Abstract
Ma’am – We read with great interest the article by He et al. [2008] describing the effects on HIV acquisition and disease progression of a single-nucleotide polymorphism (SNP, rs2814778, -46T→C) that disrupts the promoter region of the Duffy antigen receptor for chemokines (DARC) gene and abolishes gene expression in red blood cells. He et al. reported that HIV-infected African Americans have a frequency of the null homozygous genotype (-46C/C) of 70% while non-HIV infected individuals have a null genotype frequency of 60%. Based on this frequency difference they argued that the null allele confers susceptibility to infection with HIV-1. They also reported that the null genotype is associated with better outcomes amongst those who do become infected, including longer survival, slower loss of CD4+ T-lymphocytes, and delayed progression to HIV-associated dementia.
We sought to evaluate these suggested associations using a cohort of 471 HIV-1 infected African Americans with estimated seroconversion dates enrolled in the TriService AIDS Clinical Consortium (TACC) HIV Natural History Study (NHS) and 227 HIV-negative African Americans recruited in conjunction with ongoing genetic studies at Duke University in Durham, NC. A principal component based procedure implemented in the EigenSoft routines (Price et al., 2006) was used to correct for population structure. This approach has been extensively used to adjust for population stratification that would otherwise inflate association statistics (McCarthy et al., 2008).
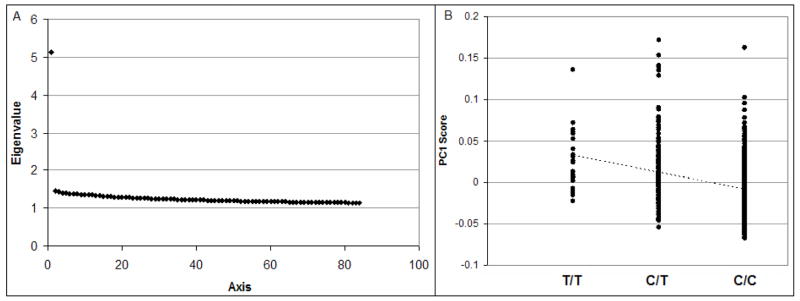
In assessing population stratification in the 698 African Americans using EIGENSTRAT (Price et al., 2006), the first axis makes a much larger contribution to the proportion of variation explained than other axes and reflects the degree of African versus European ancestry in individuals (Figure 1A). To further demonstrate the separation of African and European ancestries in the admixed African American population, we added 60 HapMap Utah residents with ancestry from northern and western Europe (CEU) and 60 HapMap Yoruba in Ibadan, Nigeria (YRI) samples into the EIGENSTRAT analysis (see Supplemental Materials). The first axis separates African and European ancestries and is highly correlated to the first axis without the seed populations (r2=0.9958, Supplemental Materials). We note that the DARC -46T→C polymorphism is strongly associated with the first axis (p = 6.14×10−23) (Figure 1B). This confirms that the DARC -46T→C polymorphism itself is highly informative about ancestry in African American populations, as expected, and that it could therefore generate strong associations due to stratification for any traits that correlate with ancestry.
Figure 1.
Population stratification of rs2814778. (A) The high Eigenvalue for the first axis indicates that this axis accounts for a large proportion of population structure in our sample. This axis represents the degree of African versus European chromosomal ancestry on a genome-wide level (see Supplementary Materials for further information). (B) The principal component (PC) score for each subject along axis one (PC1) is significantly correlated with genotype at rs2814778, highlighting the importance of stratification control for association testing at this polymorphism.
We tested for an effect of the DARC -46T→Cpolymorphism on viral load at set-point, progression to AIDS, and CD4+ T cell decline. Viral set-point was defined as previously described (Fellay et al., 2007) for 394 HIV-infected patients. A linear regression using gender, age at seroconversion and the first EIGENSTRAT axis as covariates revealed no association with the DARC -46C/C genotype and viral setpoint (p = 0.524; when not corrected for population stratification, p = 0.905).
