTION
Body fluids contain a plethora of information on the (patho)physiological state of any organism. Although this fact has been known for centuries, it is harder than anticipated to actually extract this information. Major constituents of information-containing components in body fluids are peptides and proteins. Several of these compounds have been described as biomarkers for disease, and some are in widespread use in clinical laboratories. Although those single biomarkers exhibit high sensitivity, they might lack specificity. Consequently, the field of proteomics has moved toward panels of biomarkers, as recently discussed at a conference at the FDA (Goodsaid et al., 2007).
High hopes were raised with the introduction of modern mass spectrometers, and many of those hopes have not resulted in any actual useful biomarkers. Several reasons for the failure or shortcoming have been uncovered. The biggest obstacle might lay within the proteome of the preferred target (blood) itself: The blood proteome is highly complex and variable, and its components cover several orders of magnitude in concentration (12 have been proposed by Anderson (Anderson and Anderson, 2002)) with often only a few, highly abundant proteins. This enormous dynamic range in blood poses an analytical challenge that is most likely too large to be successfully tackled with today’s technologies. As a consequence, unfortunately (almost) all attempts to define biomarkers from blood (plasma and especially serum) have failed. There is no doubt that such biomarkers must exist, but it becomes less disputed that today’s mass-spectrometry based technologies in general do not enable the discovery of proteomic biomarkers from blood.
A reduction of the aim and the complexity of the body fluid to be examined, in combination with robust technology, has proven to be quite successful. Urine and cerebrospinal fluid (CSF) seem to be more amendable to proteome analysis due to a lower dynamic range and a higher stability. Furthermore, analysis of peptides in body fluids has been successful, and gave rise to a new field, peptidomics, which was successfully applied in several recent studies, where only the low-molecular-mass proteins and peptides were analyzed from the less-variable and less-complex urine and cerebrospinal fluid (CSF). In parallel to the shift of biomarker research from blood to urine and CSF, capillary electrophoresis coupled mass spectrometry (CE-MS) combined with standardized sample preparation protocols and dedicated software appeared as a most suitable tool to study body fluid proteomes.
In this review, we discus the principal theoretical and practical obstacles encountered when analyzing body fluids for biomarker discovery. We will outline why CE-MS presents as the least-cumbersome solution for several technical challenges, and highlight the improvements made in the last few years. We will present several examples of a successful application of CE-MS for biomarker discovery, and will discuss current challenges and possible future improvements.
II. CLINICAL PROTEOMICS
A. Principal considerations
In the discovery phase, clinical proteomics represents a comparative multiparametric analysis (Mischak et al., 2007a). A successful study should combine clinical knowledge and include a clearly formulated clinically relevant question, technical expertise, and the use of adequate statistics. Comparability and reproducibility are important factors, because variability is inherently present due to:
biological variability,
variability in the sample collection and preparation, and
analytical variability.
Furthermore, the application of adequate statistical algorithms is essential. As also outlined in detail below, the use of a simple t-test is inappropriate, and will inevitably result in the definition of faulty biomarkers that cannot subsequently be validated.
As for any multidimensional assay, proteome analysis of an increasing number of variables requires more time and effort. In addition, to obtain statistically significant data, expanding the number of analyzed components requires an increase in the number of analyzed samples and, consequently, greater computing power; those increases render the task even more difficult. Although it is generally possible to determine within seconds the concentration of a single protein, it would probably require weeks or months with the parallel use of an array of high-end mass spectrometers to analyze all proteins in a single complex sample. Therefore, the desire for a maximal amount of data and information must be balanced against the time and effort required for that analysis. Any approach must ensure a reproducible analysis (including collection, sample preparation, and data evaluation) to generate comparable data for future studies. That approach would allow future use of those data in other studies to largely eliminate the necessity for hundreds or even thousands of control measurements that are otherwise essential for validation.
Due to the complexity of the proteome, a separation step is required before mass spectrometry analysis. It is beyond the scope of this review to outline the differences in the ionization processes and the modern mass spectrometers; those topics have been summarized in several reviews (Fliser et al., 2007; Kolch et al., 2005). In general, quadrupole (Q), ion-trap, time-of-flight (TOF), and Fourier–transform ion cyclotron resonance (FT-ICR) instruments or their combinations (e.g., hybrid instruments such as Q-TOF, that combine quadrupole and time-of-flight detectors) are currently used for proteome analysis to analyze proteins and peptides. Highest resolution (>100,000) and mass accuracy (<1ppm) can be obtained with FT-ICR instruments. Unfortunately, the high costs of those instruments limit their use. Furthermore, the detection limits of those instruments are considerably higher than those achieved with TOF-instruments and consequently place limitations on the analysis of lower-abundance peptides (Frommberger et al., 2007; Coon et al., 2008). In general, any mass spectrometer that can deliver high-quality data can be utilized; a mass deviation <30 ppm and resolution >5000 are the minimum requirements.
Ionization can be achieved with matrix-assisted laser desorption/ionization (MALDI) or electrospray ionization (ESI). MALDI is more susceptible to “signal suppression”, due to analytes that compete for the energy available, as shown in Figure 1 (Zürbig et al., 2006). However, MALDI generates mostly singly charged ions that greatly ease spectra interpretation. ESI, generally used on-line, generates charged droplets in a high-voltage field that give rise to multiply charged ions during subsequent Coulomb explosions. That approach is less stable, but also less susceptible to signal suppression. However, ESI spectra clearly are more complex, and contain multiply charged ions and different ionization states of the same peptide/protein that require sophisticated software solutions for data interpretation.
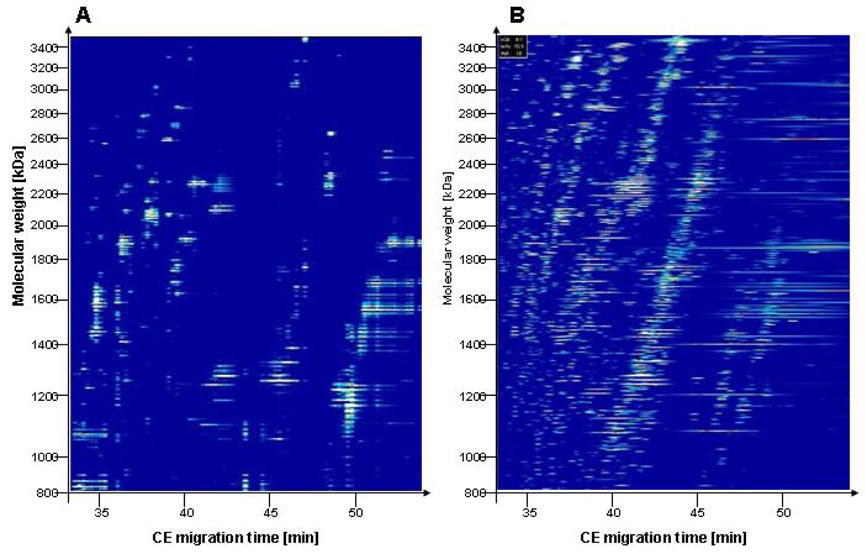
Figure 1.
Comparison of off-line and on-line CE-MS analysis of a urine sample. Data plots of CE-MALDI-TOF-MS (A) and of CE-ESI-TOF-MS (B) are shown. Molecular mass [Da] (logarithmic scale) is indicated on the left, and the migration time (in minutes) is indicated at the bottom. MALDI ionization enables detection of a low number of peptides in a complex sample with high confidence, whereas other signals appear suppressed, compared to ESI.
B Biomarker definition and assessment
A biomarker is a specific molecule used to measure or indicate the effects or progression of a disease, condition, or treatment. The biomarker is defined by the intended use and is not suitable beyond the intended use, unless proven otherwise in a study that clearly validates such broader application, as also recently outlined at a conference at the FDA (Goodsaid et al., 2007). Furthermore, the biomarker should be accurately defined. The mere description of a deduced protein based on a few tryptic peptides that were generated in the discovery phase does not represent a valid definition of a biomarker. A proteomic biomarker is, in general, defined by its C- and N-termini, as well as by post-translational modifications (Mischak et al., 2007a; Good et al., 2007). In some instances, those modifications, in fact, represent “the biomarker” [e.g., advanced glycation end-products in diabetes (Thornalley et al., 2003; Lapolla et al., 2004), or the recently described fragmentation products of abundant proteins associated with nephritic syndrome (Candiano et al., 2006)].
For a potential biomarker to be used in a clinical setting, a separate validation phase, aside from the initial discovery phase of the biomarkers, is required (deVera et al., 2006; Lescuyer et al., 2007; Mischak et al., 2007a; Good et al., 2007). However, the two phases do not necessarily require two different analytical platforms. If the discovery platform can be used in a routine clinical setting, then it should also be used for validation purposes. The common belief that biomarkers identified with mass spectrometry can and must be transformed into an immuno-based assay, and that such a development is essential for clinical application, might be a myth, and in fact might prohibit the establishment of specific and useful assays. Antibodies recognize epitopes that can be shared by multiple peptide- or protein-fragments, and even by different proteins. That fact might hinder detection of a specific peptide/protein, as shown recently (Good et al., 2007). Although a particular peptide fragment might serve as an excellent biomarker for the detection of a certain disease, an antibody that recognizes that sequence might also recognize several other peptides within the same sample due to the existence of a similar or identical epitope. That type of assay might consequently give rise to apparently insignificant data, whereas the MS analysis, due to its higher specificity, could in fact establish significance.
III. TECHNICAL ASPECTS
A Targeted material
1 Blood
A common choice of a targeted fluid for clinical proteome analysis has been blood. As described above, although a plethora of manuscripts describe proteome analysis for blood-derived serum and plasma, the potential biomarkers that were described appear not to be too useful. Several reasons might be responsible for the failure to actually identify generally useful biomarkers. One main obstacle appears to be that a few proteins in blood account for >99% of the total protein content. That fact has led to several approaches to remove these components by affinity chromatography. Whereas those techniques reduced the abundant proteins, their elimination was not quantitative. That limit is well-illustrated in a recent report (Rao et al., 2007), where one of the defined biomarkers for diabetic nephropathy (although in urine) was an abundant protein that was thought to be removed by an affinity column. Furthermore, the affinity columns also remove many other proteins and peptides, and introduce additional variability. For example, depletion of plasma for human serum albumin resulted in co-depletion of 814 other proteins (Shen et al., 2005). Another obstacle is the inherent variability of blood-derived fluids, due, in part, to the activation of proteases.
New approaches that might lead to useful biomarkers include the selection of a sub-proteome [e.g., glycosylated or phosphorylated proteins, as reviewed recently (Temporini et al., 2008)], the use of combinatorial peptide ligand beads (equalizer beads, “Proteominer”) (Guerrier et al., 2007; Sennels et al., 2007; Righetti and Boschetti, 2008), or the assessment of proteolytic fragments (Candiano et al., 2006). It is still too early to know whether those approaches will, in fact, be successful. Hence, although it is undisputed that blood must contain specific biomarkers for probably almost any disease, it appears equally correct that those biomarkers are generally not accessible with the current MS-technologies.
2. Tissue
A promising approach has been the identification of potential biomarkers in affected tissues. Certainly, if only the affected tissue is compared to “normal” tissue, then the likelihood to identify disease-indicative proteins and peptides is largely increased. Tissue is generally not easily accessible for routine clinical assessment, and even less so for monitoring purposes, which require multiple samples. However, tissue-derived biomarkers might also be found in blood (although at much lower concentrations) hence could be analyzed in blood with immunological assays (Lescuyer et al., 2007). Successful, yet still preliminary, approaches include the identification of proteasome activator complex subunit 3 (PSME3) as a biomarker for colorectal carcinoma from tissue (Roessler et al., 2006). That biomarker was subsequently validated in an independent dataset with immunological detection of PSME3 in serum. Using a similar approach, Cathepsin D was identified as a potential biomarker of lung cancer by using supernatant from a tissue culture, and was subsequently validated in serum samples (Lou et al., 2007).
3. Cerebrospinal fluid
Whereas cerebrospinal fluid (CSF) at first sight does not appear to be easily accessible, its collection for clinical diagnostic indications is not uncommon. CSF, due to its contact with the brain and the central nervous system, contains biomarkers that are indicative of neurological and neurodegenerative diseases. Furthermore, several of the obstacles observed for blood (e.g., high intrinsic proteolytic activity, highabundance proteins that obscure biomarkers, presence of large amounts of cells and lipids) generally do not apply to CSF. Consequently, CSF has been investigated by several groups, and potential biomarkers for diseases, such as Morbus Alzheimer have been described [see a recent review (Roche et al., 2008)].
4. Urine
Although urine was in the past considered an unstable body fluid that contained only a low amount of information, it has gained considerable interest, and some of the previously thought obstacles turned out to be more of a myth than actual facts. Undisputed advantages of urine are that it can be obtained in large quantities, and medically trained personnel are not required for collection. Surprisingly, the urinary proteome is quite stable. That stability might, in part, be due to the fact that urine is “stored” in the bladder for a considerable amount of time before collection, to provide sufficient time for complete proteolytic processing by endogenous proteases. In two independent sets of experiments, Schaub et al. (Schaub et al., 2004) and Theodorescu et al. (Theodorescu et al., 2006) showed that the low molecular mass urinary proteome does not undergo any significant changes for 3 days at 4°C, or 6 hours at room temperature, respectively. In addition, urine can be stored frozen at −20°C for several years, without any significant alterations of its proteome. Those reports indicate a much greater stability of the urinary proteome compared to blood. However, it is important to note that other issues will influence quality and comparability of proteomics data from urine samples. Among those are protein concentration and variability due to diet and exercise. The variation in concentration (due to, e.g., liquid intake) can be compensated for by adjustments based on urinary creatinine or abundant “urinary housekeeping peptides”; i.e., peptides present in almost every human urine sample (Theodorescu et al., 2005). Variability due to diet or exercise can, in part, be avoided by collection of the second urine of the day, to produce highly consistent proteome/peptidome data (Weissinger et al., 2004). That observation is most likely due to the fact that changes due to exercise and diet display in the urine with several hours delay; hence, urine collected in the afternoon/evening was found to show the highest degree of variability.
