Abstract
New inhibitors are urgently needed to overcome the burgeoning problem of drug resistance in the treatment of Plasmodium falciparum infection. Targeting the folate pathway has proved to be a powerful strategy for drug development against rapidly multiplying systems such as cancer cells and microorganisms. Antifolates have long been used for malaria treatment but, despite their success, much less is known about parasite folate metabolism than about that of the human host. In this article, we focus on folate enzymes used clinically as anticancer drug targets, in addition to those that have potential to be used as drug targets, for which there are inhibitors at various stages of development. We discuss how this information could lead to the identification of new targets in malaria parasites.
What do folate derivatives do?
Folate derivatives (FDs) are important cellular cofactors in the synthesis of nucleotides required for DNA replication, in the synthesis of the amino acids glycine and methionine and in the metabolism of histidine, glutamic acid and serine. These factors are also required for the initiation of protein synthesis in mitochondria through formylation of methionine. Rapidly dividing cells such as those in tumors, bacteria and malaria parasites rely heavily on the availability of FDs for growth. Folate antagonism has occupied a central position in cancer treatment for >50 years. Methotrexate (MTX), an inhibitor of dihydrofolate reductase (DHFR) developed in the late 1940s [1], remains the most widely used single therapeutic agent for cancer [2]. This success and the emergence of MTX resistance have led to sustained efforts to understand folate metabolism more completely and to develop new strategies for its disruption. Currently, four different enzymes of the folate pathway are the targets of anticancer drugs in clinical use or clinical development, and active research is underway to validate other enzyme targets.
Folate antagonism has also proved to be a good strategy for drug development against malaria. Sulfadoxine–pyrimethamine (SP, also known as Fansidar™) – the antifolate combination targeting dihydropteroate synthase (DHPS) and DHFR – has long been an affordable drug of choice for the treatment of chloroquine-resistant parasites [3]. A new combination of chlorproguanil (a biguanide-based molecule whose active metabolite inhibits DHFR) and dapsone (a DHPS inhibitor) has been developed to treat SP-resistant parasites and is now available in many African countries [4]. Proguanil (Paludrine®) is another antifolate that is commonly used for prophylaxis against Plasmodium falciparum malaria and is metabolized to its active form of cycloguanil. Proguanil, together with atovaquone, is also used in a combination known as Malarone® [5].
Despite the success of antimalarial antifolates, much less is known about folate metabolism in Plasmodium than in the human host. Moreover, all drugs in clinical use or at the experimental stage target only DHFR or, less frequently, DHPS [6,7], yet observations of the mammalian folate pathway and experience in cancer research indicate that other enzymes could be good targets for drug discovery. In this article, to gain more insight into the malaria folate pathway, we compare mammalian and malarial metabolism, focusing on folate enzymes used as anticancer drug targets, both in current clinical practice and in various stages of experimental investigation and development. Where appropriate, we have exploited malaria genome information to search for candidate enzymes that have not otherwise been described. The P. falciparum genome sequence is now finished [8] and the sequences of several other Plasmodium species are nearing completion (see http://plasmodb.org; http://www.tigr.org; and http://www.sanger.ac.uk). Where relevant, observations from bacteria, yeast and plants have been included because these can also be extremely informative.
FDs in mammalian cells
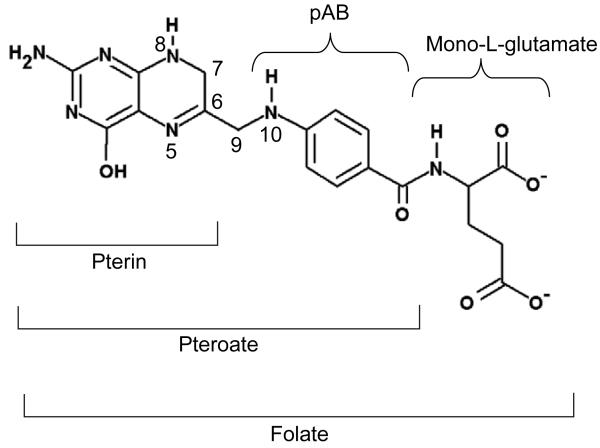
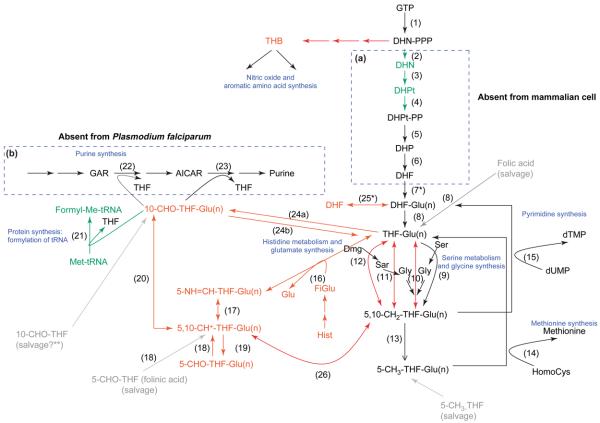
Mammalian cells do not synthesize de novo the folate moiety from which FDs are produced. FDs are acquired from dietary intake or exogenous culture medium and are modified appropriately. The folate metabolites comprise nine variants of the canonical pterin ring that can also be polyglutamated to varying degrees (Figure 1): folic acid (FA), dihydrofolate (DHF), tetrahydrofolate (THF), 5,10-methenyltetrahydrofolate (5,10-CH+-THF), 5,10-methylenetetrahydrofolate (5,10-CH2-THF), 5-methyltetrahydrofolate (5-CH3-THF), 5-formyltetrahydrofolate (5-CHO-THF), 10-formyltetrahydrofolate (10-CHO-THF) and 5-formiminotetrahydrofolate (5-NH=CH-THF). Figure 2 summarizes the biochemical relationships of these forms.
