Abstract
Meru, Kenya has watersheds which are shared by wildlife, humans and domesticated animals. These surface waters can be contaminated by the waterborne pathogen Cryptosporidium. To quantify the seasonality and prevalence of Cryptosporidium in Meru regional surface waters, we used a calcium carbonate flocculation (CCF) and sucrose floatation method, and a filtration and immunomagnetic bead separation method, each of which used PCR for Cryptosporidium detection and genotyping. Monthly water samples were collected from January through June in 2003 and 2004, bracketing two April-May rainy seasons. We detected significant seasonality with 8 of 9 positive samples from May and June (p < 0.0014), which followed peak rainy season precipitation and includes some of the subsequent dry season. Six of 9 positive samples revealed C. parvum, and 3 contained C. andersoni. None contained C. hominis. Our results indicate that Meru surface waters are Cryptosporidium-contaminated at the end of rainy seasons, consistent with the timing of human infections reported by others from East Africa and contrasting with the onset of rainy season peak incidence reported from West Africa.
Keywords: calcium carbonate flocculation, Cryptosporidium, Kenya, PCR detection, seasonality
INTRODUCTION
Cryptosporidium is a protozoan parasite which usually causes asymptomatic or mild human clinical disease, although severe dehydrating disease and death can occur in immunocompromised populations, such as HIV-infected people. No clearly effective therapy for cryptosporidiosis in immunocompromised people exists (Griffiths 1998; Tzipori & Ward 2002; Abubakar et al. 2007), and thus the avoidance of Cryptosporidium-contaminated water is important. Zoonotic and human-to-human transmission occurs, often via surface and recreational waters, and the disease is under-diagnosed except in those with persistent diarrhea (Bagley et al. 1998; Griffiths 1998; Hunter et al. 2001; Rose et al. 2002). Prevalence rates of ~ 10% to 33% in those with AIDS have been reported from developing countries (Kaplan et al. 1996; reviewed in Griffiths 1998). Malnutrition, which impairs cellular immunity, is another recognized risk factor for cryptosporidiosis (Gendrel et al. 2003).
We report initial data on Cryptosporidium in surface waters of Meru, Kenya, which lies north of Nairobi. Watersheds in Meru arise on Mt. Kenya in wildlife preserves, and water flows east through intensively farmed, densely populated areas before reaching arid, pastoral areas. Thus, Meru surface waters could be contaminated by parasite oocysts from wildlife, human, or pastoral animals, and infect people with AIDS or malnutrition as well as others in the general population. In this pilot study, we repetitively collected monthly samples from predetermined sites during January to June, which is before, during, and after the intensively rainy period of April and May. We also field tested two methods for parasite isolation and detection from surface waters during 2003 and 2004. Our approach included Cryptosporidium molecular genotyping of the positive water samples so as to better understand the potential sources of the parasites.
MATERIALS AND METHODS
Water collection sites
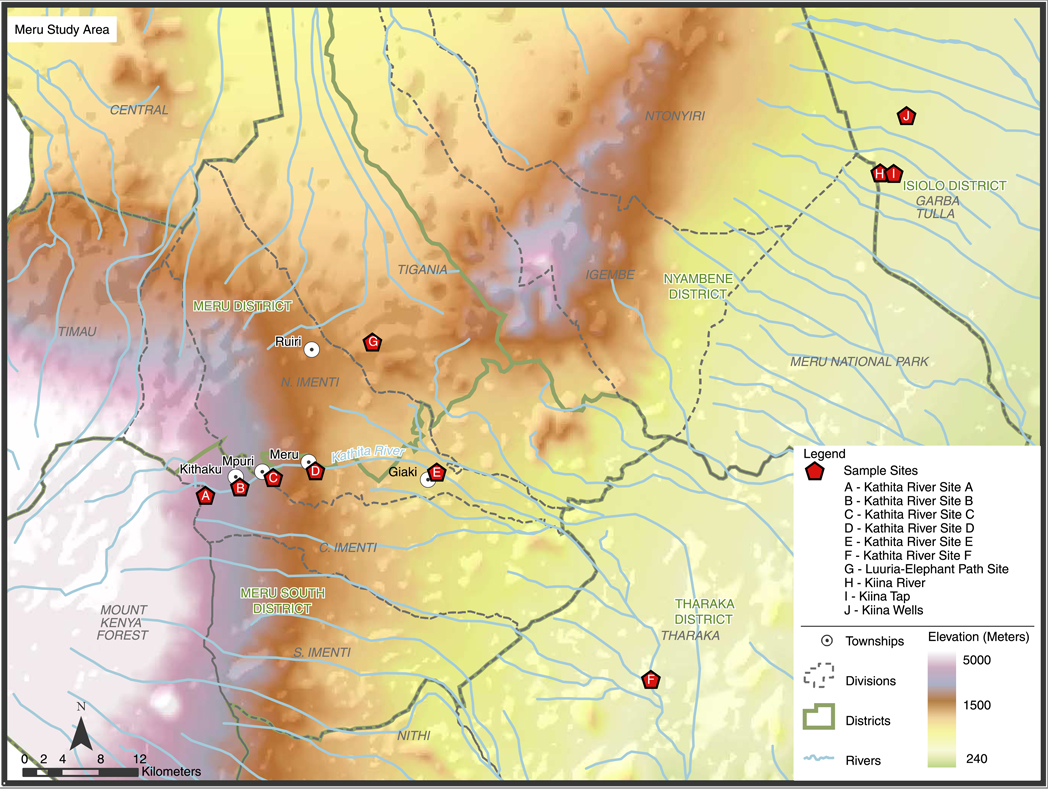