Urine likely reflects information on diseases of organs in direct contact with urine, such as the kidney and bladder, but also the vascular system. In contrast, information from essentially every organ is deposited in blood. Although that potential wealth of information in the blood proteome appears as an advantage at first sight, it might turn out to be a large problem in addition to the above-mentioned obstacles. Due to the complexity of information present, it appears very challenging to extract the minute amounts of data that are specific for a single organ/disease from blood.
Given the urinary proteome’s complexity that we can currently only estimate, relevant changes associated with differences among samples due to variations in procedures for collection, storage and, of course, processing are to be expected. Those issues must be taken into account, and standardized protocols for urine sampling and for handling of the samples must be adopted (Thongboonkerd, 2007).
B Instrumentation
In general, four types of approaches are used for clinical proteomics. Their advantages and disadvantages have been outlined in recent reviews (Fliser et al., 2007; Mischak et al., 2007b; Theodorescu and Mischak, 2007), and three methods will be mentioned briefly here before detailing CE-MS.
1. Two-dimensional gel-electrophoresis followed by mass spectrometry (2DGE-MS)
2DGE-MS still is the most commonly used method to separate and identify proteins >20 kDa. It is technically demanding and time-consuming, and could yield variable results; thus comparison of multiple datasets is rather difficult. Definition of biomarkers based on appropriate statistics is frequently difficult or even impossible, due to the low number of independent datasets and the high variability. However, 2DGE-MS enables assessment of mass of a potential biomarker in its native form, which is an important part of the definition of a potential biomarker (see above).
2. Multi-dimensional Protein Identification Technology (MudPIT)-LC-MS/MS
Similar considerations also apply in part to (Multidimensional Protein Identification Technology (MudPIT)-LC-MS/MS, as recently reviewed by (Issaq et al., 2005). The increased number of variables in comparison to 2DGE renders statistical evaluation even more challenging. Furthermore, information on the molecular mass of the actual biomarker as well as on any post-translational modifications (PTM) is generally lost. Such information, however, is critical. Specific degradation products of proteins have been described as biomarkers. PTMs (e.g., advanced glycation end-products) might even be the hallmark of a biomarker.
3. Surface-Enhanced Laser Desorption/Ionization (SELDI)
The Surface-Enhanced Laser Desorption/Ionization (SELDI) technology appeared attractive due to its ease of use and its high throughput. However, several obstacles, including the low-resolution of the mass spectrometer, and the lack of reproducibility, prevented its successful application. While the resolution of the MS has been improved upon, the value and reproducibility of the defined biomarkers could not be established in recent studies (McLerran et al., 2008b; McLerran et al., 2008a). A further limitation is the inability to rigorously characterize potential protein biomarkers with amino acid sequence data. The last problem can be potentially corrected, in some cases, with TOF/TOF instruments (Freed et al., 2008).
4. Capillary electrophoresis coupled to mass spectrometry (CE-MS)
Quite surprisingly, no vendor has yet positioned itself to sell a complete CE-MS system. This factor might be one of the largest hurdles in the further exploitation of the CE-MS approach.
4.1. CE
Mostly, capillary zone electrophoresis (CZE) has been utilized in MS coupling, and CE is often (and also here) used synonymously for CZE. Other initially quite promising approaches like capillary isoelectric focussing (C-IEF) appear to be less widely used, mostly due to sophisticated technology that requires exceptional experts to perform such analysis, and also due to technical limitations (e.g., the problem of background ampholytes that interfere with MS detection). Whereas initial manuscripts indicated that proteome analysis might be possible on a large scale with C-IEF-MS (Jensen et al., 1999), that initial optimism unfortunately has not yet been substantiated with additional reports. The different types of CE modes that can be applied towards proteome analysis have recently been described in detail in excellent reviews (Kasicka, 2008; Dolnik, 2008).
4.2. Coating
Several types of internal capillary coating are described to reduce interaction of proteins and peptides with the capillary wall, as well as the electroosmotic flow. Those types of coatings and their potential advantages were described in detail in recent manuscripts and reviews (Simo et al., 2004; Ullsten et al., 2004; Erny et al., 2006; Garza et al., 2007; Gaspar et al., 2008). However, both phenomena appear to be of little or no consideration at the very acidic pH of 2 – 2.5 that is typically used in peptide separation. As a consequence, we were unable to determine any benefit of several types of coating tested. Stable and dynamic coating, in fact, decreased the resolution of the CE and the sensitivity of MS detection at the acidic pH routinely used (Mischak, unpublished observation).
4.3. Coupling/interface
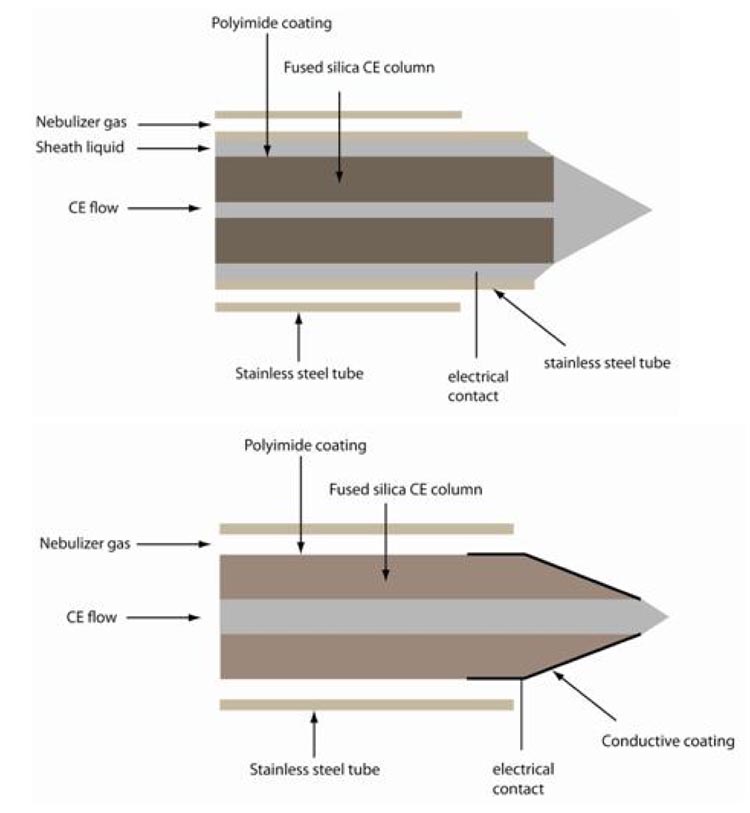
An excellent and comprehensive overview on the different methods of coupling (both ESI and MALDI) is given in a recent review by (Stutz, 2005). CE can be coupled off-line to MALDI targets, as described in several recent manuscripts (Amon et al., 2006; Zürbig et al., 2006), the setup being essentially identical to the sheath-flow coupling (see below). However, while coupling to MALDI appears to be less technically challenging and interpretation of the data is more straightforward (as outlined above), this approach also results in loss of resolution, signal suppression, and higher variability of signals due to matrix effects. Consequently, coupling to ESI appears to be the preferred option. As outlined in our previous review (Kolch et al., 2005), CE coupling to MS via sheath flow interfacing is unexpectedly stable, and also represents a sensitive detection device (in the amol range). Stability and sensitivity have also been confirmed by a number of recent articles and reviews (Gaspar et al., 2008; Tempels et al., 2007; Haselberg et al., 2007; Hernandez-Borges et al., 2007). In general, CE can be interfaced with any type of MS, similar to LC. As outlined in several reviews, two types of coupling, sheathless and sheath-flow interfacing is currently being used (Gaspar et al., 2008; Zamfir, 2007). A schematic drawing of these two types of coupling is given in Figure 2. Although sheathless coupling shows an improved detection limit (likely due to lower flow rates), it also shows reduced stability, a major disadvantage when comparing large number of samples. Consequently, the majority of reports on CE-MS in fact utilize sheath-flow interfacing (Gaspar et al., 2008).
Figure 2.
Schematic drawing of commonly used CE-ESI-MS interfacing. In general, sheath-flow and sheathless coupling is employed. Upper panel: in sheath-flow coupling, a sheath liquid is applied on the outside of the capillary, that circumflows the end of the capillary and closes the electrical circuit. Ionization is essentially comparable to the micro- or nanoflow ionspray. The detection limit is generally inferior in comparison to sheathless interfaces, but the stability of this form of coupling is a benefit that generally outweighs the lower sensitivity. Furthermore, the ionization is quite efficient and detection limits in the high attomole range can be achieved when the flowrates are in range of 200 and 500 min. Lower panel: in sheathless coupling the electrical field is established using an outer metal or graphite coating of the capillary as electrical pole. While this approach results in no dilution of sample and excellent ionization, it also results in instable spray. We were never able to obtain stable pray for several hours, a prerequisite for routine applications.
4.4. Calculation of migration time
A hallmark of CE-separation is the appearance of “streaks” of peptides, when migration time is plotted against mass (Figure 3). These “streaks” appear to be a result of the simple separation principle used. Separation is a result of the electrical force applied onto an ion. That force in turn is dependent on the charge and on the flow resistance, which is dependent on the cross-section area of the ion. At acidic pH, the amino groups are protonated, and protons, in general, are the sole source of charge under these conditions. The position of each peptide in a CE-separation can, therefore, be calculated with good accuracy if its mass and the number of basic amino acids are known (Zürbig et al., 2006).
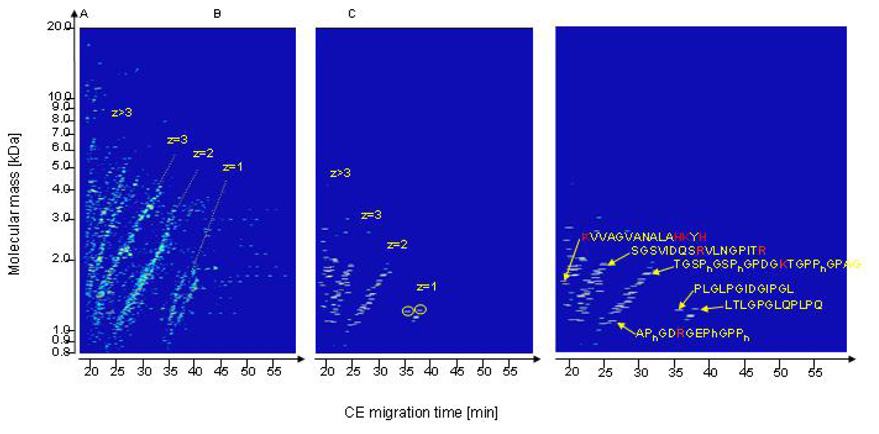
Figure 3.
Compiled CE-MS data from healthy volunteers. (A) Contour plot of the entire urine peptidome. The molecular mass (logarithmic scale) on the y-axis is plotted against normalized CE migration time on the x-axis. The arrangement of the analyzed peptides in distinct lines is obvious. (B) Contour plot of 107 identified polypeptides. The lines already observed in (A) result from the number of positive charges z (at pH 2.2). Peptides marked with circle: Collagen type VI alpha 4 fragment (PLGLPGIDGIPGL); 1217.702 Da; migration time: 34.03 min. Peptide marked with dashed circle: Insulin-like 3 fragment (LTLGPGLQPLPQ); 1232.713 Da; migration time: 36.19 min. (C) Correlation between the effective net charge, molecular mass, and the CE migration time for several examples of determined peptide sequences. Basic amino acids are colored in bold. Reprinted from (Zürbig et al., 2006) with permission.
4.5. CE-MS versus LC-MS
As already outlined in a previous review (Kolch et al., 2005), CE holds several advantages over LC. Those advantages, that were very recently confirmed in detail in several reviews (Song et al., 2008; Bakry et al., 2007), are especially beneficial when analyzing a large number of heterogeneous samples that contain interfering compounds, such as lipids, precipitates, etc. CE’s main advantages are the robustness, ability to recondition fast with NaOH, simple separating principle with high reproducibility, and, with respect to MS interfacing, a buffer that does not change its composition during analysis, as no buffer-gradient is applied. Such changes in buffer composition require a ramping of ionization parameters to maintain optimal ESI ionization.
A disadvantage of CE is the limited loading capacity. Whereas ml quantities can be loaded onto an LC column, a CE can be filled with a maximum of ca. 1 µl; and in general only 10–100 nl. Although pH-stacking can be used very effectively, a maximum of 30–50% of the total capillary volume can be filled with sample; that volume corresponds to 0.5 – 2 µl when using 50 or 75 µm ID capillaries with 80 – 100 cm length. The limited loading capacity does not seem to be much of a problem in CE-MS coupling, because the amount of information in the sample is generally extremely high. As shown in Figure 4, the information accessible is more limited by the dynamic range, and the fact that more-abundant peptides will obscure less-prominent signals. Even if the detection limits were lowered significantly or significantly more sample could be loaded, the number of detected peptides might not significantly increase. In a large fraction of the CE-MS data-space, signal is already present. Additional signals at the same or an overlapping position (due to isotopic distribution, a peptide generally covers 3–6 mass units) cannot be detected with good confidence, would be obscured by the already present stronger signal, and might even result in conflicts in data interpretation.
