Abstract
High-throughput genome sequencing techniques have now reached vector biology with an emphasis on those species that are vectors of human pathogens. The first mosquito to be sequenced was Anopheles gambiae, the vector for Plasmodium parasites that cause malaria. Further mosquitoes have followed: Aedes aegypti (Yellow fever and Dengue fever vector) and Culex pipiens (lymphatic filariasis and West Nile fever). Species that are currently in sequencing include the body louse Pediculus humanus (Typhus vector), the triatomine Rhodnius prolixus (Chagas disease vector) and the tick Ixodes scapularis (Lyme disease vector). The motivations for sequencing vector genomes are to further understand vector biology, with an eye on developing new control strategies (for example novel chemical attractants or repellents) or understanding the limitations of current strategies (for example the mechanism of insecticide resistance); to analyse the mechanisms driving their evolution; and to perform an exhaustive analysis of the gene repertory. The proliferation of genomic data creates the need for efficient and accessible storage. We present VectorBase, a genomic resource centre that is both involved in the annotation of vector genomes and act as a portal for access to the genomic information (http://www.vectorbase.org).
Keywords: insect, vector, human pathogen, genomic resources, VectorBase
INTRODUCTION
The burden of infectious diseases on the world remains a major challenge to medical science. Understanding the complex interactions between vector, pathogen and host is necessary for the comprehension of these diseases but has proved especially difficult (Aksoy et al., 2002; Aksoy, 2003; Xu et al., 2005).
Observation and biological experiments have been for many decades the source of all the data and advances in the field of infectious diseases. The last few years have seen the start of a new era in this domain: the generation and analysis of genomic data derived from the sequencing of organisms. Genomic data can be generated relatively quickly and present a broader view opening the way for a range of genome-wide studies (e.g. expression microarrays, RNAi knock-down studies) as well as giving a boost to the hypothesis-driven experimentation for these species. Consideration of size and perceived simplicity meant that pathogens were at the forefront of the genomics area with viral and bacterial genomes being among the first to be sequenced. The speed and accuracy of modern sequencing technologies has yielded essentially complete genome sequences for many species in a relatively fast and cost effective manner, quickly adding the human genome to the list of the sequenced organisms. The vector genomes came later with the publication of the genome of the malaria mosquito Anopheles gambiae (Holt et al., 2002). The sequence of a genome is in itself of limited use without the associated annotation that attempts to describe the location and function of genes, as well as the control elements active in the genome. The annotation process is often based on a similarity approach that is, in turn, reliant on information from previous annotations.
This review focuses on the genomic resources for invertebrate vectors of human pathogens. We will discuss the motivations for sequencing the genomes of these species as opposed to other strategies for producing data, such as Expressed Sequence Tag (EST) sequencing. We will then present the current state of genomics resources for vector species and finally introduce VectorBase, a resource centre which organises and stores genomic data for presentation via the World Wide Web (WWW).
THE USE OF GENOMIC DATA
The most obvious reason for sequencing the genome of a vector is to improve our understanding of the organism’s biology with a view to designing new control measures, exploiting its pharmaceutical potential or developing new molecular tools for genetic manipulation. The ability to screen on a genome-wide basis is a powerful driver for genome sequencing as a cost-effective method of understanding individual genes. A good example would be the genome-wide studies that investigate the temporal, spatial and conditional expression of all genes in a single experiment. Knowledge of a genome also facilitates the quantification of polymorphisms within a species as well as comparative investigations into the complex mechanisms driving insect evolution.
Genomics and vector biology
a. Vector biology
A bottleneck in progress toward controlling invertebrate vectors of human pathogens is the lack of knowledge of their basic molecular biology. To better control these insects, we would like to understand at a molecular level their feeding habits, their mating behavior and mode of reproduction, their choice of habitat and, more than anything else, their relationship with the host and the pathogens. Connecting this with the genome information can help to understand these processes at a molecular level. For example, the sequencing of the A. gambiae genome has proven beneficial in identifying molecular mechanisms responsible for host seeking and other odor-mediated behaviors (Biessmann et al., 2005). A better understanding of vector population structure is essential when planning intervention strategies (Cuamba et al., 2006). Access to the genome sequence can be useful to facilitate the identification of new DNA markers such as micro-satellites and single nucleotide polymorphisms (SNPs) that make the identification of groups within the population more sensitive and cost effective.
b. Vector control
The control of vectors is a key weapon in the fight against infectious diseases. A better understanding of the biology of the vector can lead to a faster identification of new targets and ultimately new control measures. For example, David et al. (2005) used the A. gambiae annotations and expression profiling using micro-arrays to identify genes involved in insecticide resistance. A potential list of 230 genes was reduced to just five that were highly regulated in insecticide resistant mosquito strains.
c. New molecular tools
The identification of new transposons and repeated sequences facilitates the development of tools for the genetic manipulation of vectors. The availability of the genomic sequence helps to identify these elements faster and on a larger scale.
