Introduction
Large libraries of peptides, cyclic peptides, and other molecules are standard tools for the discovery of drugs, molecular probes, and affinity reagents. In particular, one-bead-one-compound (OBOC) libraries,1 prepared by the split-and-mix method,2 provide access to a broad chemical space with a minimum of reagents. Once such a library has been screened against the target of interest, the chemical identity of the library elements on the hit beads is identified. For peptide libraries and their variants, mass spectrometry (MS) based peptide sequencing provides the most rapid method for such analysis. OBOC libraries are constructed in a number of ways to facilitate MS analysis,3–5 but one common feature is that the peptide must be cleaved from the bead prior to being introduced into the mass spectrometer. While a number of chemical6 and photochemical7 cleavage strategies have been developed, the most common strategy is to incorporate a CNBr-cleavable methionine-linker group at the C-terminus of the peptide.8 CNBr cleavage has also been widely used in proteomics to cleave proteins.9 Using such chemistry, up to 100 beads from an OBOC peptide library can be sequenced in a 24 hour period.10 A large fraction of that time, however, is devoted to the CNBr cleavage step. Standard literature protocols describe CNBr cleavage as requiring between 12 and 24 hours using 20 – 30 µl of 0.25 M CNBr in 70% aqueous formic acid at room temperature.11 Although the CNBr cleavage time may be reduced to 2–4 hours at elevated temperatures (~ 47°C), significant side-products may be generated.12 All reports that we have found that describe CNBr cleavage chemistry from single beads have utilized the same conditions as for proteomics, although the two chemical processes are not necessarily equivalent.
Microwave-assisted organic reactions have been widely performed with great effects on increasing reaction rates and improving yields and selectivity.13,14 However, the “specific” or “nonthermal” microwave effects13c,14a are still in dispute.15, 16 In principle, molecular rotation is induced by the interaction of molecular dipoles with the microwave fields leading to heat generation. 13a Reactions involving molecules with significant dipole moments may be more influenced by microwave irradiation, showing a great improvement in kinetic rates and product yields. Proteins and peptides have large dipole moments, and thus associated reactions have the potential for significant improvements via microwave-assistance.17 In particular, microwave irradiation has been successfully applied to solid-phase peptide synthesis, increasing the peptide coupling reaction rate and not generating appreciable racemization.18,19 Recently, microwave irradiation was also used for the rapid generation of combinatorial peptide libraries.20
Here we report on the optimization of single-bead CNBr cleavage conditions, and demonstrate that the reaction time may be reduced to 1 minute through the use of microwave assisted cleavage in an aqueous medium. We further show that microwave-assisted cleavage yields very pure cleaved peptides from single beads. We performed MS analysis of various peptides of known sequence that were cleaved using a variant of the typical CNBr cleavage chemistry, as well as a rapid, microwave-assisted protocol. The peptides ranged in length from 5-mers to 8-mers, and were prepared to represent the natural and non-natural stereoisomers of all amino acids except cysteine and methionine. The MS analysis yielded comparable results from both cleavage methods. Thus, microwave-assisted CNBr cleavage of peptides in water is significantly more efficient than the standard protocol. 21
Results and Discussion
The standard protocol for CNBr cleavage of peptides from single beads is to utilize 20 – 30 µl of 0.25 M CNBr in 70% aqueous formic acid for 12 – 16 hr at room temperature. We first investigated elevated temperature (45°C) and reduced reaction times (2 – 4 hr). Franz et al. reported that such reaction conditions generated a byproduct that exhibited a mass shift of +28 amu relative to the parent mass. To reduce such side products, other acidic media such as 70% TFA22 and HCl23,24 were tested. 0.1 – 0.2 N HCl(aq) showed the cleanest reaction by MS analysis (Scheme 1).
Scheme 1.
The CNBr cleavage of peptides bound to a single bead a) for 4 hr at 45°C and b) for 1 min under 30 Watts microwave irradiation.
High quality MS data were obtained by using 0.1 N aqueous HCl for 2 – 4 hr at 45°C without producing any noticeable side products (see supporting materials S-Fig. 2). The oxidation of tryptophan (W),25 was significantly reduced by purging the reaction vessels with Ar for a short time before and after the addition of the reaction medium. Consequently, the final optimized conditions were 5 – 20 µl of 0.25 M CNBr in 0.1 N aq. HCl for 2 – 4 hr at 45°C. This protocol was utilized as a comparison against the microwave-assisted protocol and for quantifying the amount of peptide cleaved from single beads.