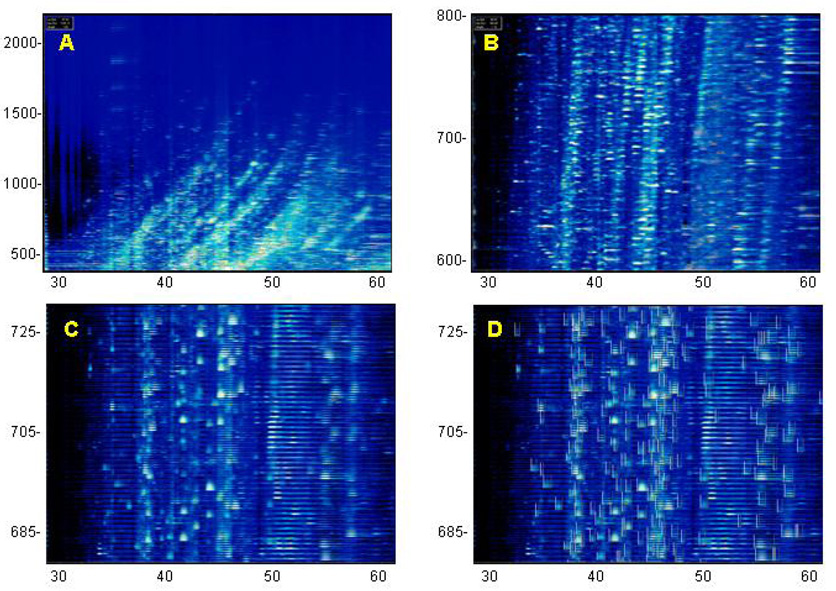
Figure 4.
CE-MS analysis from a human urine sample performed on a Beckman PACE800 CE System interfaced with a Waters LCTpremier mass spectrometer. Mass (in Da) per charge is indicated on the left, migration time (in min) is indicated on the bottom. Panel A shows all signals obtained from 30 – 60 min, m/z 400 – 2200. Panel B shows the signals in the m/z range from 500 – 800. Panel C is a further magnification, data from m/z 680–730 are shown. Signals that were identified as relevant by MosaiquesVisu (z > 1, signal-to-noise ratio >4, present in >3 consecutive spectra) are indicated by boxes in Panel D.
C. Sample preparation for CE-MS analysis
A critical issue in clinical proteomics is sample preparation. Ideally, the sample should not be manipulated at all, and the analytes in the sample should be assessed without any interference. Unfortunately, this ideal situation cannot be accomplished, and samples must undergo pre-analytical manipulation. To enable a subsequent comparison, this step should be robust, highly reproducible, and kept to a minimum
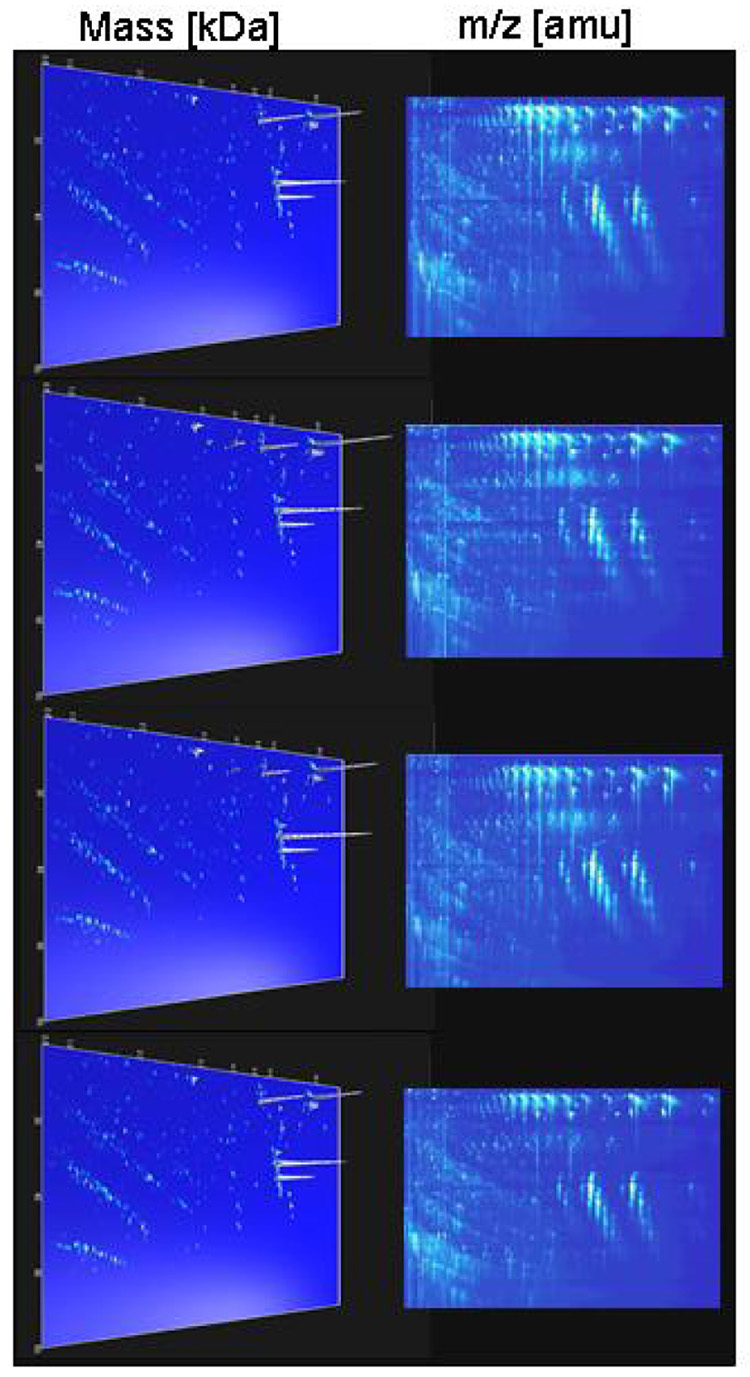
We have initially utilized ion-exchange or reversed-phase chromatography. However, both approaches have resulted in a selective loss of some peptides (e.g., small, highly charged peptides that do not bind to a reversed phase resin). Moreover, larger proteins tended to precipitate on the column, and also subsequently in the capillary. Because those problems reduced reproducibility, especially for samples with a high protein content, we have implemented an ultrafiltration step in the presence of urea and SDS, followed by a desalting step on PD-10 columns (Theodorescu et al., 2005). As also outlined in detail by Theodorescu et al., the presence of detergent and chaotropic agent efficiently inhibits protein-protein interaction to limit losses of analytes due to association with other proteins (e.g., albumin). This protocol enabled the preparation of a sample that contained the low-molecular mass proteins and peptides, and resulted in a higher comparability of data from patients with and without proteinuria. Furthermore, this protocol also enabled reproducible and comparative analysis of rat urine samples (Frommberger et al., 2007), as shown in Figure 5.
Figure 5.
Reproducibility of urinary rat polypeptide evaluation. (A) Examples of electrophoreograms from four (of 19) consecutive measurements of a single urine sample. The m/z values of the 2-D raw data plots (upper Panel, 400 – 2500 m/z in [Da]) and the molecular mass (logarithmic scale) of the deconvoluted 3-D plots (lower Panel, 800 – 25000 Da) on the y-axis are plotted against CE migration time (20 – 60 min) on the x-axis. The arrangement of the analyzed peptides in distinct lines is obvious, and can be comprehended as a result of the number of positive charges at pH 2. White arrow indicates 18.7 kDa signals that probably represent the major urine protein. (B) An average of 1300±106 polypeptides were detected in each one of the 19 replicates (left Panel). Reprinted from (Frommberger et al., 2007) with permission.
D. Sequencing
As detailed below, urinary polypeptides have been shown to serve as excellent biomarkers for diagnostic purposes. However, their (patho)physiological role remains unknown as long as their identity is not determined. Unfortunately, identification of naturally occurring peptides is quite cumbersome, and presents a unique challenge. The biomarkers cannot be easily isolated, and their sequence analysis must be thus performed from a complex mixture.
Tandem mass spectrometry – the process of ion fragmentation with subsequent m/z measurement and the connection between a selected precursor ion with its product ions – is used to determine amino acid sequence and is typically induced by isolating the peptide m/z of interest and subjecting it to several hundred collisions with rare-gas atoms. This process, collision-activated dissociation (CAD), supplies sufficient internal energy to induce covalent bond breakage. Unfortunately, CAD of peptides that contain certain PTMs (e.g., phosphorylation, glycosylation, etc.) or that are too large (~ > 20 residues) does not routinely produce sufficient backbone fragmentation to permit sequence identification. For this reason, most proteomic applications rely on enzymatic digestion to create shorter peptides that are more easily sequenced. Application of this practice towards biomarker identification is problematic because the entire peptide sequence is required. Further, identification of peptides bearing PTMs is essential because PTMs might be disease-specific and can themselves serve as biomarkers (e.g., advanced glycation end-products in diabetes mellitus (Lapolla et al., 2005)). Sequencing of unmodified biomarkers, in contrast to assigning a protein’s primary sequence based on the analysis of a tryptic digest, remains a great challenge. Consequently, many potential markers identified in peptidomic experiments have been among the abundant proteins (Schaub et al., 2005; Rossing et al., 2008a; Coon et al., 2008). New peptide-fragmentation technologies such as electron-capture dissociation (ECD) with FT-ICR MS enable localization of even labile PTMs, such as glycosylation. FT-ICR MS offers two complementary fragmentation techniques to analyze PTMs by tandem mass spectrometry; infrared multiphoton dissociation (IRMPD) and ECD (Marshall et al., 1998; Zubarev, 2003). ECD fragmentation results in a complementary cleavage of the backbone N-Cα bond with minimal loss of PTMs. ECD FT-ICR MS has been successfully used to identify urinary polypeptides larger than 8 kDa, because of the high mass accuracy of FT-ICR MS (Chalmers et al., 2005). Furthermore, localization of glycosylation sites in various glycoproteins, including human IgA1, was accomplished with ECD FT-ICR (Renfrow et al., 2005; Renfrow et al., 2007).
Another emerging technology is electron-transfer dissociation (ETD). This method offers the same dissociation as produced by ECD, but is rapid and sensitive enough to permit coupling with chromatographic separations. This methodology shows great potential (Coon et al., 2005; Good and Coon, 2006), and has demonstrated the best performance to sequence naturally occurring human peptides in our hands (Mischak, unpublished).
A major obstacle to sequence and identify potential biomarkers appears to be PTMs. Because PTMs result in a precursor-ion mass that is different from the theoretical mass of the unmodified peptide and undefined C- and N-termini (in contrast to, e.g., tryptic digests), identification with simple search algorithms often fails. Improvements can be achieved with better mass accuracy (precursor-ion mass and MS/MS spectra), the change in precursor-ion mass due to modifications (taking into account the fact that all possible modifications result in too many degrees of freedom), and a bias of the search algorithms towards high sequence coverage of an unmodified protein. In addition, MS/MS experiments frequently produce a limited number of preferred fragmentation products (at proline residues, carbohydrate side-chains, etc.).
Although CE can be interfaced with an MS/MS instrument, direct sequencing off the CE does represent a challenging undertaking, because only limited amounts of sample can be loaded onto the capillary (see above), to yield low intensity peaks in the MS that often result in no significant signal in the subsequent MS/MS analysis.
An alternative approach is the interfacing of MS/MS with LC to allow the loading of larger amounts of material. Because the theoretical migration time can be used to calculate the exact position of a peptide in CE-MS, sequences can be accurately attributed to a position in the CE-MS analysis. This approach, in combination with highly accurate precursor-ion mass determination with CE-FT-ICR analysis, has proven quite successful (Coon et al., 2008). However, a large number of peptides that produced high-quality spectra could not be identified. We attribute this failure to the inability to correctly interpret the spectra of peptides that contain unknown PTMs.
CE fractions can be collected and spotted off-line onto a MALDI target plate. Subsequently, the polypeptides of interest can be analyzed with MALDI-TOF/TOF (Rejtar et al., 2002; Kolch et al., 2005). That approach has the advantage that the signal of interest can be located in the MS mode, and optimal fragmentation conditions can be determined without repeated separation. However, sequencing of native peptides with MALDI-TOF/TOF is generally unsuccessful, mostly due to low sensitivity and insufficient mass accuracy. In our hands, more than 90% of the spectra obtained from MALDI-TOF/TOF did not allow identification of the native peptide of interest. However, MALDI-TOF/TOF represents a simple method, and unmodified peptides have been identified with MALDI-MS/MS (Kaiser et al., 2004; Mischak et al., 2004); and other approaches like MALDI-LTQ-MS/MS may yield better results.
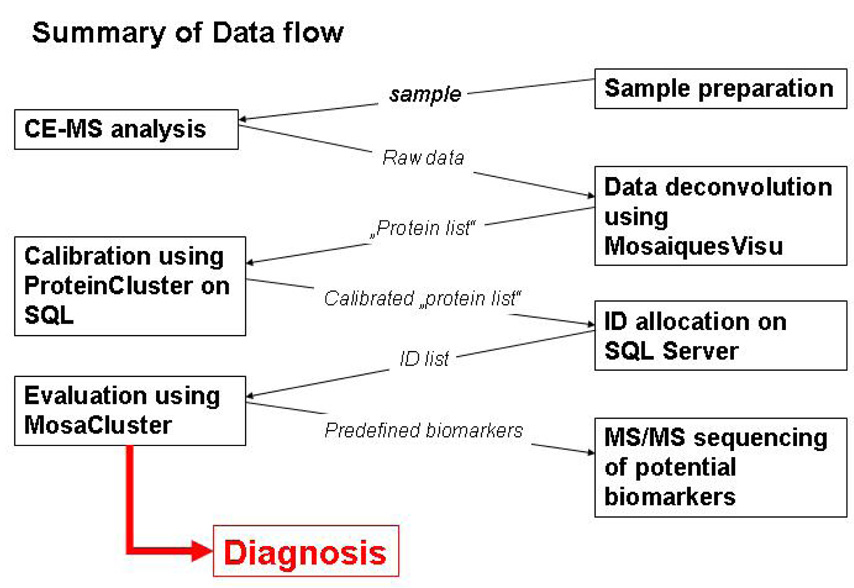
E. Data evaluation
In general, the enormous amount of information provided by a single analysis renders impossible the evaluation of the data with only the software provided with mass spectrometers. The process of data evaluation used in our studies includes several steps, all of which are reviewed briefly here and are also depicted in the flow scheme in Figure 6. The information initially required is the tentative identification of each peptide/protein (preferable via several reproducible physicochemical parameters like mass, migration time, etc., to allow for high resolution), as well a measure of its relative abundance. As a consequence, features that must be implemented in suitable software include the ability to determine the charge of a particular peak, to identify and combine peaks of the same mass but different charge-states, and to perform an efficient normalization of migration times and amplitudes to compensate for any differences between individual measurements.