Meru water supplies are untreated surface waters taken directly from rivers or transient, seasonally flooded areas, which may be piped to households or common urban water distribution points. Water collection sites are often common watering points for domestic and wild animals. Ten sites were sampled. Five sites were on the Kathita River, which supplies Meru Town and surrounding villages. The highest Kathita River site (site A, Figure 1) is where the river exits a protected wildlife forest preserve. Downstream sites, coded B, C, E, and F, reflect a graded transition of increasing human and animal influence. Site C is the intake for the Meru Town municipal water system, which is chlorinated but unfiltered. Site D is the sewage outflow from Meru Town. Four other sites were chosen because of mixed human and animal use. These included a forest pool where elephants drink (Luuria, site G), and two sites near Kiina Town northeast of Meru Town. Kiina Tap (H) and Kiina River (I) are used by pastoralists for their herded animals, and by wildlife such as elephants, as they border Meru National Park. Miriti Well (site J), one of a group known as the Kiina Wells, is further northeast and is used by both pastoralists and wildlife. Sites H – J are in Isiolo District, and are in the far eastern, lower parts of the Meru ecosystem. Sites were mapped using a global positioning satellite (GPS) monitor (Figure 1). Under ideal conditions, all sites could be visited in a 3 day period excepting Miriti Well which required a fourth day.
Figure 1.
Map of the Meru, Kenya Region.
Sampling framework and precipitation data
Samples were obtained during the first six months of 2003 and 2004, bracketing a major precipitation period which occurs in March and April. This period was chosen so that we could analyze 2003 samples and revise our 2004 sampling strategy if required. Precipitation data was obtained from the Meteorology Department, Ministry of Transport and Communication, Meru Weather Station, housed at Kenya Methodist University (KEMU) in Meru.
Sampling methods
We used a calcium carbonate flocculation (CCF) and sucrose gradient floatation technique in 2003, and a filtration method in 2004, each followed by immunomagnetic isolation of parasite oocysts and PCR analysis. Prior work (Kato et al. 2003) documented that the CCF method could successfully isolate oocysts from Meru waters.
Calcium carbonate flocculation (CCF) followed by sucrose gradient floatation
Ten litres of water were collected into plastic containers certified as free from contaminants (Fisher Scientific, Pittsburgh, USA). Samples were brought to KEMU, saturated with CaCl2 and NaHCO3, and adjusted to pH 10 with NaOH (Vesey et al. 1993). Insoluble CaCO3 crystals were allowed to settle and the supernatant (~9 L) discarded. The precipitate was dissolved in sulfamic acid. K2Cr2O7 (25 g per L) was added to kill pathogens until processing, which was typically within a week. Samples were centrifuged at 5,000 × g for 10 minutes, and the supernatant was discarded leaving ~20 ml of concentrated suspension. The suspension was thrice washed with distilled water at 5,000 × g for 10 min at 4°C with supernatant removal via vacuum leaving 3 ml of suspension and sediment. The washed sediment then had distilled water added to a total volume of 7 ml and was thoroughly mixed. Seven ml of sucrose solution (specific gravity 1.18) was layered below the sediment solution via syringe, and centrifuged at 3,000 × g for 20 minutes at 4°C in a 15 ml polystyrene tube. Two separate layers were obtained, with soil sediments at the bottom. The top 2 ml of the sucrose layer was removed using a Pasteur pipette, diluted with water in another 15 ml tube, and centrifuged at 1,800 × g for 3 min at 4°C. The supernatant was aspirated leaving 500 µl. This was repeated 3 times leaving a final volume of 500 µl which was subjected to immunomagnetic separation (IMS).