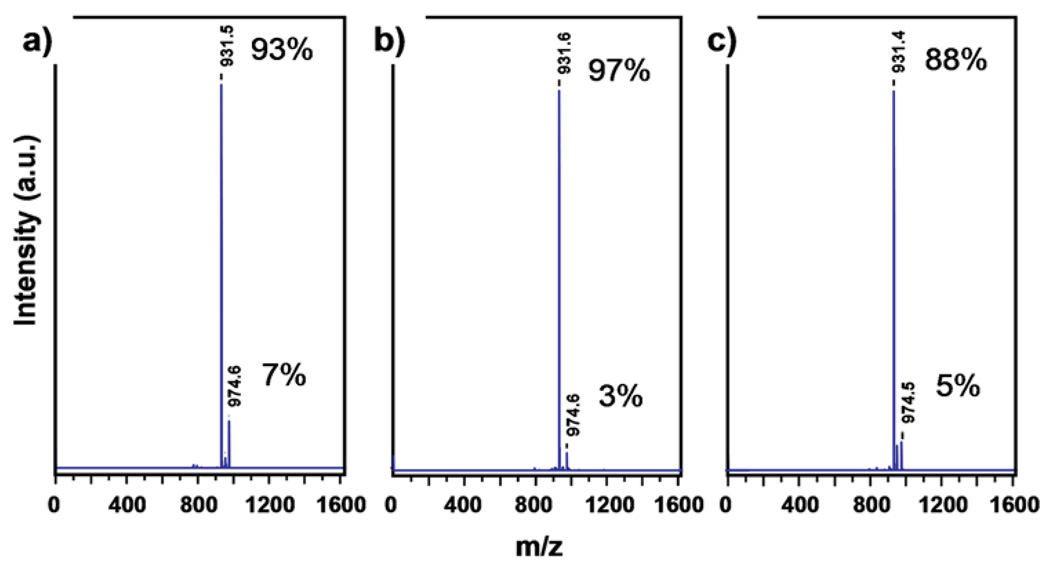
Microwave-assisted acceleration was performed using identical chemical conditions (Scheme 1), but with 1 to 4 minutes of microwave irradiation replacing the 45°C heating. Figure 1 shows MS data of the peptide HLYFLRM* (M* = homoserine lactone) from a single 90 µm TentaGel S Amino bead, in which the CNBr-cleavable M (methionine) is at the C-terminal. 10 percent of the cleaved peptide from a single bead was used as a sample for MALDI-MS. Data are shown for rapid microwave-assisted CNBr cleavage for 1 and 4 minutes (Figs 1a and 1b) using a household microwave oven. This process was calibrated against a CEM Discover microwave reactor (see supporting materials, S-Fig. 7). With the CEM reactor, a 1 minute, 30 Watt exposure produced results identical to the 1 min microwave irradiation from the household microwave oven. Fig. 1c is a plot of MS data from a peptide cleaved from a single bead using the above-described protocol of 4 hr CNBr cleavage at 45°C. In each plot, the major peak represents the primary mass of the peptide HLYFLRM* (931 amu). The data indicate that, at least for this peptide, the microwave-assisted and thermal cleavage processes are equivalent, and neither method adversely affects the peptide.
Figure 1.
MS spectra of the peptide HLYFLRM* (M* = homoserine lacton, MW = 931.5), following CNBr cleavage using 3 protocols. a. Microwave assisted cleavage in water, with a 1 minute microwave exposure. b. Microwave-assisted cleavage in water, with a 4 minute microwave exposure. c. Cleavage at 45°C in H2O/MeCN for 4 hours. Adjacent to each peak is the % of the total intensity
A calibrated optical fiber thermometer was used to measure the temperature of a 1 minute irradiation of 20 µl of water within the same microvessels utilized for bead cleavage. For both the 30 W Discover reactor and for the household microwave oven, ΔT was 13–14°C during the course of the exposure. Thus, 45°C was obtained when starting at 31°C, and 39°C was obtained when starting at 26°C (see supporting materials, S-Fig. 8).