High efficiency transformation is possible for many mosquito species. Handler (2002) describes how the piggyBac transposon allows germ-line transformation of insects, including the yellow fever and malaria vectors Aedes aegypti (Lobo et al., 2002), A. gambiae (Grossman et al., 2001), Anopheles fluviatilis (Rodrigues et al., 2006), Anopheles stephensi (Nolan et al., 2002) and Anopheles albimanus (Perera et al., 2002). The same transposon was employed by Adelman (2004) to transform somatic cells in A. aegypti. The Hermes and Mariner transposons were successfully used to transform respectively Culex pipiens quinquefasciatus germ line (Allen et al., 2001) and A. aegypti somatic cells (Coates et al., 1998). In addition, the availability of the genome sequence facilitates the development of high-throughput genome wide technologies such as high density expression micro-array, genome tiling array, or chip-chip methodology. David et al. (2005), for example, developed the first micro-array to study insecticide resistance in malaria vectors. More recently, Vontas et al. (2007) monitored gene expression in insecticide resistant and susceptible strains of A. stephensi and identify a small number of genes putatively differentially expressed between the strains. Halasz et al. (2006) developed a method to analyse tilling array data and tested it on an A. gambiae tilling array, identifying non-exonic loci that were actively transcribed. Chip-chip methodologies have been applied to Drosophila, including analyses to study the binding of transcription factors (Moses et al., 2006; Zeitlinger et al., 2007), and are likely to be applied to mosquitoes soon.
d. Biopharmaceutical potential
The transmission of infectious agents (pathogens) from a vector to the human host is usually by direct contact (biting or sucking). To make the most of the feeding interaction with the host, the vector requires a system of anti-coagulants, vaso-dilatators and other modulators of the haemodynamic process. Identifying any of these compounds is potentially of interest to the medical industry. Ribeiro and collaborators have already characterized some of these agents through studies of mosquito salivary gland gene expression (Ribeiro et al., 2006; Calvo et al., 2007; Santos et al., 2007).
Genomics and evolution
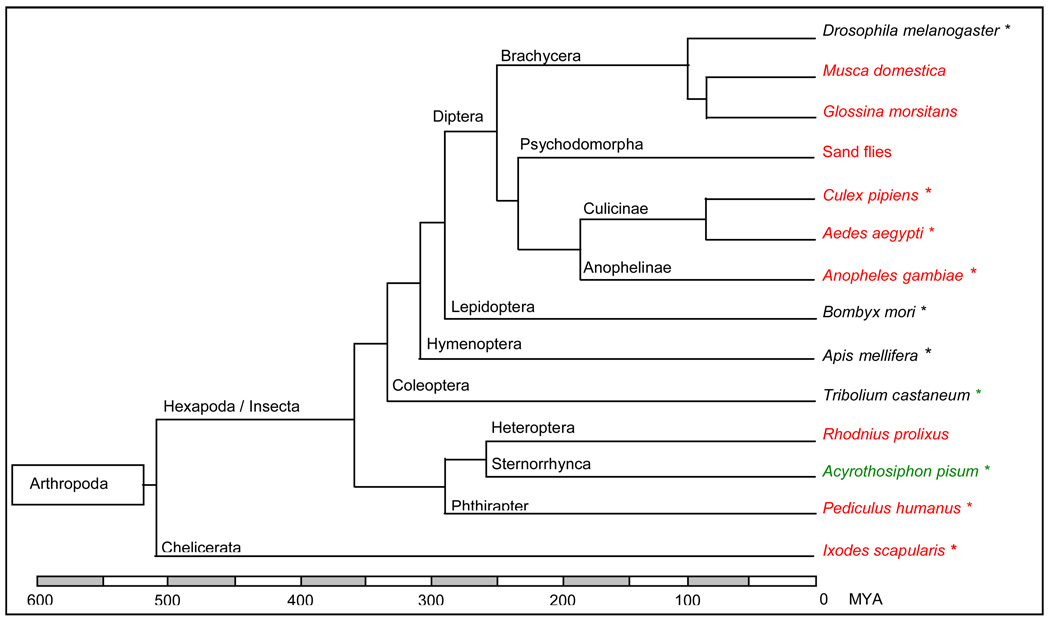
Recent sequencing efforts within the Insecta group have allowed the generation of increasingly accurate phylogenetic trees for organisms (Fig. 1). Genomic comparison helps to explain the evolutionary events linking species by identifying genes conserved across species, genes evolving quickly and in particular species-specific expansion of gene families. Such results can lead to the identification of genes linked to a given behavior (e.g. blood feeding), or involved in pathogen transmission or insecticide resistance (e.g. immunity genes). Ultimately, this could lead to new approaches in the control of disease transmission by these organisms. For example, Waterhouse et al. (2007) used the genome sequences of Drosophila melanogaster, A. gambiae and A. aegypti to compare the insect immune repertoire and identified conservative and rapidly evolving immune-related genes.
Figure 1.
Evolutionary relationships for insects with a current or planned genome project. Vectors of human pathogens are in red, vectors of non-human pathogens are in green, selected non-vectors are in black. A star (*) after the organism name indicates current projects; other projects are planned. The time scale is approximate.
Genome vs. ESTs
The sequencing of Expressed Sequenced Tags (ESTs), short fragments of expressed sequences, often precedes the sequencing of a genome, either as a complement or as a temporary alternative. The generation of EST sequences is rapid and relatively cheap and has been used for gene discovery for species where the resources are not currently available for full genome sequencing, such as the sand fly and tsetse fly. Ribeiro and collaborators have analysed the gene expression of several mosquito (Anopheline and Aedine) and tick species using small libraries, ranging from several hundred to several thousand sequences. EST sequences were clustered together and a series of bioinformatics analyses was then applied to each consensus sequence. The results of these analyses were collated into a spreadsheet that can be queried or browsed to identify the function of each transcript cluster consensus (Ribeiro et al., 2004; Santos et al., 2004; Arca et al., 2007).
