Abstract
Two novel antibiotics, neocitreamicins I and II, were isolated from a fermentation broth of a Nocardia strain. This producing strain was obtained using an in situ diffusion chamber that facilitates the cultivation of soil microorganisms. The structures of neocitreamicins I and II were elucidated spectroscopically using UV, MS, and NMR data, and found to be related to the polycyclic xanthone antibiotics of the citreamicin class. The neocitreamicins showed in vitro activity against Gram-positive bacteria including strains of methicillin-resistant Staphylococcus aureus and vancomycin-resistant Enterococcus faecalis.
Keywords: Neocitreamicin, Citreamicin, MRSA, VRE, Nocardia
Introduction
The citreamicin group of antibiotics, are members of the polycyclic xanthones, an antibiotic class that was first reported in 1989.1-3 Other structurally related polycyclic xanthone compounds include cervinomycin and simaomicin.4-7 The citreamicins were shown to have potent antibiotic activity against a variety of Gram-positive bacteria including strains of methicillin-resistant Staphylococcus aureus (MRSA) and vancomycin-resistant Enterococcus faecalis (VRE). 2 In particular, citreamicin ζ was also demonstrated to be effective in vivo at preventing Streptococcus pyogenes infection.1
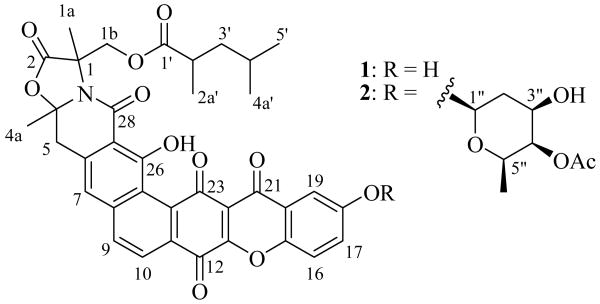
In an effort to meet the challenge of the rapid emergence of bacterial drug resistance, a collection of microorganisms was screened for the production of novel antibiotics. Each of these microorganisms was originally isolated using our proprietary in situ diffusion chamber technology.8 A novel Nocardia strain (G0655) isolated from a sandy soil sample collected in Falmouth, Massachusetts (USA) was found to produce a series of compounds with potent antibiotic activity. The two major components belong to the citreamicin group of antibiotics with intriguingly different structural moieties. In this paper we describe the producing strain and the fermentation, isolation, structure elucidation, and biological activities of neocitreamicins I (1) and II (2) (Fig. 1), the first reported additions to this class of antibiotics since their initial discovery almost two decades ago.
Fig. 1.
Structures of neocitreamicins I (1) and II (2).
Experimental
General Experimental Procedures
All NMR spectra were taken on a Bruker-DRX-500 spectrometer equipped with a 5 mm QNP probe. High resolution ESI-LC-MS data were recorded on a MicroMass Q-Tof-2 spectrometer equipped with an Agilent 1100 solvent delivery system and an online diode array detector using a Phenomenex Gemini-C18 reversed phase column (50 × 2.0 mm, 3 μm particle size). Elution was performed with a linear gradient using deionized water with 0.1% formic acid and acetonitrile with 0.1% formic acid as solvents A and B, respectively, at a flow rate of 0.2 ml/min. The gradient increased from 10% to 100% of solvent B over 20 minutes followed by an isocratic elution at 100% of solvent B for 8 minutes. Analytical and semi-preparative chromatography was performed on a Zorbax SB-C18 Reversed Phase column (250 × 9.4 mm, 5 μm particle size) using a Shimadzu SCL-10AVP HPLC system including an SPD-M10AVP diode array detector set at 254 nm. Elution was performed with a linear gradient using deionized water with 0.1% trifluoroacetic acid and acetonitrile with 0.1% trifluoroacetic acid as solvents A and B, respectively, at a flow rate of 3 ml/min. The gradient increased from 50% to 85% of solvent B over 30 minutes followed by an isocratic elution at 95% of solvent B for 8 minutes.