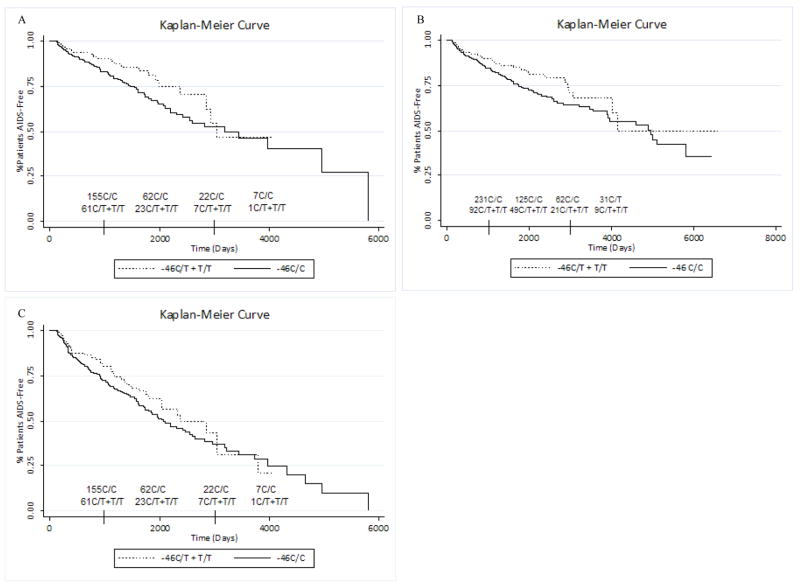
We defined HIV disease progression as time to AIDS (1993 Centers for Disease Control and Prevention (CDC) definition). Because many subjects in the cohort eventually initiated highly active antiretroviral therapy (HAART), we considered multiple methods to account for treatment initiation in our statistical models. In our primary model, subjects were censored at HAART initiation so that time to AIDS is considered only in untreated patients to rule out any effects of HAART. The Cox proportional hazards model was adjusted for gender, age at seroconversion, and the first EIGENSTRAT axis and shows no association between the -46C/C genotype and faster disease progression (HR 1.53, 95% CI 0.921–2.54, p=0.101; without correction for population stratification HR = 1.52, 95% CI 0.932–2.47, p = 0.094; Figure 2A). Censoring at January 1, 1996 (the approximate date when HAART first became available to the cohort), as opposed to HAART initiation, produced similar results (data not shown). In a separate model adjusted for the same covariates, we considered HAART as a time-updated covariate rather than censoring at HAART initiation. In this analysis there was no significant association between -46C/C genotype and disease progression (HR 1.37, 95% CI 0.854–2.21, p=0.191; without correction for population stratification HR = 1.28, 95% CI 0.835–1.97, p = 0.256; Figure 2B). Lastly, we considered an expanded definition of progression that also included as progressors those patients who started HAART with CD4+ T-cell counts of less than 350/mm3. The follow-up was censored at HAART initiation for those patients who started treatment with CD4+ T cell counts greater than 350/mm3. Single or dual treatment with nucleoside reverse transcriptase inhibitors was included in the analysis as a time-updated covariate. This model again showed no effect of the DARC -46C/C genotype on disease progression (HR = 1.16, 95% CI 0.792–1.70, p=0.446; without correction for population stratification HR = 1.13, 95% CI 0.797–1.60, p = 0.496, Figure 2C).
Figure 2.
Survival curves for progression to AIDS. Kaplan Meier curves do not indicate that the DARC -46C/C genotype is associated with slower disease progression. This is true regardless of whether progression (A) is censored at HAART initiation, (B) includes HAART as a time-varying covariate or (C) includes patients with CD4+ T-cell counts less than 350/mm3 at the time of HAART initiation. At time 0, there are 332 patients with the C/C genotype and 121 patients with the C/T or T/T genotype.
The rate of CD4+ T-cell decline prior to HAART initiation was assessed as an additional biological marker of disease progression. CD4+ counts over time were considered for all samples with ≥3 pre-HAART CD4+ counts available. The average rate of CD4+ decline in these samples (N=263) was −5.10 cells per month. For patients with the -46C/C genotype the rate of CD4+ decline was −5.32 cells per month, and for all other patients it was −4.55 cells per month. Finally, an analysis using a mixed linear model, which included as covariates gender, age at seroconversion and the first EIGENSTRAT axis, failed to demonstrate a significant effect of genotype with respect to rate of CD4+ T-cell decline (p = 0.9359).