Figure 1.
Structure of DHF. The pterin component is 2-amino-4-hydroxy-7, 8-dihydropteridine; the pAB moiety derives from p-aminobenzoic acid. Polyglutamated forms arise by amide-bond formation at the γ position of successive glutamate residues. Key positions are numbered.
Figure 2.
Comparative folate biochemistry in mammalian cells and Plasmodium falciparum. All reactions occur in mammalian cells except those in box (a). Reactions in box (b) are absent from P. falciparum. Reactions in red have not been characterized in malaria parasites, and those in green are presumptive reactions based on the existence of products downstream of these reactions or on the existence of putative candidate genes discovered by bioinformatics analysis. Folate salvage pathways are shown in gray, and the role of folate derivatives is indicated in blue. Numbers in brackets refer to the following enzymes: (1) GTP-cyclohydrolase I; (2) postulated non-specific phosphatase following non-enzymatic loss of pyrophosphate; (3) dihydroneopterin aldolase; (4) 6-hydroxymethyldihydropterin pyrophosphokinase; (5) DHPS; (6) DHFS; (7) FPGS; (8) DHFR; (9) SHMT; (10) glycine-cleavage enzyme system; (11) sarcosine dehydrogenase; (12) dimethylglycine dehydrogenase; (13) methylenetetrahydrofolate reductase; (14) methionine synthase; (15) TS; (16) glutamate formiminotransferase; (17) formiminotetrahydrofolate cyclodeaminase; (18) methenyltetrahydrofolate synthetase; (19) SHMT; (20) MTC; (21) MTFT; (22) GART; (23) AICARFT; (24a) formyltetrahydrofolate synthetase; (24b) 10-formyltetrahydrofolate dehydrogenase; (25) GH; (26) MTD. Asterisk (*) denotes enzymes (FPGS and GH) for which all FDs can be the substrate in mammals, but only THF forms are used in bacteria [56]. Double asterisk (**) indicates that the ability of the parasite to salvage 10-CHO-THF has not been formally demonstrated. Abbreviations: AICAR, 5-aminoimidazole-4-carboxamide ribotide; DHN, 7,8-dihydroneopterin; DHN-PPP, 7,8-dihydroneopterin triphosphate; DHPt, 7,8-dihydro-6-hydroxymethylpterin; DHPt-PP, 7,8-dihydro-6-hydroxymethylpterin pyrophosphate; Dmg, dimethyl glycine; FiGlu, formiminoglutamic acid; formyl-Met-tRNA, formylmethionyl tRNA; GAR, glycinamide ribotide; Glu, glutamic acid; Glu(n), polyglutamate form of FDs; Gly, glycine; His, histidine; HomoCys, homocysteine; Met, methionine; Sar, sarcosine; Ser, serine; THB, tetrahydrobiopterin.
The predominant FD in mammals is 5-CH3-THF, which represents 80–90% of total folate, followed by THF (5–10%). All other FDs together represent <10% of the total [9]. Each FD has a specific role in transferring a one-carbon (C1) group in five major biosynthetic pathways: (i) the synthesis of purines (10-CHO-THF and 5,10-CH+-THF) and the pyrimidine deoxythymidine-5′-monophosphate (dTMP) (5,10-CH2-THF); (ii) the formylation of methionyl-tRNA (10-CHO-THF); (iii) the metabolism of serine and glycine (THF and 5,10-CH2-THF); (iv) the metabolism of histidine and glutamate (5-NH=CH-THF); and (v) the synthesis of methionine (5-CH3-THF) (Figure 2). The 5-CHO-THF form, whose cellular role was unclear initially after it was excluded as a C1 donor, is now considered to be a storage molecule for folate that is crucial in the regulation of mammalian folate metabolism [10]. The predominance of 5-CH3-THF, presumably, reflects its role as a cofactor in the conversion of homocysteine to methionine, which, in turn, is the precursor of S-adenosylmethionine – the methylation cofactor involved in almost 100 reactions [2].
FDs in Plasmodium falciparum
Unlike in mammalian cells, FDs (except for 5-CH3-THF [11]) have not been directly identified and characterized in P. falciparum. FD presence is deduced from the existence of parasite enzymes that use the various compounds as substrates and, in most cases, such use has been established in vitro. For instance, because the enzyme activities of dihydrofolate synthase (DHFS), folylpolyglutamate synthase (FPGS), DHFR, thymidylate synthase (TS) and serine hydroxylmethyltransferase (SHMT) have been expressed from cloned parasite genes, it can be concluded that the following substrates are present in the parasite: dihydropteroate (DHP), DHF, THF and 5,10-CH2-THF [12-16]. Following the detection of 5-CH3-THF [11] and enzyme activities in parasite extracts for methylenetetrahydrofolate reductase and methionine synthase (the enzyme that uses 5-CH3-THF as a substrate) [17], we conclude that all reactions leading to the synthesis of dTMP and methionine occur in the parasite (reactions 7–15 in Figure 2), although some of the genes involved have yet to be identified. The presence of 10-CHO-THF can also be assumed because formylmethionyltRNA, the product of the reaction that uses 10-CHO-THF as a substrate, exists in malaria parasites [18]. However, the presence of the following FDs have never been reported: 5-NH=CH-THF, 5-CHO-THF and 5,10-CH+-THF. Absence of 5,10-CH+-THF is consistent with the observation that P. falciparum is dependent on an exogenous supply of purines (see later).