Figure 6.
Data flow of application in clinical proteome analysis
Several oftware solutions for MS data evaluation have been reported. Although we cannot give a direct comparison of these solutions, we would like to point out some of the key requirements. For MALDI-derived spectra, assignment of charge appears to be less critical (the assumption that the charge is one is generally correct). However, charge assignment is critical for the data obtained from ESI due to the multiply charged ions. Furthermore, because ESI frequently results in different charge states of the same molecule, these charge states must be combined to enable relative quantitation. In addition, any peptide can usually be found in several consecutive spectra, thus the values obtained in the different spectra must be added. MosaiquesVisu (Wittke et al., 2003; Neuhoff et al., 2004) enables these tasks by 1) identifying relevant peaks that can be interpreted with a matched filtering algorithm 2) combining signals of peaks in consecutive spectra, and 3) combining peaks of identical mass and different charge-state at the same position in time to result in the initial definition of a peptide by mass and migration time, and the combined signal intensity as a measure of relative abundance. Further, K+ and Na+ adducts can be identified based on the indicative mass shifts of an analyte at the same position in time, and the requirement that these adducts be less abundant than the protonated peptide. These adducts can subsequently be eliminated from the final list of peptides present in the sample, with their amplitude being added to the amplitude of the protonated peptide.
Migration time shows variability due mostly to the amount of ions present in a sample. However, the relative migration time (in correlation to the other peptides present in the sample) does not change considerably. Hence, internal standards can be used to subsequently calibrate migration time. Those processes enable one to assign unique and reproducible identifying parameters to each peptide: mass and migration time, with amplitude (ion counting) as a measure of relative abundance.
A limitation of proteomic methods is the lack of possibilities to directly deduce the amount of a protein or peptide from an MS spectrum, due mainly to different ionization properties of the peptides that make comparison of ion signals less reliable. As a consequence, chemically synthesized marker peptides with stable isotope labels were introduced as tools for absolute quantification (Righetti et al., 2004). Due to the isotope-specific mass differences, the synthetic peptides can be unambiguously identified in the mass spectra, even in the case of extremely complex protein samples. Unfortunately, this approach suffers from the disadvantages of being time-consuming and expensive. Moreover, systematic quantification errors can occur at each sample-processing and analysis due to, e.g., adhesion to the centrifuge tube or saturation of the MS detector (Ong and Mann, 2005). As a consequence, substantial efforts were undertaken to develop and improve strategies based on ion counting (Zybailov et al., 2005). In contrast to stable isotope-labeled internal standards (Barnidge et al., 2003), ion counting is not suitable for absolute analyte quantification. However, that approach can be optimized in such a way that it allows the relative quantification of peptides with deviation characteristics of approximately 10% (DeKeyser and Li, 2006), and personal observation). To establish an optimal standardization method for CE-MS generated urinary peptide profiles, we used several peptides that can be found with high probability and signal intensity in a urine sample, generally independent from disease. Linear regression of the average values of 30–50 of these peptides can be used to calibrate the obtained ion-counts, which can serve as a measure for relative abundance. We found that ion intensities expressed as peak counts can be used with good confidence for the quantification of urinary polypeptide levels, and that absolute quantification by the addition of isotope-labeled marker peptides offered no additional benefit (Jantos et al, manuscript submitted).
F. Comparison of Datasets
The main aim in most clinical proteomics studies is the definition of differentially present proteins/peptides. Hence, definition of identity is important. That goal can rather easily be accomplished on arrays, where the position on the array tentatively assigns identity. Such a straightforward way to assign identity does not exist in mass spectrometry-driven proteomics; certain deviations must be interpreted and taken into account. Clearly, the deviation permitted, in turn, compromises the accuracy of protein assignment. When using TOF-MS, we found a mass deviation of 50 ppm as an acceptable compromise between the need to assess identity with high accuracy, and at the same time avoid assigning a different identity to the same peptide. Based on the same thought, a migration time deviation of 1% was found to be the optimal compromise.
G. Software and Statistics
After initial processing of peak spectra to identify protein or peptide targets, the next step is to use the datasets to conduct comparative studies on the basis of multivariate statistical analyses. The classification methods can be based, e.g., on linear discriminant analysis (Shannon et al., 2003) or a support vector machine (Smola and Scholkopf, 2004). As with any classification procedure, those methods have their own advantages and drawbacks. Note, however, that neither of those supervised learning methods includes a variable selection procedure. In that context, statistical evaluation of the different peptides appears mandatory to reduce the high dimensionality. Nevertheless, a given biomarker that shows statistical significance does not automatically perform well as a class-discriminating item.
Considering the high dimensionality of the dataset, the statistical analysis must correct for any multiple testing artifacts that are inherent to such an analysis. To see why that correction is of utmost importance, the presumption is that n independent tests are performed with 0.05 as the critical significance level. The probability for a single test to come to a non-significant result (that is, a correct conclusion) is, hence, 1−0.05 = 0.95 (95%). Because the n tests are independent from each other, the probability that all of those n tests correctly reject the n null hypothesis is determined by the product of the single results: 0.95 × … × 0.95 = 0.95n. Hence, the probability to at least wrongly reject one of the n null hypotheses is given by 1−0.95n. Thus, if our experiment involves performing 200 tests on 200 biomarkers, then the experimental error probability is 1−0.95200 = 0.99996. In other words, it is almost certain that, by performing 200 tests on 200 biomarkers, at least one of the declared significant findings is a false positive. Because of the test’s independence, the probability of k such false positives among n biomarkers is simply given by the binomial distribution, with the significance level α as the probability of “success” (i.e., having a false positive). In the example of 200 biomarkers tested at the significance level of 0.05, this probability amount for k = 5 is 0.97355. Even for k = 8, the probability that the findings are rather false positives is still 0.78669. Bonferroni corrections, and their relatives such as the Holm procedure, are the most widespread approach to control the experiment-wide false positive rate (Abdi, 2007). Distribution-free resampling methods, like those from Westfall and Young (Westfall and Young, 1993), are also very powerful and strict methods to control for the experimental error rate. A major drawback of those procedures is that they may lack sufficient statistical power (no significant biomarker can be identified), especially when a limited number of datasets is available. This drawback has lead Benjamini and Hochberg in their seminal paper to introduce the elegant approach of false discovery rate (FDR), which conserves sufficient statistical power of looking for biomarkers that are differentially expressed between two samples when subjected to two different treatments (e.g. disease/no disease) (Benjamini and Hochberg, 1995).
Those reports and the application of statistical methods on a theoretical example shown above clearly underline the importance of using proper statistics. If adequate statistical methods are not employed or replaced by, e.g., a simple students-T test, then the data obtained will likely hold no value, and will be proven invalid in the next set of experiments.
IV. APPLICATION OF CE-MS
A. Urinary biomarkers for renal diseases
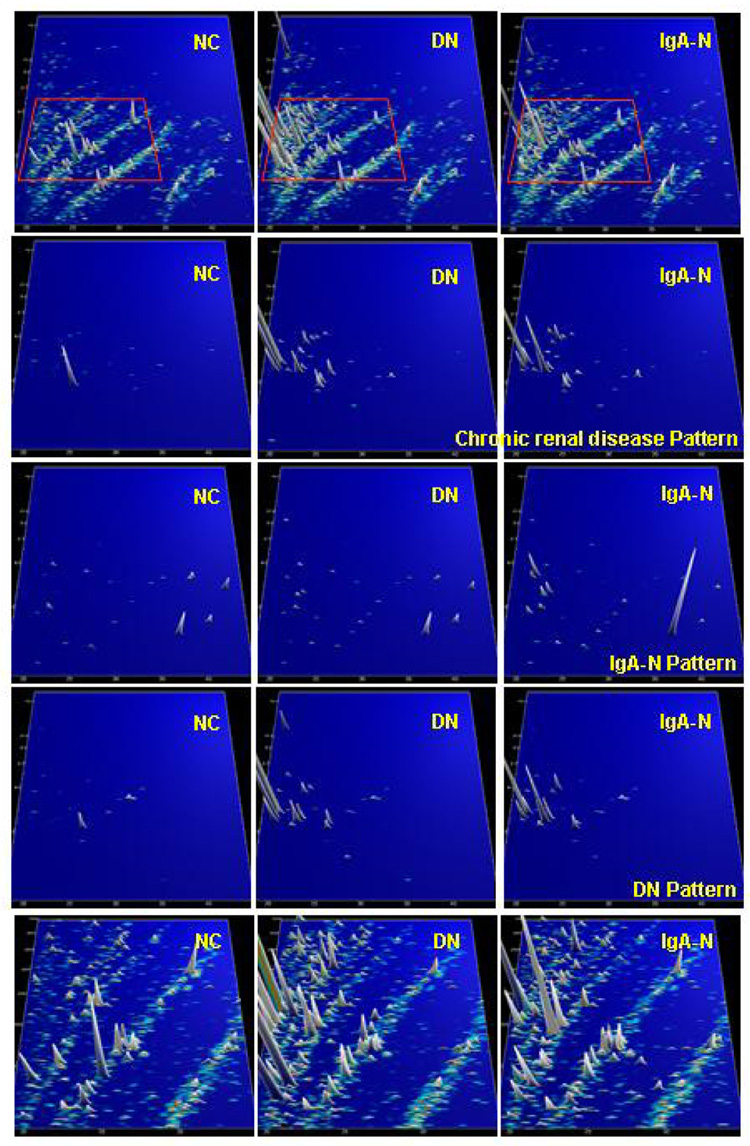
CE-MS analysis of urine samples from patients with various types of chronic renal diseases resulted in the establishment of panels that consisted of 20 to 50 urinary polypeptide markers that allowed diagnosis and discrimination of IgA nephropathy, focal-segmental glomerulosclerosis (FSGS), membranous glomerulonephritis MGN), and minimal-change disease (Weissinger et al., 2004; Haubitz et al., 2005b; Neuhoff et al., 2004). Although those initial studies showed the potential of urinary proteome analysis, they did not include a blinded validation set. In subsequent studies that used an improved and robust sample preparation protocol (Theodorescu et al., 2005) and appropriate statistical evaluation of the individual biomarkers, those initial findings were confirmed and validated. As an example, distribution of potential biomarkers for different chronic renal diseases is shown in Figure 7.
Figure 7.
Protein patterns of healthy volunteers (NC), and patients with diabetic nephropathy (DN) and IgA nephropathy (IgA-N), respectively. Upper panel: compiled patterns consisting of 20 to 100 single measurements, molecular mass (0.8–25 kDa, on a logarithmic scale) against normalized migration time (18–45 min), peak height and color encode the signal intensity. Three middle panels: only selected candidate disease-specific biomarkers are displayed on the same scale. An array of general biomarkers for kidney disease present in DN and IgA-N can be defined. In addition, biomarkers that are specific for DN or IgA-N can be identified, as indicated on the right-hand side. Lower panel: zoom of the upper patterns (all peptides analysed, 1.5–5 kDa, 19–35 min). As evident, several additional biomarkers (of mostly lesser statistical value) are present, which can be further exploited. Reprinted with permission from (Mischak et al., 2007b).
Julian et al. recently reported on the identification and validation of biomarkers for urinary polypeptide biomarkers of renal disease in patients with IgA-associated glomerulonephritides (Julian et al., 2007). In a cohort of 402 patients with various renal disorders and 207 healthy controls, specific biomarkers were defined and subsequently validated. Good et al. (manuscript submitted) identified biomarkers for chronic renal disease and markers that enabled differential diagnosis of FSGS and MGN. Haubitz et al. (manuscript submitted) identified urinary biomarkers that enabled differential diagnosis of ANCA-associated vasculitis, and assessement of therapeutic intervention. All of those studies were based on identical pre-analytical and analytical parameters that enabled comparison between the studies. One of the most striking findings was the significant change of specific collagen fragments associated with each one of those diseases. Because all of those findings were validated in independent blinded test sets, they strongly suggest that defined collagen fragments are specifically associated with the different chronic renal diseases, probably due to changes in the activity of proteases involved in extracellular matrix turnover (see also below).
Decramer et al. (Decramer et al., 2006) applied CE-MS-based urinary proteome analysis to define specific biomarker patterns for different grades of ureteropelvic junction obstruction, a frequently encountered pathology in newborns. In their blinded prospective study, the biomarker patterns could predict the clinical outcome of newborns without signs of proteinuria with 95% accuracy nine months in advance. The accuracy was increased even further to 97% after 12 months (Decramer et al., 2007). Those data not only indicated the potential of urinary proteomics to enable the diagnosis of renal disease, but also suggested the potential to gauge the prognosis.
Rossing et al. (Rossing et al., 2005) demonstrated in a randomized double-blinded study that treatment of macroalbuminuric patients with candesartan had a significant impact on the expression of 15 out of the 113 urinary peptides indicative for diabetic nephropathy (DN).
In initial studies from Mischak et al. (Mischak et al., 2004) and Meier et al. (Meier et al., 2005), CE-MS spectra from patients with diabetes type I or II with/without macroalbuminuria and healthy volunteers were analyzed to create stage-specific polypeptide patterns. In patients with type II diabetes mellitus and unchanged albumin excretion rate, the detected peptide pattern differed significantly from that in patients with high-grade albuminuria. Comparable results were obtained for patients with diabetes type I and renal involvement, diabetic nephropathy (DN). Those results were recently confirmed in a study from Rossing et al., who used the improved sample-preparation protocol (Rossing et al., 2008a). In that study, the authors demonstrated that urinary peptidome profiling by CE-MS enabled the detection of diabetes and DN, and predicted the development of DN in a blinded, prospectively collected population. Further differentiation of patients with diabetic nephropathy from other chronic renal diseases was possible in a blinded cohort with 81% sensitivity and 91% specificity. Those results were further confirmed with blinded samples in an independent study, in which diabetes and DN could be detected with high specificity and sensitivity in a blinded assessment (Snell-Bergeon et al., 2008).