We also tested for an effect of -46C/C genotype on risk of HIV acquisition. The frequency of the -46C/C genotype was not significantly different between the HIV-infected and non-HIV infected African Americans in this study (70.7% and 68.3%, respectively, Table 1). We used a logistic regression model to test the association between -46C/C and HIV acquisition using gender and the first EIGENSTRAT axis as covariates. We found no association of the -46C/C genotype with HIV acquisition (OR 0.864, 95% CI 0.534–1.41, p = 0.555; without correction for population stratification OR = 1.05, 95% CI 0.685–1.63, p = 0.809). Assuming an odds ratio of 1.5, as was reported in He et al., we calculate that our study has 60% power to detect an effect of the DARC polymorphism at the 0.05 level. Our results, however, are not only not significant, but, when correcting for population stratification, they are in the opposite direction of those reported by He et al (OR<1.0). We used a simple simulation framework to test the probability of a lower allele frequency in HIV-positive samples compared to controls (that is, an OR < 1, opposite to the direction previously reported) assuming that the He et al. estimate of an odds ratio of 1.5 is correct. Specifically we simulated random sampling of 471 individuals assuming a base allele frequency of 0.70 for the risk genotype and compared this with 227 individuals sampled from a population with a base allele genotype of 0.60. After repeating this procedure 1 million times we found that the probability that the observed odds ratio would be below 1 was p < 0.01, indicating that the observed odds ratio of 0.864 is an unlikely outcome if the real effect of the variant is in the same direction and of similar magnitude to that reported in He et al.
Table 1.
Genotypes of the HIV+ and HIV− cohorts at DARC -46T→C
| HIV+ | Expected HIV+ | HIV− | Expected HIV− | |
|---|---|---|---|---|
| -46C/C | 333 | 322 | 157 | 155 |
| -46C/T | 113 | 135 | 62 | 65 |
| -46T/T | 25 | 14 | 8 | 7 |
| Total | 471 | 471 | 227 | 227 |
| %C/C | 70.7% | 68.3% | ||
| %C/T+T/T | 29.3% | 31.7% | ||
| F(C) | 0.827 | 0.828 | ||
| F(T) | 0.173 | 0.172 | ||
| HWE P-value | 0.073 | 0.934 | ||
Genotype at the DARC -46T→C does not violate HWE in either population. The low p-value in the HIV+ population is caused by an excess of both homozygous states, as opposed to a consistent overrepresentation of one allele as would be expected for a true risk allele.
Although the previous report identified the DARC -46C/C genotype as an important risk factor for HIV acquisition and disease progression, the work presented here, corrected for population stratification, does not replicate these findings. Whereas He et al. reported an overrepresentation of the DARC -46C/C genotype in an HIV+ population, we observed similar allele frequencies in the HIV+ and HIV− populations. It is possible that this result is indicative of the modest power of our acquisition study; however, it must be emphasized that in addition to a lack of effect on HIV acquisition, a well-powered analysis of disease course indicates trends in the opposite direction of those previously published. The cohort used in our study offers several advantages. For the disease progression analyses, our cohort is larger, includes members from all 3 U.S. military services (only 1 was evaluated in the previous report), and includes only subjects with estimated dates of seroconversion for more accurate time to event analyses. Therefore, while we cannot rule out the possibility that DARC -46C/C could be associated with faster time to death, it does not appear to be associated with slower progression to AIDS or with CD4 decline amongst African Americans.
Another possible explanation for the discrepant results relates to population stratification. Stratification due to population substructure can create spurious association between alleles and traits when both differ between subpopulations (Pritchard, 2000, Reich and Goldstein, 2001). Of particular concern, the strength of the stratification effect is known to increase sharply with the magnitude of the allele frequency difference between subpopulations. Thus, the DARC null allele would be expected to have a particularly large stratification effect associated with it in African American populations. He et al. reported the use of 11 markers to develop a model to predict ancestry and to control for the effects of stratification. It appears that He et al. used the probability of assignment of individuals to one of the two population groups (African American versus European American) directly as a covariate to control for population stratification. In addition to the fact that 11 markers are insufficient to accurately estimate ancestry and control for stratification, a model that predicts the probability of membership in one group versus another (African American versus European American) is not the same as a predictor of the degree of African versus European ancestry. The latter prediction is what is required for appropriate control of stratification. For these reasons He et al. did not implement appropriate stratification controls and it seems likely that some, or all, of their association signal may be due to stratification.