Folate enzymes as drug targets in clinical use
DHFR
DHFR (EC 1.5.1.3) catalyzes the reduction of 7,8-DHF to 5,6,7,8-THF (reaction 8 in Figure 2) and has a pivotal role in two major folate-based reactions: (i) synthesis of endogenous THF (in microorganisms that can produce folate de novo) and (ii) salvage of oxidized forms of folate, particularly the recycling of DHF resulting from the synthesis of dTMP, and from exogenous sources. The central role of this enzyme has made it a major target for drug development.
The first antifolate drug used clinically against tumor cells was aminopterin (4-amino-pteroylglutamate), a potent but extremely toxic DHFR inhibitor. Since its development, the 5-methyl derivative amethopterin (commonly known as MTX) [1] has been the drug of choice for the past 50 years for treating several types of cancer [2]. The success of MTX and the emergence of resistance have led to active searches for other potent anti-DHFR agents. The following compounds have reached (at least) phase I or II clinical trials for treatment of various forms of malignancy: edatrexate, trimetrexate [19] and piritrexim [20].
The dhfr gene is well characterized in P. falciparum. It is expressed as a bifunctional protein with TS [21] and has proved to be an excellent target for antimalarial drugs. The DHFR inhibitors pyrimethamine and proguanil were the first antifolates used for the treatment of malaria [22], and the potentiation of these agents with sulfa drugs (inhibitors of DHPS) led to development of the SP combination [23]. Unfortunately, whenever SP has been used, the development and spread of resistance have been relatively rapid. A new family of potent diaminotriazine-based inhibitors of Plasmodium DHFR is now being studied [24], and several groups are carrying out in vitro screens to identify further effective agents [25-27]. With the crystallographic structure of P. falciparum DHFR–TS now resolved, more screening programs can be expected that employ a more precise approach to inhibitor design [28]. However, it is noteworthy that, since the 1960s, no new antimalarial antifolate drug has entered into clinical trials.
The rapid development of resistance that is characteristic of SP usage is associated with the selection of point mutations at codons 51, 59, 108 and 164 of dhfr and primarily at codons 437, 540 and 581 of dhps. The drug-elimination profile in the body is one parameter that determines the emergence and selection of resistance. Thus, rapid selection of pyrimethamine resistance is associated with the extremely long half-life of this drug [29,30]. Resistant parasites are selected when the concentration of pyrimethamine in vivo falls below that required for therapeutic effectiveness, leading to a strong selective pressure for mutations in the target genes [29,30]. To minimize this selection, a related DHFR inhibitor, chlorproguanil, has recently been combined with the DHPS inhibitor dapsone in a short-acting formulation known as Lapdap™ [4].
It has been suggested that the malaria parasite and its host differ fundamentally in the way that translation of DHFR–TS is regulated, so that the parasite enzyme is replenished less readily when targeted by DHFR inhibitors [31]. However, other work does not support this difference [32], and the validity of this hypothesis as a possible new paradigm for exploiting antimalarial drug selectivity is still open to debate.
TS
TS (EC 2.1.1.45) catalyzes the reductive methylation of 2′-deoxyuridine-5′-monophosphate (dUMP) to dTMP, using 5,10-CH2-THF as the C1 methyl donor (reaction 15 in Figure 2). TS is the only folate enzyme that causes oxidation of the cofactor to DHF as its C1 fragment is lost. Mammalian cells can either synthesize thymidine de novo or salvage it exogenously. However, the demand for this DNA precursor is so great in rapidly dividing cells that the salvage pathway cannot supply enough nucleotides, making TS a particularly vulnerable target in tumor cells. Major efforts have focused on the design of TS inhibitors, which fall into two main categories: folate and nucleotide analogs [33,34]. Fluorouracil (5FU) was one of the first TS inhibitors to be developed as an antineoplastic agent and has been widely used to treat various malignancies [34]. Among newly developed TS inhibitors, six folate analogs are currently being evaluated in phase II or III trials: AG337 (Thymitaq®), raltitrexed, pemetrexed, nolatrexed, ZD9331 and GS7904L [33].
As mentioned, TS is expressed in P. falciparum as a bifunctional protein with DHFR [21]. Unlike mammals, P. falciparum cannot salvage thymidine and, thus, relies completely on the de novo thymidylate cycle through TS for the synthesis of dTMP [35]; however, no anti-TS antimalarial drug has yet reached clinical studies. Derivatives of orotic acid have been tested as anti-TS nucleotide analogs [36-38]. Unlike uridine derivatives, orotate analogs are transported into the parasite and incorporated in the pyrimidine pathway that generates dUMP de novo. The synthesis of the resulting metabolite, which is toxic to TS, is specific to the parasite because mammalian cells salvage orotate derivatives poorly. The precursor of the anticancer drug 5FU, 5-fluoro-orotic acid (5FO), seemed to be the most potent, with an IC50 in the nanomolar range. In vivo animal studies were promising, and the therapeutic index of 5FO could be increased by the addition of uridine, which mammalian cells can salvage, thus bypassing the toxicity of 5FO, but which the parasite cannot. However, the poor pharmacokinetic properties of this compound have probably limited its further development.
Other anti-TS compounds tested against malaria include the anticancer analogs ICI D1694 and 1843 U89 in combination with thymidine [39,40]. Although proof of principle was demonstrated, the activity of these inhibitors did not meet expectations [39,40], and no further reports of the search for anti-TS folate analogs against malaria parasites have been published since.