We quantified both processes using liquid chromatography (LC) of cleaved peptides. To enhance the UV absorbance for LC analysis, we introduced 5-(dimethylamino)naphthalene-1-sulfonyl (Dansyl) group26 at the N-terminal of the benchmark peptide HLYFLR. The reaction proceeded smoothly in the presence of N,N-diisopropylethylamine (DIEA) to elaborate the corresponding sulfonamide. The resulting resin was treated with TFA for removal of protective groups. The dansyl group attached at the N-terminal of the peptide HLYFLRM*, as evidenced by MALDI-MS analysis which yielded a parent mass at 1064 amu (see supporting materials, S-Fig. 3). To obtain sufficient amount of peptide for HPLC quantification, peptides were separately released under both thermal conditions and microwave conditions with ~ 320 µg of the resin, respectively.
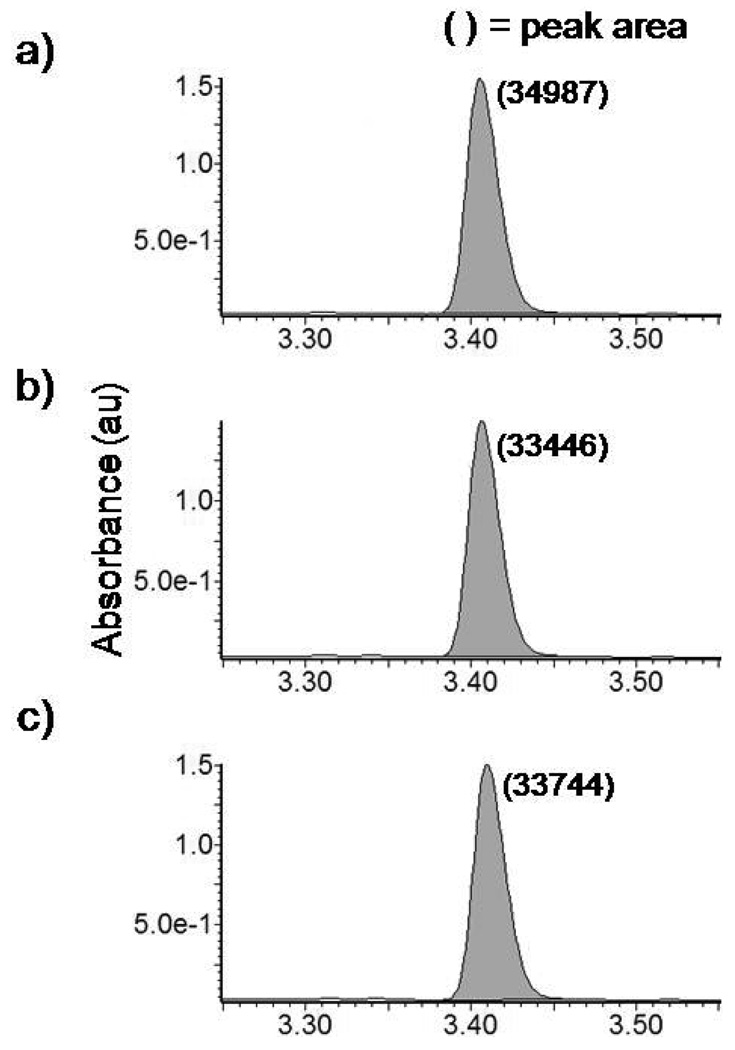
We first investigated the amount of the cleaved peptides after 30 min, 1 hr and 2hr under the thermal conditions. The 2 hr thermal reaction led to nearly complete cleavage of peptides from beads. The 30 min and 1 hr thermal reactions resulted in ~36% and ~70% cleavage of peptides, respectively (see supporting materials, S-Fig. 6). Then, samples of identical quantity for the thermal and microwave reaction conditions were prepared. The amount of the released peptide under 1 min microwave conditions was comparable to both 2 min microwave cleavage and 4 hr thermal (45°C) cleavage (Figure 2). 1 min microwave reaction led to effectively complete cleavage of peptides.
Figure 2.
HPLC spectra of a) CNBr micro-wave assisted cleavage for 2 min, b) CNBr microwave assisted cleavage for 1 min, c) CNBr cleavage for 4 hr at 45°C, which was separately shown to lead to complete cleavage. All peaks exhibit a cleaved peptide retention time of 3.40 or 3.41 minutes, and the integrated peak area for all three plots is equal to within 5%, indicating that for the 1 min microwave-assisted cleavage reaction, the reaction has proceeded to completion.