16S rDNA Sequence Analysis
16S rDNA sequence analysis was utilized to determine the taxonomic identity of isolate G0655. Chromosomal DNA was isolated from approximately 106 cells after a 5 minute vigorous agitation in the presence of 50 mg of glass beads and 100 μl of H2O in a 0.5 ml Eppendorf tube. PCR amplification of nucleotide bases 20 through 710 of the gene encoding the 16S rRNA was carried out using G0655 chromosomal DNA, GoTaq Green Master Mix (Promega M7122), and universal primers Bac8F and 1492R.9 PCR thermocycler parameters included 30 cycles of 95 °C for 30 s, 45 °C for 30 s, and 72 °C for 105 s. The amplified DNA fragment was sequenced by Macrogen (Rockville, MD) using primer 782R and compared by BLAST alignment to the GenBank nucleotide collection.
Phenotypic Analysis
Phenotypic characterization of isolate G0655 was performed using standard ISP media 1 through 410, as well as cornmeal agar11, potato dextrose agar11, R4 agar (10 g/l glucose, 1 g/l yeast extract, 0.1 g/l casamino acids, 3 g/l L-proline, 10 g/l MgCl2·6H2O, 4 g/l CaCl2·2H2O, 0.2 g/l K2SO4, 5.6 g/l TES, 1 ml Trace Metal Solution, 15 g/l agar) and SMS agar (0.125 g/l casein, 0.1 g/l potato starch, 1 g/l casamino acids, 15 g/l agar). Isolate G0655 was streaked onto the surface of the solid media, incubated at 28°C for 7 days, and observed to generate the descriptions in Table 1.
Table 1.
Cultural characteristics of strain G0655
| Medium | Descriptionab | |
|---|---|---|
| Tryptone-yeast extract agar (ISP-1) |
G: | Poor, flat, white |
| A: | Poor, white | |
| R: | Light yellow (241, 236, 126) | |
| S: | None | |
| Yeast extract–malt extract agar (ISP-2) |
G: | Abundant, raised, beige (231, 217, 135) |
| A: | Good, velvety, yellowish | |
| R: | Orange brown (174, 132, 32) | |
| S: | None | |
| Oatmeal agar (ISP-3) |
G: | Moderate, flat, white |
| A: | Poor, white | |
| R: | White | |
| S: | None | |
| Inorganic salts-starch agar (ISP-4) |
G: | Poor, flat, white |
| A: | Poor | |
| R: | White | |
| S: | None | |
| Cornmeal agar | G: | Abundant, raised, white |
| A: | Good, velvety, white | |
| R: | Orange brown (204, 123, 64) | |
| S: | None | |
| Potato Dextrose agar | G: | Moderate, flat, white |
| A: | Poor, white | |
| R: | Light brown (240, 204, 128) | |
| S: | None | |
| R4 agar | G: | Abundant, raised, light beige (253, 251, 170) |
| A: | Good, velvety, yellowish | |
| R: | Dark brown (129, 109, 41) | |
| S: | None | |
| SMS agar | G: | Abundant, raised, white |
| A: | Good, velvety, white | |
| R: | Orange brown (177, 150, 56) | |
| S: | None | |
G: growth, A: aerial mycelium, R: reverse, S: soluble pigment.
For accurate color depiction use Microsoft Windows color palette and enter the 3 listed values in the order given for red, green and blue.
Fermentation
A colony of isolate G0655 was homogenized using a sterile pestle in a 1.5 ml Eppendorf tube containing 200 μl sterile water. The bacterial homogenate was transferred to a 250 ml Erlenmeyer flask containing 40 ml of seed broth (1.5% glucose, 1.5% glycerol, 1.5% malt extract, 2.5% yeast extract, 0.5% casamino acids, and 0.1% calcium carbonate). The seed broth was incubated for 4 days at 28 °C on a rotary shaker (1 inch throw, 200 rpm) prior to production media inoculation at 2.5% (v/v). Production was performed in 500 ml aliquots of medium R4 (1.0% glucose, 0.1% yeast extract, 0.01% casamino acids, 0.3% L-proline, 1.0% MgCl2·6H2O, 0.02% K2SO4, 0.56% N-Tris-hydroxymethyl-methyl-2-aminoethanesulfonic acid) in 2 liter tri-baffled flasks at 28 °C on a rotary shaker (2.5 inch throw, 200 rpm). After 2 days of incubation, 2 g of sterile Diaion HP-20 resin (Supelco) was added to each flask. Fermentations were continued for an additional 7 days prior to harvest.