Enzymes of the purine-synthesis pathway
Folate analogs are also used as inhibitors of purine synthesis. The de novo synthesis pathway comprises ten enzymes that catalyze reactions starting with 5-phophoribosyl-1-pyrophosphate and ending with inosinic acid (inosine 5′-monophosphate), the precursor of AMP and GMP [41]. The third and ninth enzymes in this pathway – 5′-phosphoribosylglycinamide transformylase (GART) (EC 2.1.2.2) (reaction 22 in Figure 2) and 5′-phosphoribosyl-5-amino-4-imidazolecarboxamide formyltransferase (AICARFT) (EC 2.1.2.3) (reaction 23 in Figure 2), respectively – require 10-CHO-THF as a formyl donor; these enzymes are targeted by antifolates in the treatment of cancer and rheumatoid arthritis [19,42]. However, because P. falciparum cannot synthesize purines de novo [35], this pathway can be excluded as an antimalarial drug target. Consistent with this, a range of probes for GART and AICARFT sequences used in basic local alignment search tool (BLAST) searches (see www.plasmodb.org) fails to identify any candidate genes in Plasmodium.
Folate enzymes as targets at the experimental stage
Some of the folate enzymes currently being investigated as novel targets for drug development in tumor treatment have been characterized in Plasmodium and, thus, could also represent targets for new antimalarial drugs.
FPGS
FPGS (EC 6.3.2.17) catalyzes the MgATP-dependent sequential attachment of L-glutamate residues to the γ position of folates to yield polyglutamated derivatives (reaction 7 in Figure 2). These forms have been studied extensively in mammalian cells and consist of between one and nine glutamate residues attached to the p-aminobenzoate (PABA) moiety of the folate molecule [43]. Polyglutamated folates are better substrates than monoglutamated substrates for folate-dependent enzymes, exhibiting more-efficient kinetics and longer retention times, resulting in an increased concentration within cells [43]. Evidence shows that FPGS could be a good drug target for the inhibition of rapidly dividing cells [44], and investigations to identify potent inhibitors are underway [19], assisted by information about its X-ray structure [45].
In P. falciparum, pentaglutamate forms of folate occur [11], indicating that the parasite also expresses FPGS. This was confirmed by the characterization of a bifunctional fpgs–dhfs parasite gene [14,16] that, when transformed into appropriate yeast and bacterial mutants, could restore prototrophy [14]. However, exploring the potential of malarial DHFS–FPGS as a new target awaits target validation by gene knockout [46] and a robust heterologous expression system for producing sufficient amounts of soluble active enzyme. In mammals, polyglutamation of antifolate agents such as MTX that contain a free γ-glutamyl moiety also occurs, leading to increased activity and ability to target other folate enzymes [47]. The possibilities that antifolates can be polyglutamated by P. falciparum and that such metabolites could inhibit other enzymes in the pathway, such as TS, are currently being tested (A. Nzila et al., unpublished). Certainly, MTX powerfully inhibits all strains of P. falciparum, regardless of the resistance mutations in their DHFR domain, at IC50 levels of ~;40–100 nM (P. Wang and J. Hyde, unpublished) but inhibitors that are less toxic to the host must first be developed to function as prodrugs for polyglutamation.
SHMT
SHMT (also known as glycine hydroxymethyltransferase) (EC 2.1.2.1) catalyzes the conversion of THF and serine to 5,10-CH2-THF and glycine (reaction 9 in Figure 2); this provides the primary source of the C1 unit (from the β-carbon of serine) for THF-dependent reactions. Thus, SHMT inhibition is expected to affect cell growth dramatically, especially because there is a considerable increase in its activity in proliferating cells during S-phase [48]. Analogs of both serine and THF have been assayed [49], although none of the inhibitors has yet been tested in animals.
The shmt gene and its product have been investigated in P. falciparum [12,14]. Transcript levels rise rapidly in late trophozoites just before DNA synthesis, which is consistent with the key role of SHMT in this process [50]. The malarial gene can complement glycine auxotrophy in Escherichia coli, demonstrating its ability to convert serine to glycine [12]. In yeast and humans, both cytosolic and mitochondrial isoforms of SHMT occur that are encoded by separate genes. However, P. falciparum has only a single gene, and no signal peptide is identified even by prediction algorithms adapted to the parasite. Thus, the functionality of SHMT in these cell compartments awaits experimental determination, and no published study has explored its potential as a parasite drug target.
C1-THF synthase enzymes
C1-THF synthase is an enzyme complex that also occurs in both cytoplasmic and mitochondrial forms in mammals [51]. The cytoplasmic form is an NADP-dependent trifunctional protein that catalyzes three reversible reactions: (i) methylenetetrahydrofolate dehydrogenase (MTD) (EC 1.5.1.5), which is responsible for the conversion of 5,10-CH2-THF to 5,10-CH+-THF (reaction 26 in Figure 2); (ii) methenyltetrahydrofolate cyclohydrolase (MTC) (EC 3.5.4.9), which is required for synthesis of 10-CHO-THF from 5,10-CH+-THF (reaction 20 in Figure 2); and (iii) formyltetrahydrofolate synthetase (EC 6.3.4.3), which catalyzes the generation of 10-CHO-THF from THF and formate (reaction 24a in Figure 2). The mitochondrial enzyme is an NAD-dependent bifunctional protein bearing the MTD and MTC activities [52]. The overall roles of C1-THF synthase are to incorporate formate into the production of 10-CHO-THF, which is essential for purine synthesis and the production of formylmethionyl-tRNA, and to provide an alternative route for the generation of 5,10-CH2-THF (for methionine and dTMP synthesis) [51]. The inhibition of MTD by folate analogs has been reported, although their activities were insufficient to warrant studies in animal models [53]. Crystallographic structures of the MTD and MTC domains have been resolved recently and should greatly facilitate the design of active compounds against these enzyme activities [54].