EST sequences represent a sub-set of the repertoire of genes expressed in the RNA sample from which the library was generated. Analysis of the sequences can inform us about the variety and abundance of transcripts within a cell, a tissue type, a developmental stage or an organism. Depending on the experimental design, EST libraries can be normalized, a process by which abundant transcripts are removed in order to maximize novel gene discovery, or left in their native state where the abundance of each transcript is proportional to the expression level in the original RNA sample. Non-normalized libraries give information about expression levels and can be analysed with other libraries to identify differentially expressed genes between two samples or conditions; for example male and female mosquitoes or susceptible or resistant to an insecticide. Such studies can be carried out without any genomic sequence. Nisbet et al. (2006) describe in an article how the use of large scale EST projects has help the understanding of the immunology of host-parasites relationships and the potential of this knowledge in developing vaccines.
EST libraries are generated from polyA+ RNA and hence represent mainly expressed sequences from a genome. This information is very useful when predicting gene structures on a genomic sequence and genes predicted from EST data are high confidence and are more likely to be valid than ab initio predicted genes, for example, even if not necessarily full-length. Moreover, EST data remain the best way of finding and annotating alternative splice forms.
EST libraries can also be used as a resource for the community. The Malaria Research and Reference Reagent Resource Centre (MR4 - http://www.mr4.org/) is a repository for malaria related reagents and provides, free of charge, a variety of plasmid and clone vectors, antibodies, genomic and cDNA libraries, cell lines, preserved mosquitoes etc. It contains 15 EST libraries for various Plasmodium species and 4 EST libraries for A. gambiae. Reagents are collected from scientists who wish to make them available to the scientific malaria community. ESTs are a valuable tool for identifying genes and estimating their expression level, and when coupled to the genomic data, they provide additional information to improve the annotation.
SURVEY OF THE GENOMIC DATA FOR INVERTEBRATE VECTORS OF HUMAN PATHOGENS
In 2002, marking the start of the genomic area in the field of human pathogen vectors, the genome of the mosquito A. gambiae, vector of malaria, was sequenced and annotated (Holt et al., 2002). The annotation was helped by the existence of Anopheline mRNA and protein data, complemented by several EST libraries, and by the huge quantity of D. melanogaster data. A few years later, in 2005/06, the genome of the yellow fever mosquito A. aegypti was sequenced and annotated (Nene et al., 2007). More recently, the list of sequenced organisms has increased with the addition of the body louse Pediculus humanus and the mosquito C. pipiens, both gene sets being currently in preparation and expected to be released at the beginning of 2008. The genome sequence from the tick Ixodes scapularis has just been released and its annotation has started. The genome of the bug Rhodnius prolixus is planned to be released mid-2008 and should be annotated soon thereafter. Further organisms (including additional A. gambiae populations, the tsetse fly Glossina morsitans, and several sand fly species) are expected in the next few years. The tsetse and sand fly projects already have a certain amount of EST data and their analysis has started (Table 1).
Table 1. Vectors, human pathogens and diseases transmitted.
List of the invertebrate vectors of human pathogens with a genome project, and their most important human pathogens and diseases transmitted.
| Vector | Common name | Important pathogens | Human disease transmitted |
|---|---|---|---|
| Anopheles gambiae | African Malaria Mosquito | Plasmodium falciparum | Malaria |
| Aedes aegypti | Yellow Fever Mosquito | Yellow fever virus, Dengue virus, Chikungunya virus | Yellow fever, Dengue, Chikungunya |
| Culex pipiens quiquefasciatus | Southern House Mosquito | West Nile Virus, Wuchereria bancrofti | West Nile Fever, Encephalitis, Lymphatic Filariasis |
| Pediculus humanus | Body Louse | Rickettsia prowasekii | Typhus |
| Ixodes scapularis | Black-legged Tick | Borrelia burgdorferi, Rickettsiae, Babesia microsi | Lyme disease, Granulocytic ehrlichiosis, Babesiosis |
| Rhodnius prolixus | Kissing Bug | Trypanosoma cruzi | Chagas disease |
| Glossina morsitans | Tsetse fly | Trypanosomas | Sleeping sickness [Nagana in livestock] |
| Phlebotomus papatasi | Sand fly – Old World | Leishmania | Leishmaniasis |
| Lutzomyia longipalpis | Sand fly – New World | Leishmania | Leishmaniasis |
With the increase in the number of vectors sequenced and annotated, it becomes easier to analyse the subsequent ones. Data from closely related organisms are often used to annotate new genomes, allowing the maximum usage of quality prediction between species, but opening up the possibilities of propagating bad prediction if not careful. The annotation of A. aegypti, for example, was largely based on the D. melanogaster and A. gambiae data. In addition, sequencing techniques and analysis tools continue to become cheaper, faster and more accurate. Funding agencies have recognized the need for large scale data for pathogen vectors to help the understanding of human infectious diseases.