Filtration
For consistency between Boston and Meru, we used US Environmental Protection Agency (EPA) Method 1623, as our Boston laboratory is required by the EPA to consistently only use Method 1623, given its certification for Cryptosporidium and Giardia testing in US ambient surface waters (US EPA 2005). In Kenya, water samples were pumped through a Filta-Max™ (IDEXX, Westbrook, ME) filter in a polycarbonate housing, using a car-battery powered pump. Samples ranged from 30 to 40L, with the exact volume measured by an in-line meter. The filter was stored in a glass jar with 300 ml of phosphate-buffered saline (pH 7.2, 0.01% Tween 20) and transported to KEMU. Oocysts were washed out of the filter according to the manufacturer’s instructions (IDEXX 2002). In brief, a bolt is released allowing the compressed filter to expand, allowing retained oocysts to be eluted from the sponge matrix. The wash solution is filtered, and oocysts trapped on a membrane are eluted into ~10 ml of fluid. The sample is centrifuged to a pellet of <0.5 ml and then purified by IMS (see below). If the pellet was >0.5 ml the sample was split per Method 1623.
Immunomagnetic Separation (IMS) and DNA extraction
IMS was conducted using the Dynabeads GC-Combo (Dynal Biotech, Oslo, Norway) protocol. In brief, anti-Cryptosporidium antibodies attached to magnetic beads were mixed with samples prepared as described above to a volume of 1 ml. The oocysts were removed through magnetic removal of the beads, then dissociated from the beads using an acid wash step. The acid wash supernatant was aspirated and used for DNA extraction. Oocyst extracts underwent five freeze thaw cycles after the addition of 180 µl of tissue lysis buffer (50 Mm Tris-HCl [pH 8.5], 1Mm EDTA [pH 8], 0.5% sodium dodecyl sulfate) in cryovials which were incubated for 1 minute in liquid nitrogen and 1 minute in a 56°C heating block per cycle. Samples were centrifuged at 14,000 × g for 30 s and then incubated with 20 µl proteinase K (Sigma, St Louis, MO.) for 2 hours at 55°C while being rotated, incubated at 90°C to denature the proteinase K, chilled on ice for 1 minute, and centrifuged at 14,000 × g for 3 minutes. Supernatants were transferred to a new 1.5 ml tube, 200 ul of ethanol was added, then vortexed for 15 seconds. The mixture was loaded into a QIAmp spin column (Qiagen Inc, Valencia, California) and centrifuged at 6,000 × g for 1 minute. This was repeated using wash buffers described by the manufacturer. The final supernatant was stored at −20°C for use in PCR.
PCR and RFLP Protocols
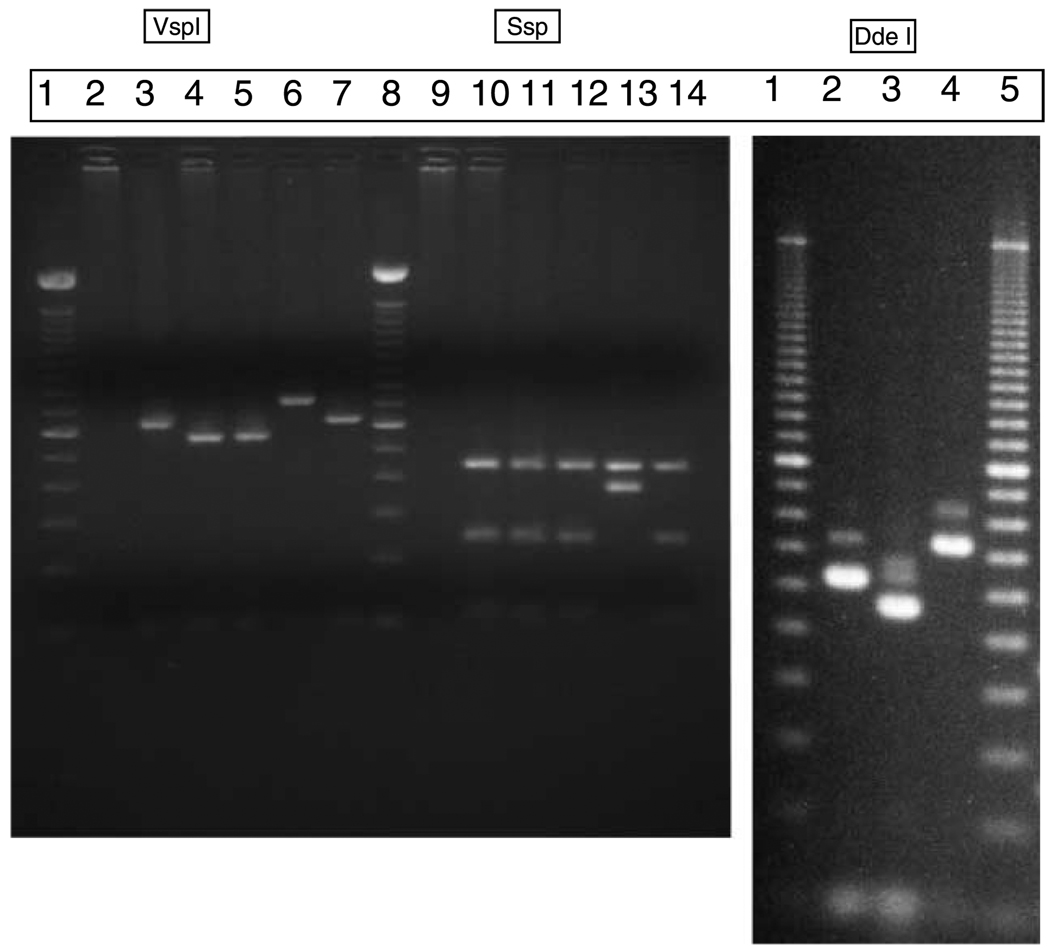
Nested PCR followed by restriction enzyme digestion was carried out using SSU primers, and VSPI and SSPI enzymes respectively (Xiao et al. 1999). Reaction volumes of 100 µl consisted of premixed reagents containing 10 mM of each of the four deoxynucleoside triphosphates, 10 × PCR buffer, 25 mM MgCL2, PCR grade water and Taq polymerase (Invitrogen). 