Isolation
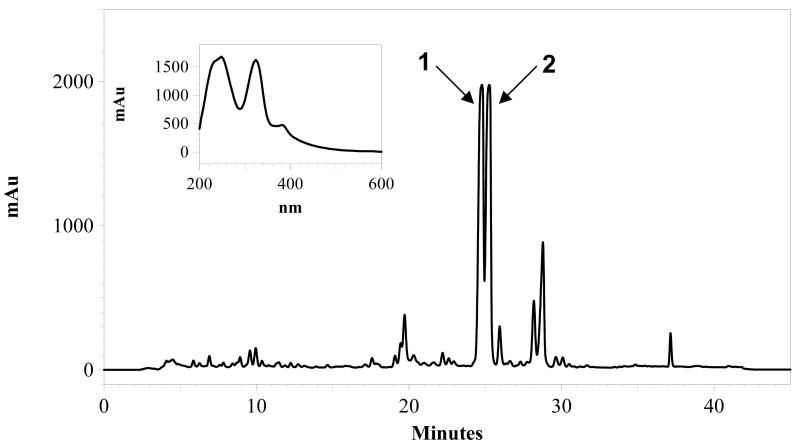
The bacterial cell mass and HP-20 resin from 5 liters of fermentation broth were collected by filtration. The mixture was washed with 500 ml of 20% acetone in water and then eluted with 500 ml 100% acetone. The 20% acetone wash was discarded and the 100% acetone eluent was evaporated to dryness, leaving a brown solid which was extracted with hexanes (2 × 40 ml). The suspension was centrifuged at 1660 g at 4 °C for 10 minutes. The hexanes extract was discarded and the remaining reddish brown pellet was dried overnight. The solid was first dissolved in 30 ml of methanol and then 70 ml of water was added, resulting in some precipitation. The suspension was adsorbed onto an Alltech Extract-Clean SPE C18-HC (10 g) cartridge. The cartridge was then sequentially washed with 40 mL portions of 20%, 30%, 50%, 70%, 90% and 100% of methanol (balance water). The 100% methanol portion contained the majority of the active components and appeared deep red in color. This solution was then injected in multiple 500 μL runs onto a semi-preparative HPLC for purification as described above. Neocitreamicins I (1) and II (2) eluted at 24.7 and 25.3 minutes, respectively. The corresponding HPLC fractions were pooled and lyophilized after removal of acetonitrile under reduced pressure to afford 1 (6 mg) and 2 (5 mg) as yellow powders.
Biological Activity Assays
Minimal inhibitory concentrations of neocitreamicins against a set of bacterial strains were determined in liquid growth medium. An 80 μg/ml stock solution of a test compound in DMSO was two-fold serially diluted with DMSO. In a 96-well microtiter plate, 10 μl of each compound solution was mixed with 90 μl of a log-phase growth cells at a dilution of OD600nm = 0.001 in Mueller-Hinton Broth or Brain Hearth infusion Broth (Enterococcus strains only). This achieved final concentrations of the test compounds from 8 μg/ml to 0.06 μg/ml. After 18 hours of incubation at 37 °C, minimal inhibitory concentrations were determined by visual inspection.
Bacterial Strains
The following isolates were obtained through the Network on Antimicrobial Resistance in Staphylococcus aureus (NARSA) Program : NRS1, NRS2, NRS71, supported under NIAID/NIH Contract No. HHSN272200700055C. Isolate G0655 has been deposited in the United States Department of Agriculture's NRRL culture collection and assigned the accession number NRRL 50106.
Results and Discussion
Taxonomy and Antibacterial Screening
The antibiotic producing strain G0655 was isolated from a sandy soil sample collected in Falmouth, Massachusetts (USA) using our proprietary in situ diffusion chamber technology.12 This technology facilitates the isolation of microorganisms by permitting them access to the chemical background present in their native soil environment. When growing on several standard laboratory media used for isolation and identification of environmental microorganisms, strain G0655 shares many morphological characteristics with members of the actinobacteria, including the ability to sporulate, the production of soluble pigments, and the formation of firm (non-mucoid) mycellial colonies (Table 1).