Although these activities have not yet been reported in P. falciparum, a gene has been identified (Mal6P1.121) whose product shows 36% identity to 79% of the bifunctional MTD–MTC of the bacterium Aquifex aeolicus. However, similarities are confined to the N- and C-terminal regions of the larger malarial protein (more than twice the size of the bacterial protein), which also lacks predicted signal sequences for both apicoplast and mitochondrial targeting. Thus, the alignment is not especially convincing and the status of this protein in the parasite remains unclear.
Methionyl-tRNA formyltransferase
Methionyl-tRNA formyltransferase (MTFT) (EC 2.1.2.9) catalyzes a reaction that is important for protein synthesis in prokaryotic cells and the mitochondria and plastids of eukaryotes. In these cases, mRNA translation is initiated with formylmethionine (compared with methionine in the eukaryotic cytoplasm) derived from formylmethionyl-tRNA, which is synthesized by transfer of the formyl moiety of 10-CHO-THF to methionyl-tRNA in the presence of MTFT (reaction 21 in Figure 2). This enzyme is a good target for antitumor agents, with several inhibitors active in the nanomolar range [55]. However, none has reached the stage of clinical evaluation.
MTFT has not yet been described in P. falciparum. However, the product of the MAL13P1.67 gene shows 26% identity to 54% of MTFT from the turnip Brassica napus, carries an apicoplast-targeting sequence and signal peptide and is annotated as a putative MTFT. Moreover, the enzyme that removes the formyl group from formylmethionyl-tRNA, peptide deformylase (EC 3.5.1.88), has also been characterized in P. falciparum and located to the apicoplast [18], which is a strong indication that methionyl-tRNA can be formylated and that MTFT is likely to exist in the parasite.
Glutamyl hydrolase
The polyglutamation process described earlier (reaction 7 in Figure 2) can be reversed by hydrolysis of the glutamate moieties at the γ-glutamyl peptide bonds of folates, reducing the number of residues and, ultimately, converting the polyglutamate to the monoglutamated form [56]. This reaction is mediated by γ-glutamyl hydrolase (GH) or folate conjugase (EC 3.4.19.9, formerly EC 3.4.22.12) (reaction 25 in Figure 2). However, this activity has not been found in P. falciparum [57], an observation supported by a failure of plant and mammalian protein probes to identify any candidate genes in the Plasmodium databases. In this respect, the parasite resembles yeast and other microorganisms that also lack this enzyme.
Concluding remarks
Sustained efforts to improve cancer treatments have led to much clarification of the folate pathway in mammalian cells, and folate antagonism continues to provide a rich source of new drugs and novel therapeutic strategies. However, although folate antagonism by inhibition of DHFR and DHPS was established long ago as a major strategy for antimalarial treatment, much less work has been dedicated to identifying other potential targets in P. falciparum. Reactions catalyzed by alternative enzymes in the folate pathway can provide a rationale for its inhibition. Moreover, this pathway presents some unique features that provide particularly attractive candidates for drug development.
Because P. falciparum cannot salvage preformed pyrimidines, it is completely reliant upon de novo synthesis, with TS catalyzing the formation of dTMP. However, because the human and parasite enzymes exhibit a strong similarity at the primary-sequence level, it is argued that compounds active against the parasite would also be toxic to the host. Nevertheless, ways of overcoming this problem could, in principle, be developed by using pyrimidine nucleotides as adjuvant therapeutic agents because the host cell can efficiently salvage them, bypassing the effect of the anti-TS compound. This concept has been tested [39] but not taken further.
Alternatively, it might be possible to develop anti-TS agents that specifically target the parasite cells. For example, ICI D1964, a potent inhibitor of mammalian TS, inhibits malaria parasites only weakly [39]. This might reflect transport properties, and one could envisage the converse situation whereby some anti-TS agents would be imported more efficiently into the parasite than into mammalian cells to enable sufficient specificity. However, ICI D1964 might bind only poorly to Plasmodium TS, and direct analysis with purified malarial enzyme will be required to evaluate a possible role for inhibitor transport. Nevertheless, TS is one of the most developed antifolate targets in cancer, with more than eight compounds reaching at least phase I clinical trials, and it should be even more attractive as an antimalarial target because of the complete reliance of the parasite on the de novo pathway. Another interesting feature of Plasmodium TS is its expression as a bifunctional protein with DHFR. The DHF generated by dTMP synthesis is likely to be directly channeled to the DHFR domain for reduction to THF [58]. As a result, inhibition of TS would lower the availability of DHF to DHFR, and the concurrent inhibition of both activities could be synergistic: a possibility that has not yet been explored in P. falciparum.
Similar considerations apply to SHMT, which is the only enzyme that mediates synthesis of 5,10-CH2-THF in P. falciparum, providing the C1 unit from serine that is required for conversion of dUMP to dTMP; however, it is also fairly well conserved relative to the human enzyme. Although clinical trials of anticancer agents directed at SHMT are less advanced than those targeting TS, this is an option for malaria that merits further exploration. Much less conserved is parasite FPGS, which has the added attraction of being expressed as a bifunctional protein with DHFS. Inhibitors are predicted to affect both activities, one being required for de novo folate synthesis and the other for polyglutamation of folates to the level necessary for efficient use. A related strategy would be to use the ability of FPGS to add glutamate residues onto folate analogs, thereby increasing their potency, which is an approach that has been exploited in the development of anticancer drugs [19]. The therapeutic potential of using the malarial enzyme in this manner has not yet been investigated because none of the antimalarial antifolates, whether in the experimental stage or in clinical use, can be biochemically modified in this way.