These sequencing and annotation projects are based on international collaborations grouping scientists from the sequencing centres (e.g. the Sanger Centre, the Broad Institute, the J.Craig Venter Institute, the Genome Sequencing Centre at Washington University, the Human Genome Sequencing Centre at the Baylor College of Medicine), experts in genome annotation (e.g. sequencing centres, VectorBase) and the larger community of scientists specialist in these organisms.
VECTORBASE, A RESOURCE CENTRE FOR GENOMIC DATA FROM INVERTEBRATE VECTORS OF HUMAN PATHOGENS
With the increasing amount of data generated by the sequencing projects and their subsequent analysis, it becomes crucial to organize the storing and the access to these data. VectorBase (Lawson et al., 2007 - http://www.vectorbase.org) is a NIH-NIAID funded Resource Centre for Invertebrate Vectors of Human Pathogens, organising information about these organisms: sequences, gene sets and related information, pictures and controlled vocabulary for mosquito and tick anatomy and physiology. A key feature of VectorBase is the Ensembl genome browser, developed at the European Bioinformatics Institute (Hubbard et al., 2007), used to display genes along the genome and to link them to related information, such as manual annotation, physical mapping data, expression data or protein and DNA similarities. Comparative data are also handled similarly to Ensembl, with homolog (ortholog/paralog) information linking genes from the various organisms and the possibility to “jump” from one genome to the other via their protein or DNA similarities. Non-comparative and non-automatic annotations are stored in a Chado database and are managed using GMOD tools. GMOD (Generic Model Organism Database - http://www.gmod.org) is a collection of software tools for creating and managing genome-scale biological databases.
Chado, developed within the FlyBase consortium, is a relational database that is part of this kit and capable of representing many of the general classes of data frequently encountered in modern biology such as sequence, sequence comparisons, phenotypes, genotypes, ontologies, publications, and phylogeny (Mungall et al., 2007). Data not handled by the classical schema are integrated via the DAS protocol, a Distributed Annotation System protocol used to exchange biological sequence annotation. It is exploited to supply data from remote databases to the VectorBase genome browser, allowing external users to map their own data to the VectorBase genomes and making them available to the whole community, but retaining the ability to update them at any time. Most of the data available through VectorBase have been generated internally, using an annotation and a genomic comparison pipelines to analyse the data. Manual annotation is provided for selected regions using approaches developed at FlyBase (Crosby et al., 2007). The expression data, while not generated in house, are collected via the BASE interface (http://base.thep.lu.se/) and mapped to the genomes internally.
VectorBase offers a number of tools to mine the data, including a BLAST server allowing the user to compare his sequences to any of the sequences (genome, traces, ESTs, transcripts or proteins) of any of the organisms hosted and a ClustalW tool to align sequences together. The search engine allows the user to enter the site from any keywords: tool or organism, gene name or protein identifier, micro-array name or Controlled Vocabulary about mosquito or tick physiologies etc. VectorBase also provides GDAV, Genome De-linked Annotation Viewer, a simple set of tools allowing the publishing of EST, gene or protein annotations generated by in a independent project. It is installed on the user’s own computer system, leaving him in full control of his data. An optional component allows, via the DAS protocol, the viewing of similarities between the user sequence and one or more genomes from the VectorBase genome browser.
The VectorBase data are available by download as flat files, either in fasta format (sequence data) or as database dumps (on request). Additionally, BioMart (Kasprzyk et al., 2004) is a more sophisticated tool for querying the databases, building MySQL queries based on simple choices from the user, and returning tabulated flat files.
Eight organisms are currently available: two with a complete gene set, two with a genome and a gene set near completion and four at various stages of sequencing, going from the newly fully sequenced tick to the on-going tsetse fly, with ESTs and traces, and the sand flies for which ESTs only are available (Table 2).
Table 2. Survey of the genomic projects for Invertebrates Vectors of Human Pathogens.
For each organism, the availability of the following data is shown: Expressed Sequence Tags (‘ESTs’), large scale genomic sequencing (‘Traces’), assembly of traces into scaffolds or chromosomes (‘Assembly’), and gene set (‘Gene set’). The organisations displaying the data or the consortium coordinating the project and the main references (genome paper or white paper) are indicated.
| Organism | ESTs | Traces | Assembly | Gene set | Coordination group(s) | References |
|---|---|---|---|---|---|---|
| Anopheles gambiae | Y [1] [5] | Y [1] [5b] | Y [1] | YY [1] | VectorBase [1] | (Holt et al., 2002) |
| Aedes aegypti | Y [1] [5] | Y [5b] | Y [1] | YY [1] | VectorBase [1] | (Nene et al., 2007) |
| Culex pipiens | Y [1] | Y [1] [5b] | Y [1] | Near completion [1– 3] | BROAD, VectorBase JCVI [1– 3] | white paper[11] |
| Pediculus humanus | Y [1] | Y [5b] | Y [1] | Near completion [1,2] | JCVI, VectorBase [1,2] | white paper[11] |
| Ixodes scapularis | Y [1] | Y [1] [5b] | Y | N | IISSC [6] | white paper[11] (Pagel Van Zee et al., 2007) |
| Anopheles gambiae M & S forms | N | Y [5b] | Near completion | N | GSC-WashU, JCVI VectorBase, AGCC [1,1,4, 10] | white paper[11] |
| Rhodnius prolixus | Y [1] | on-going [5b] | N | N | RRC [7] | white paper[11] |
| Glossina morsitans | Y [1] [5] | on-going [5b] | N | N | IGGI [8] | white paper[11] |
| Phlebotomus papatasi | Y [1] | N | N | N | SFGSC [9] | white paper[11] |
| Lutzomyia longipalpis | Y [1] [5] | N | N | N | SFGSC [9] | white paper[11] |
Y: yes - N: no - YY: update of the genome annotation.