Wittke et al. (Wittke et al., 2005) used CE-MS to analyze urinary samples from patients with different grades of subclinical or clinical acute allograft rejection, urinary tract infection, and without evidence of rejection or infection. Substantial differences were found between patients with transplanted kidneys and patients with native kidneys - most likely due to treatment with the calcineurin-inhibitor cyclosporin A. Additional biomarkers were identified that allowed differentiation between infection and acute rejection. Most importantly, the results were not confounded by acute tubular lesions, tubular atrophy, tubulointerstitial fibrosis, calcineurin inhibitor toxicity, proteinuria, hematuria, allograft function, or different immunosuppressive regimens.
B. Urinary biomarkers for urological disorders
Theodorescu et al. (Theodorescu et al., 2006) described the CE-MS detection and validation of biomarkers of urothelial carcinoma. A bladder cancer-specific biomarker pattern was established by an initial definition in a training set composed of 46 patients with urothelial carcinoma and 33 healthy subjects, and further refinement with CE-MS spectra of 366 urine samples from healthy volunteers and patients with malignant and non-malignant genitourinary diseases. With this two-step biomarker discovery approach, the authors could establish a prediction model composed of 22 urinary peptides. This model correctly classified all urothelial carcinoma patients and all healthy controls, when applied to a blinded test set that contained 31 urothelial carcinoma patients, 11 healthy individuals and 138 non-malignant genitourinary disease patients,. Differentiation between bladder cancer and other malignant and non-malignant diseases (such as renal nephrolithiasis) was accomplished with at least 86% – 100% sensitivity.
In a pilot study (Theodorescu et al., 2005), CE-MS techniques were used to define potential urinary peptide biomarkers for prostate cancer (PCa). Urine samples from 47 patients who underwent prostate biopsy were analyzed. On the basis of prostate biopsy, 26 patients in this group were diagnosed with PCa and 21 with benign prostatic hyperplasia (BPH). The data indicated that several polypeptides allowed the identification of PCa with 92% sensitivity and 96% specificity in the training set upon complete crossvalidation. However, those data could not be validated in a subsequent blinded assessment; once more the importance of a blinded test set was underlined. In a subsequent study, first-void urine was found to be a more appropriate sample to define of PCa-specific biomarkers. To enable validation of first-void urine, biomarkers specific of first-void urine were determined. The results of that study indicated that the identified biomarkers originate from secretions of the prostate into urine. After refinement of the PCa-specific biomarker pattern, using urine samples from 54 PCa and 62 BPH patients, a model with ten potential biomarkers resulted in the prediction of 88.9% (32/36) of the PCa and of 66.7% (16/24) of the BPH patients in a second blinded set of patient samples (Theodorescu et al., 2008).
C. Application of urinary proteome analysis to non-renal diseases
As described above, bodyfluids are suspected to be highly informative of the tissues with which they are directly in contact. That assumption is one of the reasons why urine was used in many studies on diseases of urogenital tract. However the fact that plasma is filtered by the kidney also stimulated researchers to look into urine for biomarkers of disease from more distant organs.
CE-MS was applied to the clinical follow-up of patients after allogeneic hematopoietic stem cell transplantation (HSCT) (Kaiser et al., 2004; Weissinger et al., 2005). Urine samples from 40 patients after HSCT (35 allogeneic, 5 autologous) and five patients with sepsis were collected during a period of 100 days (a maximum of 10 samples per patient) for CE-MS analysis. A pattern that consisted of 16 differentially excreted polypeptides indicated early graft-versus-host-disease (GVHD). The pattern of markers discriminated patients with early GVHD from patients without complications with 82% specificity and 100% sensitivity. In a subsequent study based on the improved sample-preparation protocol that included a blinded multi-center validation of 100 patients with more than 600 samples collected prospectively, the initial results were confirmed, although with reduced specificity and sensitivity (Weissinger et al., 2007). Initial results of preemptive therapy based on proteome profiling clearly indicate a benefit for the yet limited number of patients (Weissinger, unpublished).
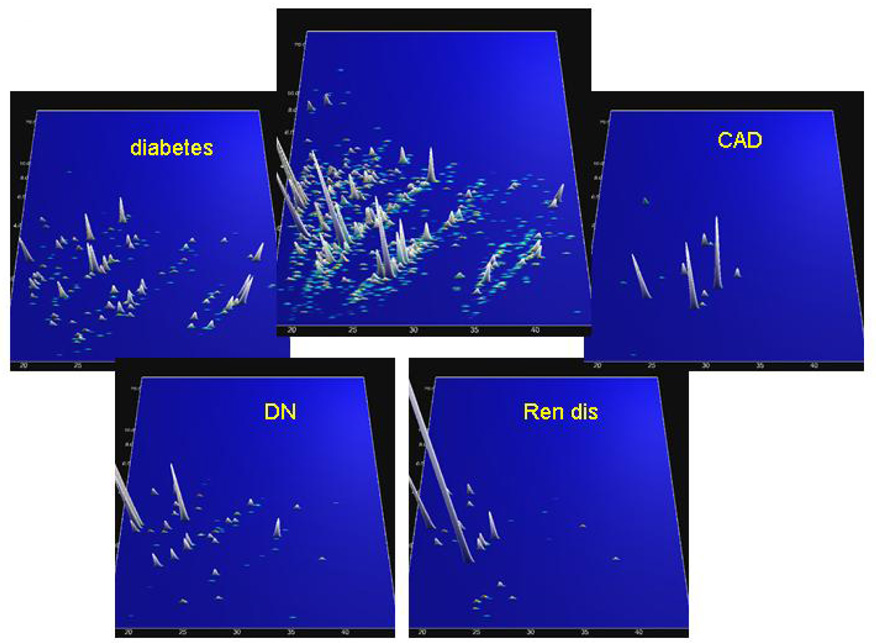
Zimmerli et al. (Zimmerli et al., 2008) examined urine from patients who underwent coronary artery bypass grafting, or from patients after acute myocardial infarction. Urine samples from patients and controls were analyzed with CE-MS to identify coronary artery disease (CAD)-specific biomarkers. In a blinded assessment, the specific urinary biomarkers identified CAD patients with greater than 90% sensitivity and specificity. In a recent study, those biomarkers could be evaluated for the prognostic potential (Snell-Bergeon et al., 2008). That study also highlighted another feature of the CE-MS analysis: one analysis can be investigated for several different biomarker panels that are indicative for different pathological conditions, as shown in Figure 8.
Figure 8.
CE-MS data from a urine sample from one patient in the CACTI study. The molecular mass (0.8–25 kDa, on a logarithmic scale) is plotted against normalized migration time (18–45 min), peak height and color encode the signal intensity. In the center plot, all relevant peptides in the sample are shown. The four panels grouped around the center panel show peptides that are statistically significantly altered in the respective diseases (diabetes, diabetic nephropathy (DN), chronic renal disease (Ren dis), and coronary artery disease (CAD)). Based on the CE-MS analysis, the patient (original label 1757) scored positive for diabetes, diabetic nephropathy, and coronary artery disease. Based on the clinical data at the time of sampling, this patient had diabetes for 34 years with urinary albumin at 105 mg/l. The patient experienced a cardiovascular event 9 months after the urine sample was collected. Reprinted with permission from (Rossing et al., 2008b).
D. Aging
Aging induces morphological changes of the kidney, and leads to a reduction of renal function. In order to gain insight into the processes of renal aging, Zurbig et al. (manuscript submitted) examined with CE-MS urine samples collected from 324 healthy individuals, aged 2–73 years. From the 5000 urinary polypeptides resolved by CE-MS that were present in at least 40% of the subjects of one age-group, 325 displayed statistically significant age-related changes. Most of the markers changed significant during puberty, and coincided with the completion of renal development. However, 49 peptides could be correlated with aging in adults. A striking observation was that some of these peptides were also found to be differentially secreted in chronic renal diseases, including DN, FSGS, MGN, and vasculitis. Association of renal aging and chronic renal disease was confirmed in a blinded evaluation of samples from healthy individuals and DN patients. Sequence information of some of those aging markers suggested that one prominent mechanisms of human aging is a reduced turnover of extracellular matrix, that results in increased fibrosis. After further refinement, the age-related polypeptide marker patterns might allow the non-invasive detection of renal lesions in healthy persons, and the testing of an individuals’ suitability for kidney donation.
E. Pathophysiological aspects of biomarkers
Even though the majority of potential urinary biomarkers described to date have not been sequenced, sequences are available for more than 500 different urinary peptides (Coon et al., 2008). Not unexpectedly, most of those peptides are derived from the most-abundant proteins in the body: albumin, beta 2-macroglobulin, uromodulin, and collagen - mainly type I, II and III. Consequently, a valid question is whether urinary peptidomics in renal disease is not just another way to measure glomerular injury, that could probably be assessed with similar precision, but less effort, by measuring albuminuria (Comper et al., 2004). Although that question cannot be answered with absolute confidence, it is certain, however, that differential diagnosis based on urinary proteome analysis is possible (Haubitz et al., 2005b; Haubitz et al., 2005a; Fliser et al., 2007; Rossing et al., 2008a). The fact that patients in complete remission without albuminuria still exhibit apparently disease-specific changes in urinary polypeptides (Weissinger et al., 2004) strongly suggests that those peptides contain clues about the pathogenesis and are not merely degradation products. It is tempting to speculate that the disease-specific peptides might be indirect indicators of the activity of disease-specific proteases, as recently suggested by Haubitz (Haubitz et al., 2005b). That hypothesis is further strengthened by the detection of specific collagen fragments that correlated with the disease-specific activity of matrix metalloproteases (Nemirovskiy et al., 2007).
Although the evidence is still scarce, it is an attractive hypothesis that urinary peptides of diagnostic value are not merely degradation products of abundant larger proteins, but a result of distinct, disease-specific processes; in many cases, due to significant changes in the activity of proteases. That assumption is supported by various findings; a) the increase of collagen and extracellular matrix is observed in patients with diabetes and DN; b) collagen fragments are significantly reduced in diabetic urine (Rossing et al., 2008a); and c) reduced activity of proteases and protection of the extracellular matrix from proteolysis by AGEs, proposed key pathological changes in diabetes mellitus (Rossing et al., 2008b).
A similar scenario might be applicable to albuminuria. Consequently, an albumin-derived biomarker is not simply “an albumin fragment”, but rather a specific fragment, defined by its specific C- and N-terminus. Unfortunately, such essential detailed information is frequently absent [e.g., see the recently published database of urinary proteins in Adachi et al. (Adachi et al., 2006)]. Once a substantial number of additional peptides are sequenced, a thorough examination of the sequences of the urinary peptides and comparison with protease specificities might provide additional support for the above hypothesis, and could lead to a better insight into the regulation and pathophysiological role of specific proteases in many diseases.
A related hypothesis can be proposed, on the urinary peptidome displaying, to a large degree, the turnover of the extracellular matrix. That hypothesis has been generated as a result of the observation that the major urinary peptides are not, as expected, the “usual suspects” like albumin or uromodulin, but rather specific collagen degradation products, and that several of those products are significantly reduced in diseases where an increase of ECM has been reported (Schena and Gesualdo, 2005). Consequently, those peptides might be derived from ECM turnover. Changes in that turnover also result in indicative changes in urinary peptides, which serve as a very specific, non-invasive indicator for alterations in ECM turnover, which in turn is likely to be disease-specific. Such changes in the ECM turnover might be due to, e.g., an invasion of tumors (ECM must be “dissolved” in order to make room for the growing tumor), fibrosis (reduced ECM degradation), increased arterial stiffness (change in ECM composition), or changes in endothelium.
V. CONCLUDING REMARKS AND OUTLOOK
CE-MS fulfills the requirements for broad application in routine clinical practice, as indicated by the validation of GvHD, renal disease, and prostate cancer-specific marker patterns in hundreds of patient samples under identical conditions that use the same CE and MS platforms (Decramer et al., 2006; Theodorescu et al., 2006; Rossing et al., 2008a; Coon et al., 2008; Theodorescu et al., 2008). It must be stated, however, that the future implementation of proteome profiling in laboratory diagnosis relies on more than just technological advancements. Of equal importance are concerted efforts in the development of global standardization procedures for the planning, execution, and reporting of clinical proteomic studies. With the adoption of standardized methods for identification of disease-specific biomarkers, the information provided by proteomic platforms will bring clinical chemists a step closer to the ultimate goal to capture all critical pieces of information of a particular disease in a single diagnostic step. These improvements will hopefully result in the integration of MS-based proteomic methods into the armamentarium of clinical laboratories.