We have utilized Scheme 1 to cleave over a hundred beads from OBOC peptide libraries, ranging in length from 5-mers to 8-mers (not including methionine), and containing all of the amino acids excepting cysteine and methionine (methionine was used only for the cleavage at the C-terminal), as well as both natural and unnatural stereoisomers. The 9-mer peptide library were synthesized with –FLRM in the C-terminal. The peptide sequences were obtained by de novo sequencing methods with their MS/MS data. Some of the obtained sequences were confirmed by synthesizing peptides with known sequence and obtaining their MS/MS data. Representative MS data from the peptides of known sequence are presented in the supporting materials, S-Fig. 9 including 4 peptides with –FKRM* in the C-terminal. After evaporation of the reaction medium, just 10 percent of the released peptides were consumed for MALDI-MS sampling after mixing with a matrix (CHCA) solution. Most of the released peptides from the beads showed high quality of MS spectra, which can lead to high quality of MS/MS sequence information.
Conclusions
We have demonstrated an efficient microwave-assisted CNBr-based cleavage of peptides from single beads. One minute cleavage times were demonstrated for 5-mer and 8-mer peptides containing both the D- and L-stereoisomers of 18 out of the 20 amino acids. Released peptides exhibited high purity, as measured by both mass spectrometry and HPLC. This simple and rapid CNBr cleavage protocol should be useful in accelerating the screening of one-bead-one-compound peptide libraries. We have not tested whether this technique can be utilized to accelerate the site-specific cleavage of proteins.
Supplementary Material
These include Detailed Experimental Methods, and supplemental data, as referred to in the text.
Acknowledgments
We acknowledge Heather Agnew and Rosemary Rohde for the assistance provided with obtaining Edman degradation peptide sequencing results. This work was supported by the Institute of Bioengineering and Nanotechnology (Biomedical Research Council, Agency for Science, Technology and Research, Singapore), with additional support (JRH) from the National Cancer Institute Grant No. 5U54 CA119347 (J.R.H., P.I.) and a subcontract from the Mitre Corporation.
References
- 1.Lam KS, Lebl M, Krchnak V. Chem. Rev. 1997:411–448. doi: 10.1021/cr9600114. [DOI] [PubMed] [Google Scholar]
- 2.Furka A, Sebestye´n F, Asgedom M, Dibo´ G. Int. J. Pept. Protein Res. 1991;37:487–493. doi: 10.1111/j.1399-3011.1991.tb00765.x. [DOI] [PubMed] [Google Scholar]
- 3.Chait BT, Wang R, Beavis RC, Kent SBH. Science. 1993;262:89–92. doi: 10.1126/science.8211132. [DOI] [PubMed] [Google Scholar]
- 4.Liu R, Markik J, Lam KS. J. Am. Chem. Soc. 2002;124:7678–7680. doi: 10.1021/ja026421t. [DOI] [PubMed] [Google Scholar]
- 5.Wang X, Zhang J, Song A, Lebrilla CB, Lam KS. J. Am. Chem. Soc. 2004;126:5740–5749. doi: 10.1021/ja049322j. [DOI] [PubMed] [Google Scholar]
- 6.Paulick MG, Hart KM, Brinner KM, Tjandra M, Charych DH, Zuckermann RN. J. Comb. Chem. 2006;8:417–426. doi: 10.1021/cc0501460. [DOI] [PubMed] [Google Scholar]
- 7.Holmes CP, Jones DG. J. Org. Chem. 1995;60:2318–2319. [Google Scholar]
- 8.Yu Z, Chu Y-H. Bioorg. Med. Chem. Lett. 1997;7:95–98. [Google Scholar]
- 9.Schroeder WA, Shelton JB, Shelton JR. Biochem. and Biophys. 1969;130:551–556. doi: 10.1016/0003-9861(69)90069-1. [DOI] [PubMed] [Google Scholar]