VectorBase: http://www.vectorbase.org/
J.Craig Venter Institute (JCVI): http://www.tigr.org/
The BROAD Institute: http://www.broad.mit.edu/
Genome Sequencing Centre – Washington University (GSC-WashU): http://genome.wustl.edu/
NCBI (dbEST): http://www.ncbi.nlm.nih.gov/dbEST
NCBI (traces): http://www.ncbi.nlm.nih.gov/Traces
International Ixodes scapularis Sequencing Committee (IISC): http://www.entm.purdue.edu/igp/overview.html
Rhodnius Research Community (RRC): no website at the time of writing.
International Glossina Genome Initiative (IGGI): http://iggi.sanbi.ac.za/
Sand Fly Genome Sequencing Consortium (SFGSC): no website at the time of writing.
Anopheles Genomes Cluster Committee (AGCC): no website at the time of writing.
White papers available at VectorBase: http://www.vectorbase.org/Docs/ShowDoc/?doc=WhitePapers
VectorBase aims to be the main resource centre for the invertebrate disease vector communities, involving the scientists and generating, updating and giving easy access to the data.
CONCLUSION
Research into invertebrate vectors of human pathogens has reached a new level since the introduction of the genomics. Many areas have benefited from the huge amount of data generated: vector biology and its consequences on population studies and vector control, molecular biology with the increased understanding of biological processes and the possibility to design new tools, biomedicine with the prospect of new biopharmaceutical targets and evolutionary biology with the addition of new organisms for comparative genomic analysis. Progress can be expected to accelerate as more invertebrate genomes are sequenced and annotated. Additional Anopheles genomes will complement the existing data about A. gambiae (Besansky, 2006) and help in understanding the relation between sub-species. The list can be extended to include animal and plant vectors: the pea aphid Acyrthosiphon pisum (Gauthier et al., 2007) is currently being sequenced and the tick Boophilus microplus, affecting cattle and horses (Guerrero et al., 2006) is awaiting funding. Other non-vector insects already sequenced contribute to our ability to annotate and understand vector genomes: the honey bee Apis mellifera (Consortium, 2006), the silk worm Bombyx mori (Mita et al., 2004) and the agricultural pest, the red flour beetle Tribolium castaneum (Brown et al., 2003; Wang et al., 2007). The future in vectors of human pathogen research looks very exciting.
ACKNOWLEDGEMENTS
The core VectorBase project is funded by contract HHSN266200400039C from the NIAID, and supported, in part, by the BioMalPar network of excellence. We would like to thank all the persons involved in the VectorBase project for developing the resources described in this review, the Ensembl team for their help when using their pipelines and the scientific community working on invertebrate vectors of human pathogens for their collaborations and support.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Adelman ZN, Jasinskiene N, Vally KJ, Peek C, Travanty EA, Olson KE, Brown SE, Stephens JL, Knudson DL, Coates CJ, James AA. Formation and loss of large, unstable tandem arrays of the piggyBac transposable element in the yellow fever mosquito, Aedes aegypti. Transgenic Res. 2004;13:411–425. doi: 10.1007/s11248-004-6067-2. [DOI] [PubMed] [Google Scholar]
- Aksoy S. Control of tsetse flies and trypanosomes using molecular genetics. Vet Parasitol. 2003;115:125–145. doi: 10.1016/s0304-4017(03)00203-6. [DOI] [PubMed] [Google Scholar]
- Aksoy S, Hao Z, Strickler PM. What can we hope to gain for trypanosomiasis control from molecular studies on tsetse biology ? Kinetoplastid Biol Dis. 2002;1:4. doi: 10.1186/1475-9292-1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen ML, O’Brochta DA, Atkinson PW, Levesque CS. Stable, germ-line transformation of Culex quinquefasciatus (Diptera: Culicidae) J Med Entomol. 2001;38:701–710. doi: 10.1603/0022-2585-38.5.701. [DOI] [PubMed] [Google Scholar]
- Arca B, Lombardo F, Francischetti IM, Pham VM, Mestres-Simon M, Andersen JF, Ribeiro JM. An insight into the sialome of the adult female mosquito Aedes albopictus. Insect Biochem Mol Biol. 2007;37:107–127. doi: 10.1016/j.ibmb.2006.10.007. [DOI] [PubMed] [Google Scholar]
- Besansky NJ. Proposal for a Twelve Genomes Cluster for Genus Anopheles. 2006 [Google Scholar]