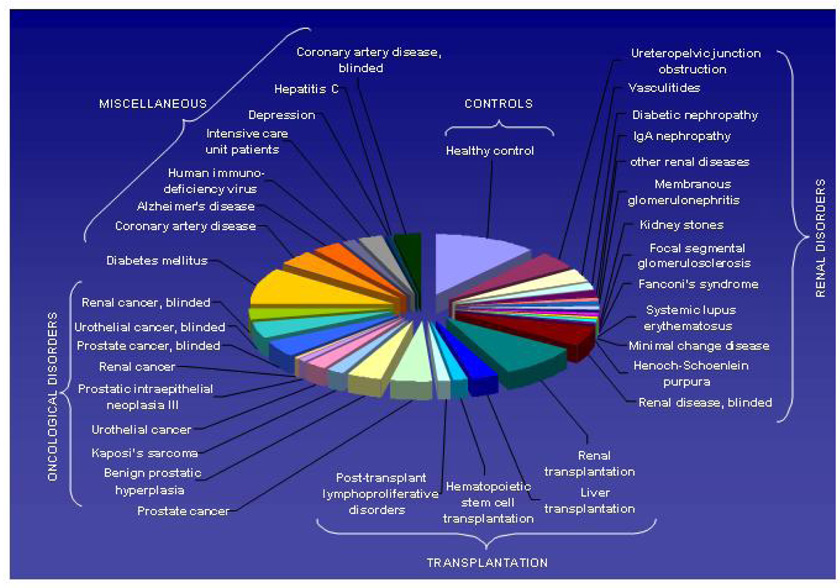
A prerequisite for the identification of valid biomarkers is a very large number of comparable datasets. A first example of such a database that contains datasets from patients with a variety of different pathological conditions and controls was recently introduced (Coon et al., 2008) (Figure 9). Such databases allow the definition of biomarkers that not only enable differentiating between case (disease) and control (healthy), but also between a specific disease and several other pathological conditions that might represent with similar symptoms, as shown in Figure 10. Hence, we are confident that publicly available databases that contain thousands of datasets from individual samples will greatly ease and expedite the definition of relevant biomarkers.
Figure 9.
Graphic depiction of comparable datasets (analyzed with identical pre-analytical preparation, instruments, and analytical parameters) obtained from subjects with different diseases/(pathological) conditions currently represented in the human urinary proteome database. Reprinted with permission from (Coon et al., 2008).
Figure 10.
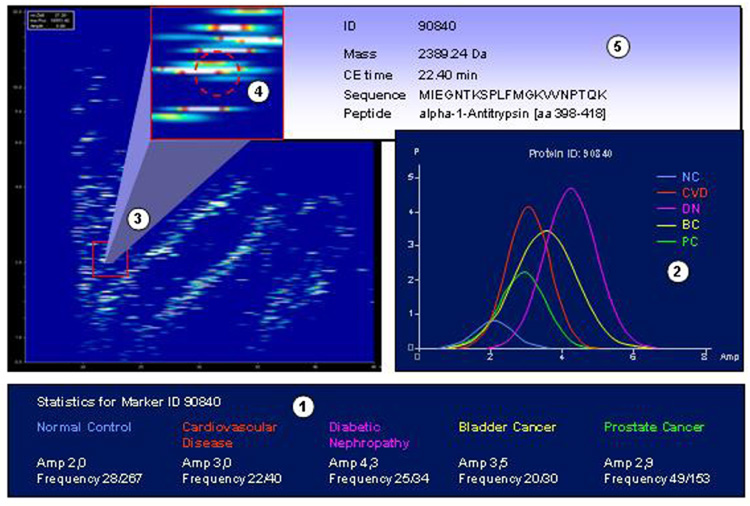
Graphic depiction of the discovery of potential biomarkers for diabetic nephropathy. CE-MS datasets from control and patients with prostate cancer, bladder cancer, and cardiovascular disease are compared to data obtained from patients with diabetic nephropathy (1) using appropriate statistics (adjustments for multiple testing as described in, e.g., (Westfall and Young, 1993; Benjamini and Hochberg, 1995; Reiner et al., 2003; Abdi, 2007)). Potential biomarkers that show significant differences in amplitude and/or distribution (2) are located in the database (3). The clustering of the biomarker (with respect to deviation) is examined in comparison to neighboring peptides (4). If found appropriate, ID, mass, normalized migration time, and, if known, sequence can be retrieved from the database (5). Reprinted with permission from (Coon et al., 2008).
A promising approach to greatly decrease the time required for a single CE-MS analysis appears to be the application of microchips for CE separation. This approach, outlined in recent reviews (Peng et al., 2008; Gaspar et al., 2008), might reduce analysis-times to less than five minutes. However, issues like the demand for higher sensitivity of the MS and very high resolution of the CE (due to the large number of analytes in the sample) still must be addressed.
The currently used approach of CE-MS analysis of human bodyfluids certainly is limited; for example, large proteins cannot be displayed. However, that limitation appears to be one of the strengths of the approach: limiting the technology to only a fraction of the proteome that can be displayed with high accuracy has enabled the identification and validation of several biomarkers in blinded, prospective, multicenter clinical trials.
Furthermore, we are certain that CE-MS does represent an excellent tool for the analysis of, for example, tryptic peptides, most likely highly complementary to the conventionally used LC-MS(MS) approaches. It is to be anticipated that CE-MS will be more widely used once a vendor can develop, and is capable of selling, a complete CE-MS system.
Acknowledgements
HM was supported in part by EUROTRANS-BIO grant ETB-2006-016 and EU Funding through InGenious HyperCare (LSHM-C7-2006-037093) and PREDICTIONS (1272568). J.J.C. acknowledges the University of Wisconsin—Madison,Thermo Fisher, the Beckman Foundation, and the NIH (1R01GM080148) for financial support. JN was supported in part by NIH grants DK078244, DK080301, DK071802, DK061525, and DK064400. EMW was supported in part by the "Deutsche-Jose-Carreras-Leukämie Stiftung" DJCLS 98/05 and by the “Deutsche Forschungsgemeinschaft”DFG Mi-658/01 to. The work of JPS was supported by Inserm, the “Direction Régional Clinique” (CHU de Toulouse, France) under the Interface program, by the Fondation pour la Recherche Médicale and by the ANR grant: ANR-07-PHYSIO-004-01). AFD was supported by British Heart Foundation Chair and Programme Grant (BHF/RG/07/005/23633), Wellcome Trust Cardiovascular Functional Genomics Initiative (066780/2/012), EURATools (LSHG-CT-2005-019015) and InGenious HyperCare (LSHM-C7-2006-037093). We are grateful to Mohammed Dakna for critically reviewing the manuscript and help with the chapter on statistics and data evaluation.
Biographies

Harald Mischak received his PhD from the Technical University of Vienna, Austria, in 1986. After postdoctoral work at the University of Vienna and subsequently at the NCI, Bethesda, Maryland, USA, he became a Group Leader at the GSF Research Centre, Munich, Germany. After a sabbatical at the NIDDK, Bethesda, Maryland, aiming at the elucidation of the structure of phorbol-ester receptors, he accepted a position as Professor in the Dep. of Nephrology at the Medical School of Hannover. He is a founder and chief scientific officer of Mosaiques diagnostics, which was started with the aim to identify disease-specific polypeptides. His main interest lies in the identification of disease-specific extracellular peptides and proteins, aiming towards improvement in clinical diagnosis.

Joshua Coon received his PhD in 2002 from the University of Florida. Immediately following he became an NIH-NRSA Postdoctoral Fellow at the University of Virginia where he co-invented electron transfer dissociation. In 2005 he started a faculty position in the Departments of Chemistry and Biomolecular Chemistry at the University of Wisconsin-Madison. His research interests are the development of new protein sequencing technologies and their application to developmental and human biology.

Jan Novak, MS, PhD, has received his MS in Biology in 1987 from Charles University in Prague and PhD in Cell and Molecular Biology from Czech Academy of Sciences in 1990. After his postdoctoral work on post-translationally modified bacterially produced peptides at the University of Alabama in Birmingham, he has worked on the role of aberrant post-translational modifications in renal diseases. Currently, he is an Associate Professor at the Department of Microbiology at the University of Alabama in Birmingham and leads a group focused on studies of glycoimmunobiology in health and disease and characterization of biomarkers for renal diseases.

Eva Weissinger, M.D. received her medical degree from the University of Vienna, Austria, and was then a clinical and research fellow in the department of cardiothoracic surgery. She received her postdoctoral training at the NIH/NCI in Bethesda, MD, USA and returned to Europe to work in the Department of Clinical Hematology at the Ludwig Maximilians University in Munich, Germany. After a sabbatical at the NIH/NHLBI in Bethesda, MD, she became Senior Scientist in the Department of Hematology, Hemostasis, Oncology, and Stem Cell Transplantation at the Hannover Medical School, where she is now an associate professor and heads the Laboratory of Transplantation Biology. Dr. Weissinger’s main research focus is allogeneic hematopoietic stem cell transplantation, particularly in refractory acute myeloid leukemia, and the application of proteomics to post-transplantation follow-up.

Joost Schanstra received his PhD in Life Sciences (1996) from the University of Groningen, The Netherlands. From 1996 to 1999 he worked as a post-doctoral researcher at Inserm U388 in Toulouse, France. In 1999 he obtained a staff research position in the same laboratory. Since 2004 he is group-leader at Inserm U858, Toulouse, France. His main interest is in paediatric renal disease. His research focuses on the identification of non-invasive biomarkers for the early detection of renal disease in the paediatric population and on the identification of new molecules/therapeutic-approaches able to slow down the progression of renal fibrosis.

Anna F. Dominiczak, OBE, MD, FRCP, FRSE, FAHA, FMedSci is a British Heart Foundation Professor of Cardiovascular Medicine and a Director of the BHF Glasgow Cardiovascular Research Centre. She graduated in Medicine at the Medical School of Gdansk, Poland and completed her postgraduate clinical and research training in Glasgow and Michigan, USA. Her major research interests are in cardiovascular genomics and vascular biology. Anna is a co-ordinator of the Wellcome Trust funded Cardiovascular Functional Genomics Consortium and one of principal investigators in the MRC-funded BRIGHT Study into the genetics of essential hypertension. She is a Fellow of the Royal College of Physicians, the Academy of Medical Sciences, the Royal Society of Edinburgh and the American Heart Association as well as a member of the Scientific Council of the European Society of Hypertension. Anna has also been invited to give numerous lectures including the Canadian Hypertension Society Presidential Lecture in 2003, the Arthur C. Corcoran Memorial Lecture of the American Heart Association in 2004 and the R. D. Wright Lecture of the Australian Council for High Blood Pressure Research in 2005.
Footnotes
Statement of Competing Financial Interests
HM is the founder and co-owner of Mosaiques Diagnostics, who developed the CE-MS technology for clinical application.