The 16S rRNA gene sequence similarity studies (first half of the gene, from base pair 20 to base pair 710) using nucleotide sequence comparison (BLAST) to the GenBank database preliminarily placed isolate G0655 within the genus Nocardia. The three most closely related species returned by this search were Nocardia acidovorans (GenBank accession no. AM402972; 98.7%), Nocardia nova (GenBank accession no. AB162790; 97.8%), and Nocardia pseudobrasiliensis (GenBank accession no. AF430042; 97.8%).
Isolate G0655 was screened for antibiotic production in a series of fermentation media by removal of aliquots of the crude broths after 4 and 14 days of growth. The samples were concentrated to dryness and reconstituted in DMSO. These DMSO extracts were applied onto the surface of actively growing lawns of Bacillus subtilis. After an overnight incubation at 37 °C, antibiotic activity was scored by measuring the zone of inhibition of growth for each extract. Antibiotic activity was detected at the 14 day time point.
Antibacterial activities of 1 and 2 were determined by serial dilution method in liquid media (Table 2). Both compounds showed good antibacterial activity against Gram-positive bacteria, including methicillin-resistant Staphylococcus aureus and vancomycin-resistant Enterococcus faecalis. This compound showed little or no activity against Gram-negative bacteria.
Table 2.
Antimicrobial activities of 1 and 2
| Microbroth MIC (μg/ml | ||
|---|---|---|
| Test Organism | 1 | 2 |
| Bacillus subtilis 1A1 | 0.06 | 0.12 |
| Staphylococcus aureus (MRSA NRS1) | 0.50 | 1.0 |
| S. aureus (MRSA NRS2) | 0.12 | 0.50 |
| S. aureus (MRSA NRS71) | 0.12 | 0.50 |
| Enterococcus faecalis (VRE 51299) | 0.06 | 0.06 |
| E. faecalis (VRE 51575) | 0.12 | 0.25 |
| Escherichia coli K-12 | >8.0 | >8.0 |
Neocitreamicin Isolation and Structural Determination
The fermentation was scaled up to a 5 liter volume to facilitate structure elucidation studies. Neocitreamicins I (1) and II (2) were isolated from the production fermentation broth by solid phase extraction and reversed phase chromatography (Fig. 2). The molecular formula of 1 was determined as C36H31NO11 on the basis of positive ion HR ESI-MS ([M+H]+ m/z 654.1993, calcd. for 654.1975) and NMR spectra data, indicating the presence of 22 double bond equivalents. Similarly, the molecular formula of 2 was established as C44H43NO15 ([M+H]+ m/z 826.2714, calcd. for 826.2711), indicating the presence of 24 double bond equivalents. The UV spectra of 1 (Fig. 2) and 2 are virtually identical to those reported for the original citreamicins.2 The structure of 1 was elucidated by interpretation of 2D NMR spectra (COSY, HSQC, and HMBC) and comparison with reported citreamicin data. The 1H and 13C NMR signals of 1 are in good accordance to those of citreamicins for the common moieties of the structure. In contrast to citreamicins, the 1H NMR spectrum of 1 lacks any signals of methoxy groups on the G ring. A COSY experiment revealed a meta coupling (3.0 Hz) between H-19 and H-17 as well as a vicinal coupling between H-16 and H-17. Thus, the G ring of 1 is less substituted when compared to citreamicins. The proton chemical shift at 7.53 ppm (H-19) helped to place the hydroxyl group at C-18 instead of C-17. Locating the hydroxyl group at C-17 should result in H-16 to resonate further upfield than 7.53 ppm. The 2,4-dimethylvalerate side chain was determined using data from COSY and HMBC experiments. Strong HMBC correlations were observed between all the three methyl groups and the C-3′ methylene. The attachment of this side chain to position C-1b was confirmed by an HMBC correlation between H-1b and the carbonyl carbon (C-1′) of the side chain. To our best knowledge, the only other natural products with the 2,4-dimethylvalerate units belong to the milbemycins.13-14
Fig 2.
HPLC analysis of a solid-phase extract of the Nocardia strain G0655 monitored at 254 nm, and UV-visible spectrum of 1.