Of the other enzymes considered in this article, the presence in P. falciparum of the key purine-synthesis enzymes GART and AICARFT can be safely excluded, whereas the status of a possible C1-THF synthase complex remains unresolved. Only MTFT seems to be a likely component of parasite folate metabolism. From the point of view of developing new antimalarial drugs, the enzymes already targeted in clinical or experimental cancer therapy have the advantage that considerable effort has gone into developing lead compounds, understanding structure–function relationships and assessing host toxicity. However, there are several other potential targets in the folate pathway that have not yet advanced to this stage, in addition to targets that are specific to parasite metabolism. These enzyme groups will be examined in part II of this article [46], in which we evaluate their potential for chemotherapeutic inhibition.
Acknowledgements
This work was supported by the Wellcome Trust (grants 056769, 056845, 062372, 067201 and 073896), the Biotechnology and Biological Sciences Research Council (grant 36/JE616379) and the NIH (Fogarty International grant TW 01186). A.N. and K.M. thank the Wellcome Trust for personal support. S.A.W. thanks the Wellcome Trust for institutional support. We apologize to those authors whose work was not cited because of space limitations.
References
- 1.Seeger DR, et al. Analogs of pteroylglutamate acid. III 4-Amino derivatives. J. Am. Chem. Soc. 1949;71:1753–1758. [Google Scholar]
- 2.Priest DG, Bunni MA. Folate and folate antagonists in cancer chemotherapy. In: Bailey LB, editor. Folate in health and disease. Marcel Dekker; 1995. pp. 379–403. [Google Scholar]
- 3.Sibley CH, et al. Pyrimethamine–sulfadoxine resistance in Plasmodium falciparum: what next? Trends Parasitol. 2001;17:582–588. doi: 10.1016/s1471-4922(01)02085-2. [DOI] [PubMed] [Google Scholar]
- 4.Bukirwa H, et al. Chlorproguanil–dapsone for treating uncomplicated malaria. Cochrane Database Syst. Rev. 2004:CD004387. doi: 10.1002/14651858.CD004387.pub2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kain KC. Current status and replies to frequently posed questions on atovaquone plus proguanil (Malarone) for the prevention of malaria. BioDrugs. 2003;17(Suppl. 1):23–28. doi: 10.2165/00063030-200317001-00006. [DOI] [PubMed] [Google Scholar]
- 6.Sirichaiwat C, et al. Target guided synthesis of 5-benzyl-2,4-diamonopyrimidines: their antimalarial activities and binding affinities to wild type and mutant dihydrofolate reductases from Plasmodium falciparum. J. Med. Chem. 2004;47:345–354. doi: 10.1021/jm0303352. [DOI] [PubMed] [Google Scholar]
- 7.Fidock DA, et al. Antimalarial drug discovery: efficacy models for compound screening. Nat. Rev. Drug Discov. 2004;3:509–520. doi: 10.1038/nrd1416. [DOI] [PubMed] [Google Scholar]
- 8.Gardner MJ, et al. Genome sequence of the human malaria parasite Plasmodium falciparum. Nature. 2002;419:498–511. doi: 10.1038/nature01097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Belz S, Nau H. Determination of folate patterns in mouse plasma, erythrocytes, and embryos by HPLC coupled with a microbiological assay. Anal. Biochem. 1998;265:157–166. doi: 10.1006/abio.1998.2865. [DOI] [PubMed] [Google Scholar]
- 10.Stover P, Schirch V. The metabolic role of leucovorin. Trends Biochem. Sci. 1993;18:102–106. doi: 10.1016/0968-0004(93)90162-g. [DOI] [PubMed] [Google Scholar]
- 11.Krungkrai J, et al. De novo and salvage biosynthesis of pteroylpentaglutamates in the human malaria parasite, Plasmodium falciparum. Mol. Biochem. Parasitol. 1989;32:25–37. doi: 10.1016/0166-6851(89)90126-6. [DOI] [PubMed] [Google Scholar]
- 12.Alfadhli S, Rathod PK. Gene organization of a Plasmodium falciparum serine hydroxymethyltransferase and its functional expression in Escherichia coli. Mol. Biochem. Parasitol. 2000;110:283–291. doi: 10.1016/s0166-6851(00)00282-6. [DOI] [PubMed] [Google Scholar]
- 13.Hall SJ, et al. Functional expression of the dihydrofolate reductase and thymidylate synthetase activities of the human malaria parasite Plasmodium falciparum in Escherichia coli. Mol. Biochem. Parasitol. 1991;45:317–330. doi: 10.1016/0166-6851(91)90100-k. [DOI] [PubMed] [Google Scholar]
- 14.Salcedo E, et al. A bifunctional dihydrofolate synthetase–folylpolyglutamate synthetase in Plasmodium falciparum identified by functional complementation in yeast and bacteria. Mol. Biochem. Parasitol. 2001;112:239–252. doi: 10.1016/s0166-6851(00)00370-4. [DOI] [PubMed] [Google Scholar]
- 15.Sirawaraporn W, et al. Antifolate-resistant mutants of Plasmodium falciparum dihydrofolate reductase. Proc. Natl. Acad. Sci. U. S. A. 1997;94:1124–1129. doi: 10.1073/pnas.94.4.1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee CS, et al. Characterization of three genes encoding enzymes of the folate biosynthetic pathway in Plasmodium falciparum. Parasitology. 2001;122:1–13. doi: 10.1017/s0031182000006946. [DOI] [PubMed] [Google Scholar]