- Biessmann H, Nguyen QK, Le D, Walter MF. Microarray-based survey of a subset of putative olfactory genes in the mosquito Anopheles gambiae. Insect Mol Biol. 2005;14:575–589. doi: 10.1111/j.1365-2583.2005.00590.x. [DOI] [PubMed] [Google Scholar]
- Brown SJ, Denell RE, Beeman RW. Beetling around the genome. Genet Res. 2003;82:155–161. doi: 10.1017/s0016672303006451. [DOI] [PubMed] [Google Scholar]
- Calvo E, Dao A, Pham VM, Ribeiro JM. An insight into the sialome of Anopheles funestus reveals an emerging pattern in anopheline salivary protein families. Insect Biochem Mol Biol. 2007;37:164–175. doi: 10.1016/j.ibmb.2006.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coates CJ, Jasinskiene N, Miyashiro L, James AA. Mariner transposition and transformation of the yellow fever mosquito, Aedes aegypti. Proc Natl Acad Sci U S A. 1998;95:3748–3751. doi: 10.1073/pnas.95.7.3748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Consortium. Insights into social insects from the genome of the honeybee Apis mellifera. Nature. 2006;443:931–949. doi: 10.1038/nature05260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crosby MA, Goodman JL, Strelets VB, Zhang P, Gelbart WM. FlyBase: genomes by the dozen. Nucleic Acids Res. 2007;35:D486–D491. doi: 10.1093/nar/gkl827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuamba N, Choi KS, Townson H. Malaria vectors in Angola: distribution of species and molecular forms of the Anopheles gambiae complex, their pyrethroid insecticide knockdown resistance (kdr) status and Plasmodium falciparum sporozoite rates. Malar J. 2006;5:2. doi: 10.1186/1475-2875-5-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- David JP, Strode C, Vontas J, Nikou D, Vaughan A, Pignatelli PM, Louis C, Hemingway J, Ranson H. The Anopheles gambiae detoxification chip: a highly specific microarray to study metabolic-based insecticide resistance in malaria vectors. Proc Natl Acad Sci U S A. 2005;102:4080–4084. doi: 10.1073/pnas.0409348102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gauthier JP, Legeai F, Zasadzinski A, Rispe C, Tagu D. AphidBase: a database for aphid genomic resources. Bioinformatics. 2007;23:783–784. doi: 10.1093/bioinformatics/btl682. [DOI] [PubMed] [Google Scholar]
- Grossman GL, Rafferty CS, Clayton JR, Stevens TK, Mukabayire O, Benedict MQ. Germline transformation of the malaria vector, Anopheles gambiae, with the piggyBac transposable element. Insect Mol Biol. 2001;10:597–604. doi: 10.1046/j.0962-1075.2001.00299.x. [DOI] [PubMed] [Google Scholar]
- Guerrero FD, Nene VM, George JE, Barker SC, Willadsen P. Sequencing a new target genome: the Boophilus microplus (Acari: Ixodidae) genome project. J Med Entomol. 2006;43:9–16. doi: 10.1603/0022-2585(2006)043[0009:santgt]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- Halasz G, van Batenburg F, Perusse J, Hua S, Lu XJ, White KP, Bussemaker HJ. Detecting transcriptionally active regions using genomic tiling arrays. Genome Biol. 2006;7:R59. doi: 10.1186/gb-2006-7-7-r59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handler AM. Use of the piggyBac transposon for germ-line transformation of insects. Insect Biochem Mol Biol. 2002;32:1211–1220. doi: 10.1016/s0965-1748(02)00084-x. [DOI] [PubMed] [Google Scholar]
- Holt RA, Subramanian GM, Halpern A, Sutton GG, Charlab R, Nusskern DR, Wincker P, Clark AG, Ribeiro JM, Wides R, Salzberg SL, Loftus B, Yandell M, Majoros WH, Rusch DB, Lai Z, Kraft CL, Abril JF, Anthouard V, Arensburger P, Atkinson PW, Baden H, de Berardinis V, Baldwin D, Benes V, Biedler J, Blass C, Bolanos R, Boscus D, Barnstead M, Cai S, Center A, Chaturverdi K, Christophides GK, Chrystal MA, Clamp M, Cravchik A, Curwen V, Dana A, Delcher A, Dew I, Evans CA, Flanigan M, Grundschober-Freimoser A, Friedli L, Gu Z, Guan P, Guigo R, Hillenmeyer ME, Hladun SL, Hogan JR, Hong YS, Hoover J, Jaillon O, Ke Z, Kodira C, Kokoza E, Koutsos A, Letunic I, Levitsky A, Liang Y, Lin JJ, Lobo NF, Lopez JR, Malek JA, McIntosh TC, Meister S, Miller J, Mobarry C, Mongin E, Murphy SD, O’Brochta DA, Pfannkoch C, Qi R, Regier MA, Remington K, Shao H, Sharakhova MV, Sitter CD, Shetty J, Smith TJ, Strong R, Sun J, Thomasova D, Ton LQ, Topalis P, Tu Z, Unger MF, Walenz B, Wang A, Wang J, Wang M, Wang X, Woodford KJ, Wortman JR, Wu M, Yao A, Zdobnov EM, Zhang H, Zhao Q, Zhao S, Zhu SC, Zhimulev I, Coluzzi M, della Torre A, Roth CW, Louis C, Kalush F, Mural RJ, Myers EW, Adams MD, Smith HO, Broder S, Gardner MJ, Fraser CM, Birney E, Bork P, Brey PT, Venter JC, Weissenbach J, Kafatos FC, Collins FH, Hoffman SL. The genome sequence of the malaria mosquito Anopheles gambiae. Science. 2002;298:129–149. doi: 10.1126/science.1076181. [DOI] [PubMed] [Google Scholar]