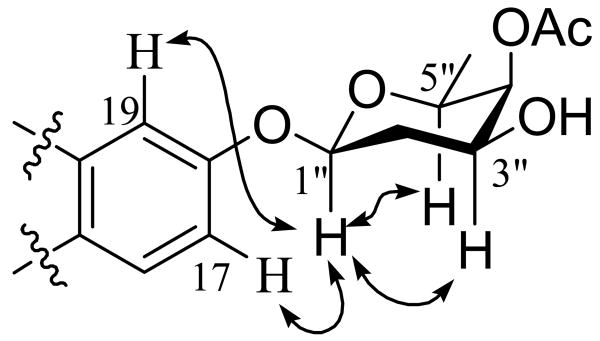
Neocitreamicin II (2) was found to be a derivative of 1 by comparing its NMR spectra (1H, 13C, COSY, HSQC, and HMBC) to those of 1. The 2,4-dimethylvalerate side chain was readily confirmed in this case through a COSY experiment as well as 23 HMBC correlations (Table 3). An HMBC correlation was also observed between H-19 and the C-21 carbonyl carbon, indicating the G ring substitution is at C-18 instead of C-17. The remaining 1H and 13C NMR signals of 2 were easily attributed to a deoxysugar moiety through COSY, HSQC, and HMBC correlations. A clear HMBC correlation between H-4″ and the carbonyl carbon of the acetyl group indicates that the deoxysugar is acetylated at 4″ position, which is also consistent with the downfield chemical shift of H-4″ (4.94 ppm). Although no HMBC correlation was observed between the anomeric proton and C-18, a ROESY experiment showed correlations from the anomeric proton to H-17 and H-19, supporting the deoxysugar is attached to position 18 (Fig. 3). The anomeric proton occupies an axial position as indicated by the large coupling constant (J = 9.7 Hz). ROESY correlations were also observed between H-1″ and H-3″ and between H-1″ and H-5″, indicating the hydroxyl and the methyl groups are in equatorial positions. Furthermore, because only a small coupling constant (2.9 Hz) was observed for H-4″, the acetoxy group must occupy an axial position. Thus, the deoxysugar moiety was determined to be an acetylated oliose derivative. Two additional stereocenters are located on the A ring of both neocitreamicins and remain undefined despite attempts to resolve them by ROESY. The stereochemistry at these positions in the original citreamicin compounds were also left undefined.2
Table 3.
1H and 13C NMR data of 1 and 2 (500 and 125 MHz, δ in ppm, J in Hz)
| 1 (CD3OD) | 2 (DMSO-dδ) | |||||
|---|---|---|---|---|---|---|
| No. | δCa | δH | HMBC | δCa | δH | HMBC |
| 1 | 63.8 | 62.1 | ||||
| 1a | 20.3 | 1.86 (3H, s) | C-1, 1b, 2 | 19.1 | 1.81 (3H, s) | 1, 1b, 2 |
| 1b | 66.5 | 4.79 (1H, d, 11.6) | C-1, 2 | 64.9 | 4.66 (1H, d, 11.6) | 1, 2 |
| 4.45 (1H, d, 11.6) | C-1, 1′, 2 | 4.40 (1H, d, 11.6) | ||||
| 2 | 173.0 | 171.3 | ||||
| 4 | 95.1 | 93.8 | ||||
| 4a | 26.2 | 1.77 (3H, s) | C-4, 5 | 25.4 | 1.71 (3H, s) | 4, 5 |
| 5 | 42.1 | 3.62 (1H, d, 14.8) | C-4, 6, 7, 27 | 39.8 | 3.74 (1H, d, 14.8) | 4, 6, 7, 27 |
| 3.52 (1H, d, 14.8) | 3.66 (1H, d, 14.8) | 4, 6 | ||||
| 6 | 136.8 | 135.4 | ||||
| 7 | 119.3 | 7.38 (1H, br s) | C-25, 27 | 118.0 | 7.51 (1H, br s) | 5, 25, 27 |