In conclusion, we have found no association between DARC genotype and progression to AIDS or risk for HIV acquisition. This highlights the importance of strict control for population stratification in genetic association studies.
Supplementary Material
Acknowledgments
Funding for this work was provided in part by the NIH-funded Center for HIV/AIDS Vaccine Immunology.
This project has also been funded in part with federal funds from the National Cancer Institute, National Institutes of Health, under Contract No. HHSN261200800001E. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government. This Research was supported in part by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research.
The Tri-Service AIDS Clinical Consortium (TACC) is a component of the Infectious Disease Clinical Research Program (IDCRP) of the Uniformed Services University of the Health Sciences (USUHS). The IDCRP is a Department of Defense tri-service program executed through USUHS and the Henry M. Jackson Foundation for the Advancement of Military Medicine (HJF), in collaboration with HHS/NIH/NIAID/DCR through Interagency Agreement HU0001-05-2-0011. The centers (investigators) participating in this work include: Uniformed Services University of the Health Sciences (Scott Wegner), Walter Reed Army Medical Center (Amy Weintrob and Glenn Wortmann), National Naval Medical Center (Aunradha Ganesan and Timothy Whitman), Naval Medical Center Portsmouth (Vince Barthel), San Antonio Military Medical Center, Naval Medical Center San Diego (Nancy Crum-Cianflone and Braden Hale), and Tripler Army Medical Center (Tomas Ferguson). The opinions or assertions contained herein are the private views of the authors, and are not to be construed as official, or as reflecting the views of the Departments of the Army, Navy, Air Force, or the Department of Defense.
Footnotes
The opinions or assertions contained herein are the private views of the authors, and are not to be construed as official, or as reflecting the views of the Departments of the Army, Navy, Air Force, or the Department of Defense.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- FELLAY J, SHIANNA KV, GE D, COLOMBO S, LEDERGERBER B, WEALE M, ZHANG K, GUMBS C, CASTAGNA A, COSSARIZZA A, COZZI-LEPRI A, DE LUCA A, EASTERBROOK P, FRANCIOLI P, MALLAL S, MARTINEZ-PICADO J, MIRO JM, OBEL N, SMITH JP, WYNIGER J, DESCOMBES P, ANTONARAKIS SE, LETVIN NL, MCMICHAEL AJ, HAYNES BF, TELENTI A, GOLDSTEIN DB. A whole-genome association study of major determinants for host control of HIV-1. Science. 2007;317:944–7. doi: 10.1126/science.1143767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HE W, NEIL S, KULKARNI H, WRIGHT E, AGAN BK, MARCONI VC, DOLAN MJ, WEISS RA, AHUJA SK. Duffy antigen receptor for chemokines mediates trans-infection of HIV-1 from red blood cells to target cells and affects HIV-AIDS susceptibility. Cell Host Microbe. 2008;4:52–62. doi: 10.1016/j.chom.2008.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MCCARTHY MI, ABECASIS GR, CARDON LR, GOLDSTEIN DB, LITTLE J, IOANNIDIS JP, HIRSCHHORN JN. Genome-wide association studies for complex traits: consensus, uncertainty and challenges. Nat Rev Genet. 2008;9:356–69. doi: 10.1038/nrg2344. [DOI] [PubMed] [Google Scholar]
- PRITCHARD JK, STEPHENS M, ROSENBERG NA, DONNELLY P. Association mapping in structured populations. Am J Hum Genet. 2000;67:170–81. doi: 10.1086/302959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PRICE AL, PATTERSON NJ, PLENGE RM, WEINBLATT ME, SHADICK NA, REICH D. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38:904–9. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- REICH DE, GOLDSTEIN DB. Detecting association in a case-control study while correcting for population stratification. Genet Epidemiol. 2001;20:4–16. doi: 10.1002/1098-2272(200101)20:1<4::AID-GEPI2>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.