Reference List
- Abdi H. In: Bonferroni and Sidak corrections for multiple comparisons. Salkind NJ, editor. Thousand Oaks (CA): Sage; 2007. [Google Scholar]
- Adachi J, Kumar C, Zhang Y, Olsen JV, Mann M. The human urinary proteome contains more than 1500 proteins including a large proportion of membranes proteins. Genome Biol. 2006;7:R80. doi: 10.1186/gb-2006-7-9-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amon S, Plematl A, Rizzi A. Capillary zone electrophoresis of glycopeptides under controlled electroosmotic flow conditions coupled to electrospray and matrix-assisted laser desorption/ionization mass spectrometry. Electrophoresis. 2006;27:1209–1219. doi: 10.1002/elps.200500725. [DOI] [PubMed] [Google Scholar]
- Anderson NL, Anderson NG. The human plasma proteome: history, character, and diagnostic prospects. Mol. Cell Proteomics. 2002;1:845–867. doi: 10.1074/mcp.r200007-mcp200. [DOI] [PubMed] [Google Scholar]
- Bakry R, Huck CW, Najam-ul-Haq M, Rainer M, Bonn GK. Recent advances in capillary electrophoresis for biomarker discovery. J. Sep. Sci. 2007;30:192–201. doi: 10.1002/jssc.200600323. [DOI] [PubMed] [Google Scholar]
- Barnidge DR, Dratz EA, Martin T, Bonilla LE, Moran LB, Lindall A. Absolute quantification of the G protein-coupled receptor rhodopsin by LC/MS/MS using proteolysis product peptides and synthetic peptide standards. Anal. Chem. 2003;75:445–451. doi: 10.1021/ac026154+. [DOI] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J Royal Stat Soc B (Methodological) 1995;57:125–133. [Google Scholar]
- Candiano G, Musante L, Bruschi M, Petretto A, Santucci L, Del BP, Pavone B, Perfumo F, Urbani A, Scolari F, Ghiggeri GM. Repetitive fragmentation products of albumin and alpha1-antitrypsin in glomerular diseases associated with nephrotic syndrome. J. Am. Soc. Nephrol. 2006;17:3139–3148. doi: 10.1681/ASN.2006050486. [DOI] [PubMed] [Google Scholar]
- Chalmers MJ, Mackay CL, Hendrickson CL, Wittke S, Walden M, Mischak H, Fliser D, Just I, Marshall AG. Combined top-down and bottom-up mass spectrometric approach to characterization of biomarkers for renal disease. Anal. Chem. 2005;77:7163–7171. doi: 10.1021/ac050983o. [DOI] [PubMed] [Google Scholar]
- Comper WD, Osicka TM, Clark M, MacIsaac RJ, Jerums G. Earlier detection of microalbuminuria in diabetic patients using a new urinary albumin assay. Kidney Int. 2004;65:1850–1855. doi: 10.1111/j.1523-1755.2004.00585.x. [DOI] [PubMed] [Google Scholar]
- Coon JJ, Ueberheide B, Syka JE, Dryhurst DD, Ausio J, Shabanowitz J, Hunt DF. Protein identification using sequential ion/ion reactions and tandem mass spectrometry. Proc. Natl. Acad. Sci. U. S. A. 2005;102:9463–9468. doi: 10.1073/pnas.0503189102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coon JJ, Zürbig P, Dakna M, Dominiczak AF, Decramer S, Fliser D, Frommberger M, Golovko I, Good DM, Herget-Rosenthal S, Jankowski J, Julian BA, Kellmann M, Kolch W, Massy Z, Novak J, Rossing K, Schanstra JP, Schiffer E, Theodorescu D, Vanholder R, Weissinger EM, Mischak H, Schmitt-Kopplin P. CE-MS analysis of the human urinary proteome for biomarker discovery and disease diagnostics. Proteomics Clin. Appl. 2008;2:964–973. doi: 10.1002/prca.200800024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decramer S, Bascands JL, Schanstra JP. Non-invasive markers of ureteropelvic junction obstruction. World J Urol. 2007;25:457–465. doi: 10.1007/s00345-007-0201-8. [DOI] [PubMed] [Google Scholar]
- Decramer S, Wittke S, Mischak H, Zurbig P, Walden M, Bouissou F, Bascands JL, Schanstra JP. Predicting the clinical outcome of congenital unilateral ureteropelvic junction obstruction in newborn by urinary proteome analysis. Nat. Med. 2006;12:398–400. doi: 10.1038/nm1384. [DOI] [PubMed] [Google Scholar]
- DeKeyser SS, Li L. Matrix-assisted laser desorption/ionization Fourier transform mass spectrometry quantitation via in cell combination. Analyst. 2006;131:281–290. doi: 10.1039/b510831d. [DOI] [PubMed] [Google Scholar]
- deVera IE, Katz JE, Agus DB. Clinical proteomics: the promises and challenges of mass spectrometry-based biomarker discovery. Clin. Adv. Hematol. Oncol. 2006;4:541–549. [PubMed] [Google Scholar]
- Dolnik V. Capillary electrophoresis of proteins 2005–2007. Electrophoresis. 2008;29:143–156. doi: 10.1002/elps.200700584. [DOI] [PubMed] [Google Scholar]
- Erny GL, Elvira C, San RJ, Cifuentes A. Capillary electrophoresis using copolymers of different composition as physical coatings: a comparative study. Electrophoresis. 2006;27:1041–1049. doi: 10.1002/elps.200500692. [DOI] [PubMed] [Google Scholar]
- Fliser D, Novak J, Thongboonkerd V, Argiles A, Jankowski V, Girolami M, Jankowski J, Mischak H. Advances in urinary proteome analysis and biomarker discovery. J Am Soc Nephrol. 2007;18:1057–1071. doi: 10.1681/ASN.2006090956. [DOI] [PubMed] [Google Scholar]
- Freed GL, Cazares LH, Fichandler CE, Fuller TW, Sawyer CA, Stack BC, Jr, Schraff S, Semmes OJ, Wadsworth JT, Drake RR. Differential capture of serum proteins for expression profiling and biomarker discovery in pre- and posttreatment head and neck cancer samples. Laryngoscope. 2008;118:61–68. doi: 10.1097/MLG.0b013e31814cf389. [DOI] [PubMed] [Google Scholar]
- Frommberger M, Zürbig P, Jantos J, Krahn T, Mischak M, Pich A, Jus I, Schmitt-Kopplin P, Schiffer E. Peptidomic analysis of rat urine using capillary electrophoresis coupled to mass spectrometry. Proteomics Clin. Appl. 2007;1:650–660. doi: 10.1002/prca.200700195. [DOI] [PubMed] [Google Scholar]
- Garza S, Chang S, Moini M. Simplifying capillary electrophoresis-mass spectrometry operation: eliminating capillary derivatization by using self-coating background electrolytes. J Chromatogr. A. 2007;1159:14–21. doi: 10.1016/j.chroma.2007.02.038. [DOI] [PubMed] [Google Scholar]
- Gaspar A, Englmann M, Fekete A, Harir M, Schmitt-Kopplin P. Trends in CE-MS 2005–2006. Electrophoresis. 2008;29:66–79. doi: 10.1002/elps.200700721. [DOI] [PubMed] [Google Scholar]
- Good DM, Coon JJ. Advancing proteomics with ion/ion chemistry. Biotechniques. 2006;40:783–789. doi: 10.2144/000112194. [DOI] [PubMed] [Google Scholar]
- Good DM, Thongboonkerd V, Novak J, Bascands JL, Schanstra JP, Coon JJ, Dominiczak A, Mischak H. Body Fluid Proteomics for Biomarker Discovery: Lessons from the Past Hold the Key to Success in the Future. J Proteome Res. 2007;6:4549–4555. doi: 10.1021/pr070529w. [DOI] [PubMed] [Google Scholar]
- Goodsaid F, Bandow JE, Mischak H. Grand Rounds in Proteomics at the FDA. Proteomics Clin. Appl. 2007;1:1526–1531. doi: 10.1002/prca.200700575. [DOI] [PubMed] [Google Scholar]
- Guerrier L, Claverol S, Fortis F, Rinalducci S, Timperio AM, Antonioli P, Jandrot-Perrus M, Boschetti E, Righetti PG. Exploring the platelet proteome via combinatorial, hexapeptide ligand libraries. J Proteome Res. 2007;6:4290–4303. doi: 10.1021/pr0703371. [DOI] [PubMed] [Google Scholar]
- Haselberg R, de Jong GJ, Somsen GW. Capillary electrophoresis-mass spectrometry for the analysis of intact proteins. J Chromatogr. A. 2007;1159:81–109. doi: 10.1016/j.chroma.2007.05.048. [DOI] [PubMed] [Google Scholar]
- Haubitz M, Fliser D, Rupprecht H, Floege J, Haller H, Rossing K, Walden M, Wittke S, Mischak H. Defining renal diseases based on proteom analysis. Nephrology Dialysis Transplantation. 2005a;20:V20. [Google Scholar]
- Haubitz M, Wittke S, Weissinger EM, Walden M, Rupprecht HD, Floege J, Haller H, Mischak H. Urine protein patterns can serve as diagnostic tools in patients with IgA nephropathy. Kidney Int. 2005b;67:2313–2320. doi: 10.1111/j.1523-1755.2005.00335.x. [DOI] [PubMed] [Google Scholar]
- Hernandez-Borges J, Borges-Miquel TM, Rodriguez-Delgado MA, Cifuentes A. Sample treatments prior to capillary electrophoresis-mass spectrometry. J Chromatogr. A. 2007;1153:214–226. doi: 10.1016/j.chroma.2006.10.070. [DOI] [PubMed] [Google Scholar]
- Issaq HJ, Chan KC, Janini GM, Conrads TP, Veenstra TD. Multidimensional separation of peptides for effective proteomic analysis. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 2005;817:35–47. doi: 10.1016/j.jchromb.2004.07.042. [DOI] [PubMed] [Google Scholar]
- Jensen PK, Pasa-Tolic L, Anderson GA, Horner JA, Lipton MS, Bruce JE, Smith RD. Probing proteomes using capillary isoelectric focusing-electrospray ionization Fourier transform ion cyclotron resonance mass spectrometry. Anal. Chem. 1999;71:2076–2084. doi: 10.1021/ac990196p. [DOI] [PubMed] [Google Scholar]
- Julian BA, Wittke S, Novak J, Good DM, Coon JJ, Kellmann M, Zurbig P, Schiffer E, Haubitz M, Moldoveanu Z, Calcatera SM, Wyatt RJ, Sykora J, Sladkova E, Hes O, Mischak H, McGuire BM. Electrophoretic methods for analysis of urinary polypeptides in IgA-associated renal diseases. Electrophoresis. 2007;28:4469–4483. doi: 10.1002/elps.200700237. [DOI] [PubMed] [Google Scholar]
- Kaiser T, Kamal H, Rank A, Kolb HJ, Holler E, Ganser A, Hertenstein B, Mischak H, Weissinger EM. Proteomics applied to the clinical follow-up of patients after allogeneic hematopoietic stem cell transplantation. Blood. 2004;104:340–349. doi: 10.1182/blood-2004-02-0518. [DOI] [PubMed] [Google Scholar]
- Kasicka V. Recent developments in CE and CEC of peptides. Electrophoresis. 2008;29:179–206. doi: 10.1002/elps.200700550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolch W, Neususs C, Pelzing M, Mischak H. Capillary electrophoresis-mass spectrometry as a powerful tool in clinical diagnosis and biomarker discovery. Mass Spectrom Rev. 2005;24:959–977. doi: 10.1002/mas.20051. [DOI] [PubMed] [Google Scholar]
- Lapolla A, Fedele D, Reitano R, Arico NC, Seraglia R, Traldi P, Marotta E, Tonani R. Enzymatic digestion and mass spectrometry in the study of advanced glycation end products/peptides. J. Am. Soc. Mass Spectrom. 2004;15:496–509. doi: 10.1016/j.jasms.2003.11.014. [DOI] [PubMed] [Google Scholar]
- Lapolla A, Tubaro M, Fedele D, Reitano R, Arico NC, Ragazzi E, Seraglia R, Vogliardi S, Traldi P. A matrix-assisted laser desorption/ionization mass spectrometry study of the non-enzymatic glycation products of human globins in diabetes. Rapid Commun. Mass Spectrom. 2005;19:162–168. doi: 10.1002/rcm.1759. [DOI] [PubMed] [Google Scholar]
- Lescuyer P, Hochstrasser D, Rabilloud T. How Shall We Use the Proteomics Toolbox for Biomarker Discovery? J. Proteome Res. 2007;6:3371–3376. doi: 10.1021/pr0702060. [DOI] [PubMed] [Google Scholar]
- Lou X, Xiao T, Zhao K, Wang H, Zheng H, Lin D, Lu Y, Gao Y, Cheng S, Liu S, Xu N. Cathepsin D is secreted from M-BE cells: its potential role as a biomarker of lung cancer. J Proteome Res. 2007;6:1083–1092. doi: 10.1021/pr060422t. [DOI] [PubMed] [Google Scholar]
- Marshall AG, Hendrickson CL, Jackson GS. Fourier transform ion cyclotron resonance mass spectrometry: a primer. Mass Spectrom. Rev. 1998;17:1–35. doi: 10.1002/(SICI)1098-2787(1998)17:1<1::AID-MAS1>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- McLerran D, Grizzle WE, Feng Z, Bigbee WL, Banez LL, Cazares LH, Chan DW, Diaz J, Izbicka E, Kagan J, Malehorn DE, Malik G, Oelschlager D, Partin A, Randolph T, Rosenzweig N, Srivastava S, Srivastava S, Thompson IM, Thornquist M, Troyer D, Yasui Y, Zhang Z, Zhu L, Semmes OJ. Analytical validation of serum proteomic profiling for diagnosis of prostate cancer: sources of sample bias. Clin. Chem. 2008a;54:44–52. doi: 10.1373/clinchem.2007.091470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLerran D, Grizzle WE, Feng Z, Thompson IM, Bigbee WL, Cazares LH, Chan DW, Dahlgren J, Diaz J, Kagan J, Lin DW, Malik G, Oelschlager D, Partin A, Randolph TW, Sokoll L, Srivastava S, Srivastava S, Thornquist M, Troyer D, Wright GL, Zhang Z, Zhu L, Semmes OJ. SELDI-TOF MS whole serum proteomic profiling with IMAC surface does not reliably detect prostate cancer. Clin. Chem. 2008b;54:53–60. doi: 10.1373/clinchem.2007.091496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meier M, Kaiser T, Herrmann A, Knueppel S, Hillmann M, Koester P, Danne T, Haller H, Fliser D, Mischak H. Identification of urinary protein pattern in type 1 diabetic adolescents with early diabetic nephropathy by a novel combined proteome analysis. J. Diabetes Complications. 2005;19:223–232. doi: 10.1016/j.jdiacomp.2004.10.002. [DOI] [PubMed] [Google Scholar]
- Mischak H, Apweiler R, Banks RE, Conaway M, Coon JJ, Dominizak A, Ehrich JH, Fliser D, Girolami M, Hermjakob H, Hochstrasser DF, Jankowski V, Julian BA, Kolch W, Massy Z, Neususs C, Novak J, Peter K, Rossing K, Schanstra JP, Semmes OJ, Theodorescu D, Thongboonkerd V, Weissinger EM, Van Eyk JE, Yamamoto T. Clinical Proteomics: a need to define the field and to begin to set adequate standards. Proteomics Clin. Appl. 2007a;1:148–156. doi: 10.1002/prca.200600771. [DOI] [PubMed] [Google Scholar]
- Mischak H, Julian BA, Novak J. High-resolution proteome/peptidome analysis of peptides and low-molecular-weight proteins in urine. Proteomics Clin. Appl. 2007b;1:792–804. doi: 10.1002/prca.200700043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mischak H, Kaiser T, Walden M, Hillmann M, Wittke S, Herrmann A, Knueppel S, Haller H, Fliser D. Proteomic analysis for the assessment of diabetic renal damage in humans. Clin. Sci. (Lond) 2004;107:485–495. doi: 10.1042/CS20040103. [DOI] [PubMed] [Google Scholar]