| 8 | 138.4 | 136.7 | ||||
| 9 | 133.8 | 8.07 (1H, d, 8.5) | C-7, 25 | 132.5 | 8.19 (1H, AB) | 11, 12, 24 |
| 10 | 125.3 | 8.21 (1H, d, 8.5) | C-8, 12, 24 | 124.3 | 8.21 (1H, AB) | |
| 11 | 131.8 | 130.3 | ||||
| 12 | 179.1 | 177.6 | ||||
| 13 | 156.4 | 155.1 | ||||
| 15 | 157.8 | 154.6 | ||||
| 16 | 121.8 | 7.67 (1H, d, 9.0) | C-15, 18, 20 | 120.9 | 7.81 (1H, d, 9.0) | 15, 18, 20 |
| 17 | 125.9 | 7.36 (1H, dd, 9.0, 3.0) | C-18 | 125.3 | 7.58 (1H, dd, 9.0, 3.0) | 18 |
| 18 | 150.3 | 149.5 | ||||
| 18-OH | exchanged | exchanged | ||||
| 19 | 109.7 | 7.53 (1H, d, 3.0) | C-17, 18 | 109.5 | 7.65 (1H, d, 3.0) | 18, 21 |
| 20 | 127.8 | 126.0 | ||||
| 21 | 175.7 | 173.1 | ||||
| 22 | 120.4 | 119.3 | ||||
| 23 | 183.2 | 180.5 | ||||
| 24 | 142.2 | 140.2 | ||||
| 25 | 120.7 | 118.6 | ||||
| 26 | 162.9 | 161.1 | ||||
| 26-OH | exchanged | 13.41 (1H, s) | 25, 26, 27 | |||
| 27 | 108.6 | 106.9 | ||||
| 28 | 167.7 | 166.1 | ||||
| 1′ | 177.5 | 175.4 | ||||
| 2′ | 38.8 | 2.56 (1H, m) | 36.7 | 2.51 (under DMSO) | 1′, 2a', 3′ | |
| 2a' | 17.7 | 1.12 (3H, d, 7.0) | C-3′ | 16.9 | 1.05 (3H, d, 6.8) | 1′, 2′, 3′ |
| 3′ | 44.5 | 1.53-1.61 (2H, m) | 42.7 | 1.46 (1H, m) | 1′, 2′, 2a', 4′, 4a' | |
| 1.21-1.30 (1H, m) | 1.18-1.25 (1H, m) | 2′, 2a', 4a' | ||||
| 4′ | 27.1 | 1.53-1.61 (2H, m) | 25.3 | 1.52 (1H, m) | 4a' | |
| 4a' | 23.0 | 0.87 (3H, d, 6.3) | C-3′, 4′, 5′ | 22.3 | 0.81 (3H, d, 6.3) | 3′, 4′, 4a', 5′ |
| 5′ | 22.7 | 0.91 (3H, d, 6.3) | C-3′, 4′, 4a' | 22.2 | 0.85 (3H, d, 6.3) | 3′, 4′, 4a', 5′ |
| 1″ | 96.8 | 5.49 (1H, dd, 9.8, 2.0) | ||||
| 2″ | 34.5 | 2.02-2.07 (1H, m) | ||||
| 1.81 (1H, m) | ||||||
| 3″ | 65.6 | 3.96 (1H, m) | 4″, 5″ | |||
| 4″ | 71.3 | 4.94 (1H, d, 2.9) | 3″, 1-Ac | |||
| 5″ | 69.1 | 3.99 (1H, m) | 6″ | |||
| 6″ | 16.5 | 1.07 (3H, d, 6.4) | 4″, 5″ | |||
| 1-Ac | 170.3 | |||||
| 2-Ac | 20.8 | 2.11 (3H, s) | 1-Ac | |||
Assignments confirmed by HSQC correlations
Fig. 3.
Selected ROESY correlations for 2.
Summary
In summary, we have described two new members of the citreamicin group of antibiotics. It is important to note that all previously reported citreamicins bear a methoxy group at the 17 position. In contrast, the neocitreamicins lack any substitution at the 17 position. While position 18 is occupied by a free hydroxyl group in 1, it was glycosylated in 2. The fact that 2 shows similar antibacterial activity suggests that the G ring end of the structure can be modified without loss of activity. The free hydroxyl group at position 18 makes 1 more amenable to medicinal chemistry efforts for the improvement of drug properties.