- 10.Joo SH, Xiao Q, Ling Y, Gopishetty B, Pei D. J. Am. Chem. Soc. 2006;128:13000–13009. doi: 10.1021/ja063722k. [DOI] [PubMed] [Google Scholar]
- 11.(a) Wang P, Arabaci G, Pei D. J. Comb. Chem. 2001;3:251–254. doi: 10.1021/cc000102l. [DOI] [PubMed] [Google Scholar]; (b) Sweeney MC, Pei D. J. Comb. Chem. 2003;5:218–222. doi: 10.1021/cc020113+. [DOI] [PubMed] [Google Scholar]; (c) Wang X, Peng L, Liu R, Gill SS, Lam KS. J. Comb. Chem. 2005;7:197–209. doi: 10.1021/cc049887b. [DOI] [PubMed] [Google Scholar]
- 12.Franz AH, Liu R, Song A, Lam KS, Lebrilla CB. J. Comb. Chem. 2003;5:125–137. doi: 10.1021/cc020083a. [DOI] [PubMed] [Google Scholar]
- 13.(a) Loupy A, editor. Microwaves in Organic Synthesis. 2nd ed. Weinheim, Germany: Wiley-VCH; 2006. [Google Scholar]; (b) Lidström P, Tierney JP, editors. Microwave-Assisted Organic Synthesis. Oxford, U.K.: Blackwell Publishing; 2005. [Google Scholar]; (c) Kappe CO, Stadler A, editors. Microwaves in Organic And Medicinal Chemistry. Weinheim, Germany: Wiley-VCH; 2005. [Google Scholar]
- 14.(a) Kappe CO. Angew. Chem., Int. Ed. 2004;43:6250–6284. doi: 10.1002/anie.200400655. [DOI] [PubMed] [Google Scholar]; (b) Larhed M, Moberg C, Hallberg A. Acc. Chem. Res. 2002;35:717–727. doi: 10.1021/ar010074v. [DOI] [PubMed] [Google Scholar]; (c) Lew A, Krutzik PO, Hart ME, Chamberlin AR. J. Comb. Chem. 2002;4:95–105. doi: 10.1021/cc010048o. [DOI] [PubMed] [Google Scholar]
- 15.(a) De La Hoz A, Diaz-Ortiz A, Moreno A. Chem. Soc. Rev. 2005;34:164–178. doi: 10.1039/b411438h. [DOI] [PubMed] [Google Scholar]; (b) Perreux L, Loupy A. Tetrahedron. 2001;57:9199–9223. [Google Scholar]
- 16.Herrero MA, Kremsner JM, Kappe CO. J. Org. Chem. 2008;73:36–47. doi: 10.1021/jo7022697. [DOI] [PubMed] [Google Scholar]
- 17.(a) Young DD, Nichols J, Kelly RM, Deiters A. J. Am. Chem. Soc. 2008;130:10048–10049. doi: 10.1021/ja802404g. [DOI] [PubMed] [Google Scholar]; (b) Rejasse B, Lamare S, Legoy MD, Besson T. J. Enzym. Inhib. Med. Chem. 2007;22:519–527. doi: 10.1080/14756360701424959. [DOI] [PubMed] [Google Scholar]; (c) Collins JM, Leadbeater NE. Org. Biomol. Chem. 2007;5:1141–1150. doi: 10.1039/b617084f. [DOI] [PubMed] [Google Scholar]
- 18.Collins JM, Collins MJ. Chapter 20. In: Loupy A, editor. Microwaves in Organic synthesis. 2nd ed. Weinheim, Germany: Wiley-VCH; 2006. pp. 898–930. [Google Scholar]
- 19.(a) Yu H-M, Chen S-T, Wang K-T. J. Org. Chem. 1992;57:4781–4784. [Google Scholar]; (b) Erdélyi M, Gogoll A. Synthesis. 2002;11:1592–1596. [Google Scholar]
- 20.Murray JK, Gellman SH. Nat. Protoc. 2007;2:624–631. doi: 10.1038/nprot.2007.23. [DOI] [PubMed] [Google Scholar]
- 21.Dallinger D, Kappe CO. Chem. Rev. 2007;107:2563–2591. doi: 10.1021/cr0509410. [DOI] [PubMed] [Google Scholar]
- 22.Sorenson MK, Darst SA. Proc. Nat. Acad. Sci. U.S.A. 2006;103:16722–16727. doi: 10.1073/pnas.0606482103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kaiser R, Metzka L. Anal. Biochem. 1999;266:1–8. doi: 10.1006/abio.1998.2945. [DOI] [PubMed] [Google Scholar]
- 24.Rodríguez JC, Wong L, Jennings P. Protein Expression and Purification. 2003;28:224–231. doi: 10.1016/s1046-5928(02)00700-3. [DOI] [PubMed] [Google Scholar]
- 25.Taylor SW, Fahy E, Murray J, Capaldi RA, Ghosh SS. J. Bio. Chem. 2003;278:19587–19590. doi: 10.1074/jbc.C300135200. [DOI] [PubMed] [Google Scholar]
- 26.Price NPJ, Firmin JL, Robins RJ, Gray DO. J. Chromat. 1993;653:161–166. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
These include Detailed Experimental Methods, and supplemental data, as referred to in the text.