- Hubbard TJ, Aken BL, Beal K, Ballester B, Caccamo M, Chen Y, Clarke L, Coates G, Cunningham F, Cutts T, Down T, Dyer SC, Fitzgerald S, Fernandez-Banet J, Graf S, Haider S, Hammond M, Herrero J, Holland R, Howe K, Johnson N, Kahari A, Keefe D, Kokocinski F, Kulesha E, Lawson D, Longden I, Melsopp C, Megy K, Meidl P, Ouverdin B, Parker A, Prlic A, Rice S, Rios D, Schuster M, Sealy I, Severin J, Slater G, Smedley D, Spudich G, Trevanion S, Vilella A, Vogel J, White S, Wood M, Cox T, Curwen V, Durbin R, Fernandez-Suarez XM, Flicek P, Kasprzyk A, Proctor G, Searle S, Smith J, Ureta-Vidal A, Birney E. Ensembl 2007. Nucleic Acids Res. 2007;35:D610–D617. doi: 10.1093/nar/gkl996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasprzyk A, Keefe D, Smedley D, London D, Spooner W, Melsopp C, Hammond M, Rocca-Serra P, Cox T, Birney E. EnsMart: a generic system for fast and flexible access to biological data. Genome Res. 2004;14:160–169. doi: 10.1101/gr.1645104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawson D, Arensburger P, Atkinson P, Besansky NJ, Bruggner RV, Butler R, Campbell KS, Christophides GK, Christley S, Dialynas E, Emmert D, Hammond M, Hill CA, Kennedy RC, Lobo NF, MacCallum MR, Madey G, Megy K, Redmond S, Russo S, Severson DW, Stinson EO, Topalis P, Zdobnov EM, Birney E, Gelbart WM, Kafatos FC, Louis C, Collins FH. VectorBase: a home for invertebrate vectors of human pathogens. Nucleic Acids Res. 2007;35:D503–D505. doi: 10.1093/nar/gkl960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobo NF, Hua-Van A, Li X, Nolen BM, Fraser MJ., Jr Germ line transformation of the yellow fever mosquito, Aedes aegypti, mediated by transpositional insertion of a piggyBac vector. Insect Mol Biol. 2002;11:133–139. doi: 10.1046/j.1365-2583.2002.00317.x. [DOI] [PubMed] [Google Scholar]
- Mita K, Kasahara M, Sasaki S, Nagayasu Y, Yamada T, Kanamori H, Namiki N, Kitagawa M, Yamashita H, Yasukochi Y, Kadono-Okuda K, Yamamoto K, Ajimura M, Ravikumar G, Shimomura M, Nagamura Y, Shin IT, Abe H, Shimada T, Morishita S, Sasaki T. The genome sequence of silkworm, Bombyx mori. DNA Res. 2004;11:27–35. doi: 10.1093/dnares/11.1.27. [DOI] [PubMed] [Google Scholar]
- Moses AM, Pollard DA, Nix DA, Iyer VN, Li XY, Biggin MD, Eisen MB. Large-scale turnover of functional transcription factor binding sites in Drosophila. PLoS Comput Biol. 2006;2:e130. doi: 10.1371/journal.pcbi.0020130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mungall CJ, Emmert DB. A Chado case study: an ontology-based modular schema for representing genome-associated biological information. Bioinformatics. 2007;23:i337–i346. doi: 10.1093/bioinformatics/btm189. [DOI] [PubMed] [Google Scholar]
- Nene V, Wortman JR, Lawson D, Haas B, Kodira C, Tu ZJ, Loftus B, Xi Z, Megy K, Grabherr M, Ren Q, Zdobnov EM, Lobo NF, Campbell KS, Brown SE, Bonaldo MF, Zhu J, Sinkins SP, Hogenkamp DG, Amedeo P, Arensburger P, Atkinson PW, Bidwell S, Biedler J, Birney E, Bruggner RV, Costas J, Coy MR, Crabtree J, Crawford M, Debruyn B, Decaprio D, Eiglmeier K, Eisenstadt E, El-Dorry H, Gelbart WM, Gomes SL, Hammond M, Hannick LI, Hogan JR, Holmes MH, Jaffe D, Johnston JS, Kennedy RC, Koo H, Kravitz S, Kriventseva EV, Kulp D, Labutti K, Lee E, Li S, Lovin DD, Mao C, Mauceli E, Menck CF, Miller JR, Montgomery P, Mori A, Nascimento AL, Naveira HF, Nusbaum C, O’Leary S, Orvis J, Pertea M, Quesneville H, Reidenbach KR, Rogers YH, Roth CW, Schneider JR, Schatz M, Shumway M, Stanke M, Stinson EO, Tubio JM, Vanzee JP, Verjovski-Almeida S, Werner D, White O, Wyder S, Zeng Q, Zhao Q, Zhao Y, Hill CA, Raikhel AS, Soares MB, Knudson DL, Lee NH, Galagan J, Salzberg SL, Paulsen IT, Dimopoulos G, Collins FH, Birren B, Fraser-Liggett CM, Severson DW. Genome sequence of Aedes aegypti, a major arbovirus vector. Science. 2007;316:1718–1723. doi: 10.1126/science.1138878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nisbet AJ, Huntley JF. Progress and opportunities in the development of vaccines against mites, fleas and myiasis-causing flies of veterinary importance. Parasite Immunol. 2006;28:165–172. doi: 10.1111/j.1365-3024.2006.00803.x. [DOI] [PubMed] [Google Scholar]