Acknowledgments
This work was supported by grant 2 R44 AI063616-03 from the National Institute of Health of the United States of America. The authors would like to recognize and thank Dr. Kim Lewis and Dr. Slava S. Epstein of Northeastern University Department of Biological Sciences for their contributions to this work.
References
- 1.Maiese WM, Lechevalier MP, Lechevalier HA, Korshalla J, Goodman J, Wildey MJ, Kuck N, Greenstein M. LL-E19085 alpha, a novel antibiotic from Micromonospora citrea: taxonomy, fermentation and biological activity. J Antibiot. 1989;42:846–851. doi: 10.7164/antibiotics.42.846. [DOI] [PubMed] [Google Scholar]
- 2.Carter GT, Nietsche JA, Williams DR, Borders DB. Citreamicins, novel antibiotics from Micromonospora citrea: isolation, characterization, and structure determination. J Antibiot. 1990;43:504–512. doi: 10.7164/antibiotics.43.504. [DOI] [PubMed] [Google Scholar]
- 3.Pearce CJ, Carter GT, Nietsche JA, Borders DB, Greenstein M, Maiese WM. The effect of methylation inhibitors on citreamicin biosynthesis in Micromonospora citrea. J Antibiot. 1991;44:1247–1251. doi: 10.7164/antibiotics.44.1247. [DOI] [PubMed] [Google Scholar]
- 4.Omura S, Iwai Y, Hinotozawa K, Takahashi Y, Kato J, Nakagawa A, Hirano A, Shimizu H, Haneda K. Cervinomycin A1 and A2, new antibiotics active against anaerobes, produced by Streptomyces cervinus sp nov. J Antibiot. 1982;35:645–652. doi: 10.7164/antibiotics.35.645. [DOI] [PubMed] [Google Scholar]
- 5.Lee T, Carter G, Borders DB. Structure determination of simaomicin α and β, extremely potent, novel anticoccidial agents produced by Actinomadura. J Chem Soc Commun. 1989;1989:1771–1772. [Google Scholar]
- 6.Kobayashi K, Nishino C, Ohya J, Sato S, Mikawa T, Shiobara Y, Kodama M. Actinoplanones A and B, new cytotoxic polycyclic xanthones from Actinoplanes sp. J Antibiot. 1988;41:502–11. doi: 10.7164/antibiotics.41.502. [DOI] [PubMed] [Google Scholar]
- 7.Kobayashi K, Nishino C, Ohya J, Sato S, Mikawa T, Shiobara Y, Kodama M. Actinoplanones C, D, E, F, and G, new cytotoxic polycyclic xanthones from Actinoplanes sp. J Antibiot. 1988;41:502–511. doi: 10.7164/antibiotics.41.502. [DOI] [PubMed] [Google Scholar]
- 8.Kaeberlein T, Lewis K, Epstein SS. Isolating “uncultivable” microorganisms in pure culture in a simulated natural environment. Science. 2002;296:1127–1129. doi: 10.1126/science.1070633. [DOI] [PubMed] [Google Scholar]
- 9.Baker GC, Smith JJ, Cowan DA. Review and re-analysis of domain-specific 16S primers. J Microbiol Meth. 2203;55:541–555. doi: 10.1016/j.mimet.2003.08.009. [DOI] [PubMed] [Google Scholar]
- 10.Shirling EB, Gottlieb D. Methods for characterizing Streptomyces species. Int J Syst Bacteriol. 16:313–340. [Google Scholar]
- 11.Atlas RM. In: Handbook of Microbiological Media. Parks LC, editor. CRC Press; Boca Raton: 1997. [Google Scholar]
- 12.Lewis K, Epstein SS, Kaeberlein S. Isolation and cultivation of microorganisms from natural environments and drug discovery based thereon. United States of America patent US 7011957 B2. 2006
- 13.Blanchflower SE, Dorgan RJJ, Everett JR, Readshaw SA. An investigation of the stereochemistry of a 22,23-dioxygenated milbemycin. Tetrahedron Lett. 1988;29:6645–6648. [Google Scholar]
- 14.Goegelman RT, Munguira E, Diez MMT, Sykes RS, Kong YL, Liesch JM, Helms GL, Turner JET. Anthelmintic milbemycin analogs of novel microorganisms. United States of America patent US 5290804 A. 1994