- Nemirovskiy OV, Dufield DR, Sunyer T, Aggarwal P, Welsch DJ, Mathews WR. Discovery and development of a type II collagen neoepitope (TIINE) biomarker for matrix metalloproteinase activity: From in vitro to in vivo. Anal. Biochem. 2007;369:41–53. doi: 10.1016/j.ab.2006.10.034. [DOI] [PubMed] [Google Scholar]
- Neuhoff N, Kaiser T, Wittke S, Krebs R, Pitt A, Burchard A, Sundmacher A, Schlegelberger B, Kolch W, Mischak H. Mass spectrometry for the detection of differentially expressed proteins: a comparison of surface-enhanced laser desorption/ionization and capillary electrophoresis/mass spectrometry. Rapid Commun. Mass Spectrom. 2004;18:149–156. doi: 10.1002/rcm.1294. [DOI] [PubMed] [Google Scholar]
- Ong SE, Mann M. Mass spectrometry-based proteomics turns quantitative. Nat. Chem. Biol. 2005;1:252–262. doi: 10.1038/nchembio736. [DOI] [PubMed] [Google Scholar]
- Peng Y, Pallandre A, Tran NT, Taverna M. Recent innovations in protein separation on microchips by electrophoretic methods. Electrophoresis. 2008;29:157–178. doi: 10.1002/elps.200700347. [DOI] [PubMed] [Google Scholar]
- Rao PV, Lu X, Standley M, Pattee P, Neelima G, Girisesh G, Dakshinamurthy KV, Roberts CT, Jr, Nagalla SR. Proteomic identification of urinary biomarkers of diabetic nephropathy. Diabetes Care. 2007;30:629–637. doi: 10.2337/dc06-2056. [DOI] [PubMed] [Google Scholar]
- Reiner A, Yekutieli D, Benjamini Y. Identifying differentially expressed genes using false discovery rate controlling procedures. Bioinformatics. 2003;19:368–375. doi: 10.1093/bioinformatics/btf877. [DOI] [PubMed] [Google Scholar]
- Rejtar T, Hu P, Juhasz P, Campbell JM, Vestal ML, Preisler J, Karger BL. Off-line coupling of high-resolution capillary electrophoresis to MALDI-TOF and TOF/TOF MS. J. Proteome. Res. 2002;1:171–179. doi: 10.1021/pr015519o. [DOI] [PubMed] [Google Scholar]
- Renfrow MB, Cooper HJ, Tomana M, Kulhavy R, Hiki Y, Toma K, Emmett MR, Mestecky J, Marshall AG, Novak J. Determination of aberrant O-glycosylation in the IgA1 hinge region by electron capture dissociation fourier transform-ion cyclotron resonance mass spectrometry. J. Biol. Chem. 2005;280:19136–19145. doi: 10.1074/jbc.M411368200. [DOI] [PubMed] [Google Scholar]
- Renfrow MB, Mackay CL, Chalmers MJ, Julian BA, Mestecky J, Kilian M, Poulsen K, Emmett MR, Marshall AG, Novak J. Analysis of O-glycan heterogeneity in IgA1 myeloma proteins by Fourier transform ion cyclotron resonance mass spectrometry: implications for IgA nephropathy. Anal. Bioanal. Chem. 2007;389:1397–1407. doi: 10.1007/s00216-007-1500-z. [DOI] [PubMed] [Google Scholar]
- Righetti PG, Boschetti E. The ProteoMiner and the FortyNiners: Searching for gold nuggets in the proteomic arena. Mass Spectrom. Rev. 2008 doi: 10.1002/mas.20178. [DOI] [PubMed] [Google Scholar]
- Righetti PG, Campostrini N, Pascali J, Hamdan M, Astner H. Quantitative proteomics: a review of different methodologies. Eur. J. Mass Spectrom. (Chichester, Eng) 2004;10:335–348. doi: 10.1255/ejms.600. [DOI] [PubMed] [Google Scholar]
- Roche S, Gabelle A, Lehmann S. Clinical proteomics of the cerebrospinal fluid: Towards the discovery of new biomarkers. Proteomics Clin. Appl. 2008;2:428–436. doi: 10.1002/prca.200780040. [DOI] [PubMed] [Google Scholar]
- Roessler M, Rollinger W, Mantovani-Endl L, Hagmann ML, Palme S, Berndt P, Engel AM, Pfeffer M, Karl J, Bodenmueller H, Ruschoff J, Henkel T, Rohr G, Rossol S, Rosch W, Langen H, Zolg W, Tacke M. Identification of PSME3 as a novel serum tumor marker for colorectal cancer by combining two-dimensional polyacrylamide gel electrophoresis with a strictly mass spectrometry-based approach for data analysis. Mol. Cell Proteomics. 2006;5:2092–2101. doi: 10.1074/mcp.M600118-MCP200. [DOI] [PubMed] [Google Scholar]
- Rossing K, Mischak H, Dakna M, Zurbig P, Novak J, Julian BA, Good DM, Coon JJ, Tarnow L, Rossing P. Proteomic discovery and validation of urinary biomarkers for diabetes and chronis renal disease. J Am Soc Nephrol. 2008a doi: 10.1681/ASN.2007091025. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossing K, Mischak H, Parving HH, Christensen PK, Walden M, Hillmann M, Kaiser T. Impact of diabetic nephropathy and angiotensin II receptor blockade on urinary polypeptide patterns. Kidney Int. 2005;68:193–205. doi: 10.1111/j.1523-1755.2005.00394.x. [DOI] [PubMed] [Google Scholar]
- Rossing K, Mischak H, Rossing P, Schanstra JP, Wiseman A, Maahs DM. The urinary proteome in diabetes and diabetes-associated complications: new ways to assess disease progression and evaluate therapy. Proteomics Clin. Appl. 2008b;2:997–1007. doi: 10.1002/prca.200780166. [DOI] [PubMed] [Google Scholar]
- Schaub S, Wilkins J, Weiler T, Sangster K, Rush D, Nickerson P. Urine protein profiling with surface-enhanced laser-desorption/ionization time-of-flight mass spectrometry. Kidney Int. 2004;65:323–332. doi: 10.1111/j.1523-1755.2004.00352.x. [DOI] [PubMed] [Google Scholar]
- Schaub S, Wilkins JA, Antonovici M, Krokhin O, Weiler T, Rush D, Nickerson P. Proteomic-based identification of cleaved urinary beta2-microglobulin as a potential marker for acute tubular injury in renal allografts. Am. J. Transplant. 2005;5:729–738. doi: 10.1111/j.1600-6143.2005.00766.x. [DOI] [PubMed] [Google Scholar]
- Schena FP, Gesualdo L. Pathogenetic mechanisms of diabetic nephropathy. J. Am. Soc. Nephrol. 2005;16 Suppl 1:S30–S33. doi: 10.1681/asn.2004110970. [DOI] [PubMed] [Google Scholar]
- Sennels L, Salek M, Lomas L, Boschetti E, Righetti PG, Rappsilber J. Proteomic analysis of human blood serum using peptide library beads. J Proteome Res. 2007;6:4055–4062. doi: 10.1021/pr070339l. [DOI] [PubMed] [Google Scholar]
- Shannon W, Culverhouse R, Duncan J. Analyzing microarray data using cluster analysis. Pharmacogenomics. 2003;4:41–52. doi: 10.1517/phgs.4.1.41.22581. [DOI] [PubMed] [Google Scholar]
- Shen Y, Kim J, Strittmatter EF, Jacobs JM, Camp DG, Fang R, Tolie N, Moore RJ, Smith RD. Characterization of the human blood plasma proteome. Proteomics. 2005;5:4034–4045. doi: 10.1002/pmic.200401246. [DOI] [PubMed] [Google Scholar]
- Simo C, Elvira C, Gonzalez N, San Roman J, Barbas C, Cifuentes A. Capillary electrophoresis-mass spectrometry of basic proteins using a new physically adsorbed polymer coating. Some applications in food analysis. Electrophoresis. 2004;25:2056–2064. doi: 10.1002/elps.200305790. [DOI] [PubMed] [Google Scholar]
- Smola AJ, Scholkopf B. A tutorial on support vector regression. Statistics and Computing. 2004;14:199–222. [Google Scholar]
- Snell-Bergeon JK, Maahs DM, Ogden LG, Kinney G, Hokanson JE, Schiffer E, Rewers M, Mischak H. Evaluation of urinary biomarkers for coronary artery disease, diabetes, and diabetic kidney disease. Diabetes Technol. Ther. 2008 doi: 10.1089/dia.2008.0040. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song EJ, Babar SM, Oh E, Hasan MN, Hong HM, Yoo YS. CE at the omics level: Towards systems biology - An update. Electrophoresis. 2008;29:129–142. doi: 10.1002/elps.200700467. [DOI] [PubMed] [Google Scholar]
- Stutz H. Advances in the analysis of proteins and peptides by capillary electrophoresis with matrix-assisted laser desorption/ionization and electrospray-mass spectrometry detection. Electrophoresis. 2005;26:1254–1290. doi: 10.1002/elps.200410130. [DOI] [PubMed] [Google Scholar]
- Tempels FW, Underberg WJ, Somsen GW, de Jong GJ. On-line coupling of SPE and CE-MS for peptide analysis. Electrophoresis. 2007;28:1319–1326. doi: 10.1002/elps.200600403. [DOI] [PubMed] [Google Scholar]
- Temporini C, Calleri E, Massolini G, Caccialanza G. Integrated analytical strategies for the study of phosphorylation and glycosylation in proteins. Mass Spectrom. Rev. 2008;27:207–236. doi: 10.1002/mas.20164. [DOI] [PubMed] [Google Scholar]
- Theodorescu D, Fliser D, Wittke S, Mischak H, Krebs R, Walden M, Ross M, Eltze E, Bettendorf O, Wulfing C, Semjonow A. Pilot study of capillary electrophoresis coupled to mass spectrometry as a tool to define potential prostate cancer biomarkers in urine. Electrophoresis. 2005;26:2797–2808. doi: 10.1002/elps.200400208. [DOI] [PubMed] [Google Scholar]
- Theodorescu D, Mischak H. Mass spectrometry based proteomics in urine biomarker discovery. World J Urol. 2007;25:435–443. doi: 10.1007/s00345-007-0206-3. [DOI] [PubMed] [Google Scholar]
- Theodorescu D, Schiffer E, Bauer HW, Douwes F, Eichhorn F, Polley R, Schmidt T, Schofer W, Zuerbig P, Good DM, Coon JJ, Mischak H. Discovery and validation of urinary biomarkers for prostate cancer. Proteomics Clin. Appl. 2008;2:556–570. doi: 10.1002/prca.200780082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theodorescu D, Wittke S, Ross MM, Walden M, Conaway M, Just I, Mischak H, Frierson HF. Discovery and validation of new protein biomarkers for urothelial cancer: a prospective analysis. Lancet Oncol. 2006;7:230–240. doi: 10.1016/S1470-2045(06)70584-8. [DOI] [PubMed] [Google Scholar]
- Thongboonkerd V. Practical Points in Urinary Proteomics. J Proteome. Res. 2007;6:3881–3890. doi: 10.1021/pr070328s. [DOI] [PubMed] [Google Scholar]
- Thornalley PJ, Battah S, Ahmed N, Karachalias N, Agalou S, Babaei-Jadidi R, Dawnay A. Quantitative screening of advanced glycation endproducts in cellular and extracellular proteins by tandem mass spectrometry. Biochem. J. 2003;375:581–592. doi: 10.1042/BJ20030763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ullsten S, Zuberovic A, Wetterhall M, Hardenborg E, Markides KE, Bergquist J. A polyamine coating for enhanced capillary electrophoresis-electrospray ionization-mass spectrometry of proteins and peptides. Electrophoresis. 2004;25:2090–2099. doi: 10.1002/elps.200305787. [DOI] [PubMed] [Google Scholar]
- Weissinger EM, Hertenstein B, Mischak H, Ganser A. Online coupling of capillary electrophoresis with mass spectrometry for the identification of biomarkers for clinical diagnosis. Expert. Rev. Proteomics. 2005;2:639–647. doi: 10.1586/14789450.2.5.639. [DOI] [PubMed] [Google Scholar]
- Weissinger EM, Schiffer E, Hertenstein B, Ferrara JL, Holler E, Stadler M, Kolb HJ, Zander A, Zurbig P, Kellmann M, Ganser A. Proteomic patterns predict acute graft-versus-host disease after allogeneic hematopoietic stem cell transplantation. Blood. 2007;109:5511–5519. doi: 10.1182/blood-2007-01-069757. [DOI] [PubMed] [Google Scholar]
- Weissinger EM, Wittke S, Kaiser T, Haller H, Bartel S, Krebs R, Golovko I, Rupprecht HD, Haubitz M, Hecker H, Mischak H, Fliser D. Proteomic patterns established with capillary electrophoresis and mass spectrometry for diagnostic purposes. Kidney Int. 2004;65:2426–2434. doi: 10.1111/j.1523-1755.2004.00659.x. [DOI] [PubMed] [Google Scholar]
- Westfall PH, Young SS. Resampling-based Multiple Testing: Examples and Methods for P-Value Adjustment. New York: Wiley; 1993. [Google Scholar]
- Wittke S, Fliser D, Haubitz M, Bartel S, Krebs R, Hausadel F, Hillmann M, Golovko I, Koester P, Haller H, Kaiser T, Mischak H, Weissinger EM. Determination of peptides and proteins in human urine with capillary electrophoresis–mass spectrometry, a suitable tool for the establishment of new diagnostic markers. J Chromatogr. A. 2003;1013:173–181. doi: 10.1016/s0021-9673(03)00713-1. [DOI] [PubMed] [Google Scholar]
- Wittke S, Haubitz M, Walden M, Rohde F, Schwarz A, Mengel M, Mischak H, Haller H, Gwinner W. Detection of acute tubulointerstitial rejection by proteomic analysis of urinary samples in renal transplant recipients. Am. J. Transplant. 2005;5:2479–2488. doi: 10.1111/j.1600-6143.2005.01053.x. [DOI] [PubMed] [Google Scholar]
- Zamfir AD. Recent advances in sheathless interfacing of capillary electrophoresis and electrospray ionization mass spectrometry. J Chromatogr. A. 2007;1159:2–13. doi: 10.1016/j.chroma.2007.03.115. [DOI] [PubMed] [Google Scholar]
- Zimmerli LU, Schiffer E, Zurbig P, Kellmann M, Mouls L, Pitt A, Coon JJ, Schmiederer RE, Mischak H, Peter K, Kolch W, Delles C, Dominiczak AF. Urinary proteomics biomarkers in coronary artery disease. Mol Cell Proteomics. 2008;7:290–298. doi: 10.1074/mcp.M700394-MCP200. [DOI] [PubMed] [Google Scholar]
- Zubarev RA. Reactions of polypeptide ions with electrons in the gas phase. Mass Spectrom. Rev. 2003;22:57–77. doi: 10.1002/mas.10042. [DOI] [PubMed] [Google Scholar]
- Zürbig P, Renfrow MB, Schiffer E, Novak J, Walden M, Wittke S, Just I, Pelzing M, Neususs C, Theodorescu D, Root C, Ross M, Mischak H. Biomarker discovery by CE-MS enables sequence analysis via MS/MS with platform-independent separation. Electrophoresis. 2006;27:2111–2125. doi: 10.1002/elps.200500827. [DOI] [PubMed] [Google Scholar]
- Zybailov B, Coleman MK, Florens L, Washburn MP. Correlation of relative abundance ratios derived from peptide ion chromatograms and spectrum counting for quantitative proteomic analysis using stable isotope labeling. Anal. Chem. 2005;77:6218–6224. doi: 10.1021/ac050846r. [DOI] [PubMed] [Google Scholar]