- Nolan T, Bower TM, Brown AE, Crisanti A, Catteruccia F. piggyBac-mediated germline transformation of the malaria mosquito Anopheles stephensi using the red fluorescent protein dsRED as a selectable marker. J Biol Chem. 2002;277:8759–8762. doi: 10.1074/jbc.C100766200. [DOI] [PubMed] [Google Scholar]
- Pagel Van Zee J, Geraci NS, Guerrero FD, Wikel SK, Stuart JJ, Nene VM, Hill CA. Tick genomics: The Ixodes genome project and beyond. Int J Parasitol. 2007;37:1297–1305. doi: 10.1016/j.ijpara.2007.05.011. [DOI] [PubMed] [Google Scholar]
- Perera OP, Harrell IR, Handler AM. Germ-line transformation of the South American malaria vector, Anopheles albimanus, with a piggyBac/EGFP transposon vector is routine and highly efficient. Insect Mol Biol. 2002;11:291–297. doi: 10.1046/j.1365-2583.2002.00336.x. [DOI] [PubMed] [Google Scholar]
- Ribeiro JM, Alarcon-Chaidez F, Francischetti IM, Mans BJ, Mather TN, Valenzuela JG, Wikel SK. An annotated catalog of salivary gland transcripts from Ixodes scapularis ticks. Insect Biochem Mol Biol. 2006;36:111–129. doi: 10.1016/j.ibmb.2005.11.005. [DOI] [PubMed] [Google Scholar]
- Ribeiro JM, Topalis P, Louis C. AnoXcel: an Anopheles gambiae protein database. Insect Mol Biol. 2004;13:449–457. doi: 10.1111/j.0962-1075.2004.00503.x. [DOI] [PubMed] [Google Scholar]
- Rodrigues FG, Oliveira SB, Rocha BC, Moreira LA. Germline transformation of Aedes fluviatilis (Diptera:Culicidae) with the piggyBac transposable element. Mem Inst Oswaldo Cruz. 2006;101:755–757. doi: 10.1590/s0074-02762006000700008. [DOI] [PubMed] [Google Scholar]
- Santos A, Ribeiro JM, Lehane MJ, Gontijo NF, Veloso AB, Sant’Anna MR, Araujo Nascimento R, Grisard EC, Pereira MH. The sialotranscriptome of the blood-sucking bug Triatoma brasiliensis (Hemiptera, Triatominae) Insect Biochem Mol Biol. 2007;37:702–712. doi: 10.1016/j.ibmb.2007.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos IK, Valenzuela JG, Ribeiro JM, de Castro M, Costa JN, Costa AM, da Silva R, Neto OB, Rocha C, Daffre S, Ferreira BR, da Silva S, Szabo MP, Bechara GH. Gene discovery in Boophilus microplus, the cattle tick: the transcriptomes of ovaries, salivary glands, and hemocytes. Ann N Y Acad Sci. 2004;1026:242–246. doi: 10.1196/annals.1307.037. [DOI] [PubMed] [Google Scholar]
- Vontas J, David JP, Nikou D, Hemingway J, Christophides GK, Louis C, Ranson H. Transcriptional analysis of insecticide resistance in Anopheles stephensi using cross-species microarray hybridization. Insect Mol Biol. 2007;16:315–324. doi: 10.1111/j.1365-2583.2007.00728.x. [DOI] [PubMed] [Google Scholar]
- Wang L, Wang S, Li Y, Paradesi MS, Brown SJ. BeetleBase: the model organism database for Tribolium castaneum. Nucleic Acids Res. 2007;35:D476–D479. doi: 10.1093/nar/gkl776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterhouse RM, Kriventseva EV, Meister S, Xi Z, Alvarez KS, Bartholomay LC, Barillas-Mury C, Bian G, Blandin S, Christensen BM, Dong Y, Jiang H, Kanost MR, Koutsos AC, Levashina EA, Li J, Ligoxygakis P, Maccallum RM, Mayhew GF, Mendes A, Michel K, Osta MA, Paskewitz S, Shin SW, Vlachou D, Wang L, Wei W, Zheng L, Zou Z, Severson DW, Raikhel AS, Kafatos FC, Dimopoulos G, Zdobnov EM, Christophides GK. Evolutionary dynamics of immune-related genes and pathways in disease-vector mosquitoes. Science. 2007;316:1738–1743. doi: 10.1126/science.1139862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X, Dong Y, Abraham EG, Kocan A, Srinivasan P, Ghosh AK, Sinden RE, Ribeiro JM, Jacobs-Lorena M, Kafatos FC, Dimopoulos G. Transcriptome analysis of Anopheles stephensi-Plasmodium berghei interactions. Mol Biochem Parasitol. 2005;142:76–87. doi: 10.1016/j.molbiopara.2005.02.013. [DOI] [PubMed] [Google Scholar]
- Zeitlinger J, Zinzen RP, Stark A, Kellis M, Zhang H, Young RA, Levine M. Whole-genome ChIP-chip analysis of Dorsal, Twist, and Snail suggests integration of diverse patterning processes in the Drosophila embryo. Genes Dev. 2007;21:385–390. doi: 10.1101/gad.1509607. [DOI] [PMC free article] [PubMed] [Google Scholar]