- 17.Krungkrai J, et al. Characterization of cobalamin-dependent methionine synthase purified from the human malarial parasite, Plasmodium falciparum. Parasitol. Res. 1989;75:512–517. doi: 10.1007/BF00931158. [DOI] [PubMed] [Google Scholar]
- 18.Bracchi-Ricard V, et al. Characterization of an eukaryotic peptide deformylase from Plasmodium falciparum. Arch. Biochem. Biophys. 2001;396:162–170. doi: 10.1006/abbi.2001.2631. [DOI] [PubMed] [Google Scholar]
- 19.McGuire JJ. Anticancer antifolates: current status and future directions. Curr. Pharm. Des. 2003;9:2593–2613. doi: 10.2174/1381612033453712. [DOI] [PubMed] [Google Scholar]
- 20.Liu G, et al. Gemcitabine, paclitaxel, and piritrexim: a phase I study. Am. J. Clin. Oncol. 2003;26:280–284. doi: 10.1097/01.coc.0000081607.16287.6f. [DOI] [PubMed] [Google Scholar]
- 21.Bzik DJ, et al. Molecular cloning and sequence analysis of the Plasmodium falciparum dihydrofolate reductase-thymidylate synthase gene. Proc. Natl. Acad. Sci. U. S. A. 1987;84:8360–8364. doi: 10.1073/pnas.84.23.8360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Peters W, editor. Chemotherapy and drug resistance in malaria. Academic Press; 1987. [Google Scholar]
- 23.Depinay A, et al. Treatment of malaria attack with a single injection of sulfadoxine–pyrimethamine. Bull. Soc. Pathol. Exot. Filiales. 1972;65:409–417. [PubMed] [Google Scholar]
- 24.Jensen NP, et al. Phenoxypropoxybiguanides, prodrugs of DHFR-inhibiting diaminotriazine antimalarials. J. Med. Chem. 2001;44:3925–3931. doi: 10.1021/jm010089z. [DOI] [PubMed] [Google Scholar]
- 25.Lau H, et al. Efficacies of lipophilic inhibitors of dihydrofolate reductase against parasitic protozoa. Antimicrob. Agents Chemother. 2001;45:187–195. doi: 10.1128/AAC.45.1.187-195.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ommeh S, et al. In vitro activities of 2,4-diaminoquinazoline and 2,4-diaminopteridine derivatives against Plasmodium falciparum. Antimicrob. Agents Chemother. 2004;48:3711–3714. doi: 10.1128/AAC.48.10.3711-3714.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kamchonwongpaisan S, et al. Inhibitors of multiple mutants of Plasmodium falciparum dihydrofolate reductase and their antimalarial activities. J. Med. Chem. 2004;47:673–680. doi: 10.1021/jm030165t. [DOI] [PubMed] [Google Scholar]
- 28.Chitnumsub P, et al. Characterization, crystallization and preliminary X-ray analysis of bifunctional dihydrofolate reductasethymidylate synthase from Plasmodium falciparum. Acta Crystallogr. D Biol. Crystallogr. 2004;60:780–783. doi: 10.1107/S0907444904001544. [DOI] [PubMed] [Google Scholar]
- 29.Kublin JG, et al. Molecular markers for failure of sulfadoxine–pyrimethamine and chlorproguanil–dapsone treatment of Plasmodium falciparum malaria. J. Infect. Dis. 2002;185:380–388. doi: 10.1086/338566. [DOI] [PubMed] [Google Scholar]
- 30.Nzila AM, et al. Molecular evidence of greater selective pressure for drug resistance exerted by the long-acting antifolate pyrimethamine/sulfadoxine compared with the shorter-acting chlorproguanil/dapsone on Kenyan Plasmodium falciparum. J. Infect. Dis. 2000;181:2023–2028. doi: 10.1086/315520. [DOI] [PubMed] [Google Scholar]
- 31.Zhang K, Rathod PK. Divergent regulation of dihydrofolate reductase between malaria parasite and human host. Science. 2002;296:545–547. doi: 10.1126/science.1068274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nirmalan N, et al. Translational up-regulation of antifolate drug targets in the human malaria parasite Plasmodium falciparum upon challenge with inhibitors. Mol. Biochem. Parasitol. 2004;136:63–70. doi: 10.1016/j.molbiopara.2004.02.013. [DOI] [PubMed] [Google Scholar]
- 33.Chu E, et al. Thymidylate synthase inhibitors as anticancer agents: from bench to bedside. Cancer Chemother. Pharmacol. 2003;52(Suppl 1):S80–S89. doi: 10.1007/s00280-003-0625-9. [DOI] [PubMed] [Google Scholar]
- 34.Pinedo HM, Peters GF. Fluorouracil: biochemistry and pharmacology. J. Clin. Oncol. 1988;6:1653–1664. doi: 10.1200/JCO.1988.6.10.1653. [DOI] [PubMed] [Google Scholar]
- 35.Reyes P, et al. Enzymes of purine and pyrimidine metabolism from the human malaria parasite, Plasmodium falciparum. Mol. Biochem. Parasitol. 1982;5:275–290. doi: 10.1016/0166-6851(82)90035-4. [DOI] [PubMed] [Google Scholar]
- 36.Krungkrai J, et al. Antimalarial activity of orotate analogs that inhibit dihydroorotase and dihydroorotate dehydrogenase. Biochem. Pharmacol. 1992;43:1295–1301. doi: 10.1016/0006-2952(92)90506-e. [DOI] [PubMed] [Google Scholar]
- 37.Hekmat-Nejad M, Rathod PK. Kinetics of Plasmodium falciparum thymidylate synthase: interactions with high-affinity metabolites of 5-fluoroorotate and D1694. Antimicrob. Agents Chemother. 1996;40:1628–1632. doi: 10.1128/aac.40.7.1628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rathod PK, Gomez ZM. Plasmodium yoelii: oral delivery of 5-fluoroorotate to treat malaria in mice. Exp. Parasitol. 1991;73:512–514. doi: 10.1016/0014-4894(91)90075-8. [DOI] [PubMed] [Google Scholar]
- 39.Rathod PK, Reshmi S. Susceptibility of Plasmodium falciparum to a combination of thymidine and ICI D1694, a quinazoline antifolate directed at thymidylate synthase. Antimicrob. Agents Chemother. 1994;38:476–480. doi: 10.1128/aac.38.3.476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jiang L, et al. Potent and selective activity of a combination of thymidine and 1843U89, a folate-based thymidylate synthase inhibitor, against Plasmodium falciparum. Antimicrob. Agents Chemother. 2000;44:1047–1050. doi: 10.1128/aac.44.4.1047-1050.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Allison AC, Eugui EM. Mycophenolate mofetil and its mechanisms of action. Immunopharmacology. 2000;47:85–118. doi: 10.1016/s0162-3109(00)00188-0. [DOI] [PubMed] [Google Scholar]
- 42.Chan ES, Cronstein BN. Molecular action of methotrexate in inflammatory diseases. Arthritis Res. 2002;4:266–273. doi: 10.1186/ar419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gangjee A, et al. Recent advances in the chemistry and biology of folypoly-γ-glutamate synthetase substrates and inhibitors. Curr. Med. Chem. Anti-Canc. Agents. 2002;2:331–355. doi: 10.2174/1568011024606352. [DOI] [PubMed] [Google Scholar]
- 44.Lowe KE, et al. Regulation of folate and one-carbon metabolism in mammalian cells. II. Effect of folylpoly-γ-glutamate synthetase substrate specificity and level on folate metabolism and folylpoly-γ-glutamate specificity of metabolic cycles of one-carbon metabolism. J. Biol. Chem. 1993;268:21665–21673. [PubMed] [Google Scholar]
- 45.Sun X, et al. Structural homologies with ATP- and folate-binding enzymes in the crystal structure of folylpolyglutamate synthetase. Proc. Natl. Acad. Sci. U. S. A. 1998;95:6647–6652. doi: 10.1073/pnas.95.12.6647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nzila A, et al. Comparative folate metabolism in humans and malaria parasites (part II): activities as yet untargeted or specific to Plasmodium. Trends Parasitol. doi: 10.1016/j.pt.2005.05.008. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Barnes MJ, et al. Impact of polyglutamation on sensitivity to raltitrexed and methotrexate in relation to drug-induced inhibition of de novo thymidylate and purine biosynthesis in CCRF-CEM cell lines. Clin. Cancer Res. 1999;5:2548–2558. [PubMed] [Google Scholar]
- 48.Snell K. Enzymes of serine metabolism in normal, developing and neoplastic rat tissues. Adv. Enzyme Regul. 1984;22:325–400. doi: 10.1016/0065-2571(84)90021-9. [DOI] [PubMed] [Google Scholar]
- 49.Matthews RG, et al. Cobalamin-dependent methionine synthase and serine hydroxymethyltransferase: targets for chemotherapeutic intervention? Adv. Enzyme Regul. 1998;38:377–392. doi: 10.1016/s0065-2571(97)00006-x. [DOI] [PubMed] [Google Scholar]
- 50.Nirmalan N, et al. Transcriptional analysis of genes encoding enzymes of the folate pathway in the human malaria parasite Plasmodium falciparum. Mol. Microbiol. 2002;46:179–190. doi: 10.1046/j.1365-2958.2002.03148.x. [DOI] [PubMed] [Google Scholar]
- 51.Pawelek PD, MacKenzie RE. Methenyltetrahydrofolate cyclohydrolase is rate limiting for the enzymatic conversion of 10-formyltetrahydrofolate to 5,10-methylenetetrahydrofolate in bifunctional dehydrogenase–cyclohydrolase enzymes. Biochemistry. 1998;37:1109–1115. doi: 10.1021/bi971906t. [DOI] [PubMed] [Google Scholar]
- 52.Pelletier JN, MacKenzie RE. Binding and interconversion of tetrahydrofolates at a single site in the bifunctional methylenetetrahydrofolate dehydrogenase/cyclohydrolase. Biochemistry. 1995;34:12673–12680. doi: 10.1021/bi00039a025. [DOI] [PubMed] [Google Scholar]
- 53.Tonkinson JL, et al. The antiproliferative and cell cycle effects of 5,6,7, 8-tetrahydro-N5,N10-carbonylfolic acid, an inhibitor of methylenetetrahydrofolate dehydrogenase, are potentiated by hypoxanthine. J. Pharmacol. Exp. Ther. 1998;287:315–321. [PubMed] [Google Scholar]
- 54.Allaire M, et al. Crystallization of the bifunctional methylenetetrahydrofolate dehydrogenase/methenyltetrahydrofolate cyclohydrolase domain of the human trifunctional enzyme. Proteins. 1996;26:479–480. doi: 10.1002/(SICI)1097-0134(199612)26:4<479::AID-PROT9>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- 55.Vaughan MD, et al. Methionine in and out of proteins: targets for drug design. Curr. Med. Chem. 2002;9:385–409. doi: 10.2174/0929867023371102. [DOI] [PubMed] [Google Scholar]
- 56.Green JM, et al. Folate biosynthesis, reduction, and polyglutamation. In: Neidhardt FC, editor. Escherichia coli and Salmonella. ASM Press; 1996. pp. 665–673. [Google Scholar]
- 57.Krungkrai J, et al. High-performance liquid chromatographic assay for pteroylpolyglutamate hydrolase. J. Chromatogr. 1987;417:47–56. doi: 10.1016/0378-4347(87)80090-7. [DOI] [PubMed] [Google Scholar]
- 58.O'Neil RH, et al. Phylogenetic classification of protozoa based on the structure of the linker domain in the bifunctional enzyme, dihydrofolate reductase–thymidylate synthase. J. Biol. Chem. 2003;278:52980–52987. doi: 10.1074/jbc.M310328200. [DOI] [PubMed] [Google Scholar]