2µl of template DNA was used. The primary run was done using 5’-TTCTAGAGCTAATACATGCG-3’ and 5’-CCCATTTCCTTCGAAACAGGA-3’ primers. The samples were incubated in the thermocycler at 94°C for 3 minutes to denature template DNA. This was followed by 35 cycles, each consisting of 94°C for 45 s, 55°C for 45 s and 72°C for 1 minute and a final extension at 72°C for 10 minutes. A secondary amplification using the primary products the primers 5’-GGAAGGGTTGTATTTATTAGATAAAG-3’ and 5’-AAGGAGTAAGGAACAACCTCCA-3’ was performed using cycling conditions identical to those in the primary PCR. Amplicons were fractionated on a 2% agarose gel and visualized by ethidium bromide staining to confirm the presence of characteristic Cryptosporidium products (Figure 2). To differentiate genotypes, 10µl of positive secondary PCR products were digested in a 40 µl (total volume) reaction mixture containing 20 U of SspI (New England Biolabs, Beverly, Mass.) or 20 U of VspI (GIBCO BRL, Grand Island, N.Y) and 4µl of the appropriate restriction buffer at 37°C for 1 hr (Xiao et al. 1999). As C. andersoni and C. muris have identical SspI and VspI restriction patterns, they were differentiated by digesting secondary products with 20 U of DdeI (New England Biolabs) at 37°C for 1h under conditions recommended by the supplier. Digested products were fractionated on 2% agarose gel and visualized by ethidium bromide staining. All positive detections were confirmed in duplicate by RFLP analysis of additional, independent PCR products from the same water sample. Positive controls included TU502 C. hominis (Akiyoshi et al. 2002), obtained from Dr. Saul Tzipori, Tufts University Cummings School of Veterinary Medicine, North Grafton, MA and the Iowa C. parvum, obtained from Pleasant Farms, Troy, ID (Abrahamsen et al. 2004).
Figure 2.
The results of restriction fragment length polymorphism (RFLP) analysis using Ssp1, Vsp1, and Dde1 for species and genotype determination respectively. Panel A: Lane 1: 100 bp ladder; 2: negative control; 3: TU502 (C.hominis); 4: IOWA (C. parvum); 5: Water sample (Karinda); 6: Water sample (Kiina river); 7: C. hominis spiked stool sample; 8: 100 bp ladder; 9: negative control; 10: TU502 (C.hominis); 11: IOWA (C. parvum); 12: Water sample (Karinda); 13: Water sample (Kiina river); 14: C. parvum spiked stool sample. Panel B: Lane 1: 100 bp ladder; 2: C. parvum; 3: C. hominis; 4: C. andersoni; 5: 100 bp ladder.
Statistical analysis
Data were analyzed using the Statistical Package for the Social Sciences (SPSS) version 15. Specific comparisons (t-tests and chi-square analyses) are denoted in the text.
RESULTS AND DISCUSSION
Oocyst detection: by method and by season
Monthly samples were collected and analyzed for January through June 2003, and January through June of 2004. In 2003 we used the CCF method, and in 2004 the filtration method. All sample concentrates underwent PCR for the detection of oocysts. We thus sampled 6 months of both low and high rainfall in two sequential years. In 2003, all 10 sites were sampled monthly. In 2004, sampling at Kiina Wells became dangerous and was discontinued, as drought had increased the chance of interactions with wild elephants. In January and June of 2004, adverse road conditions made it impossible to visit several other sites.
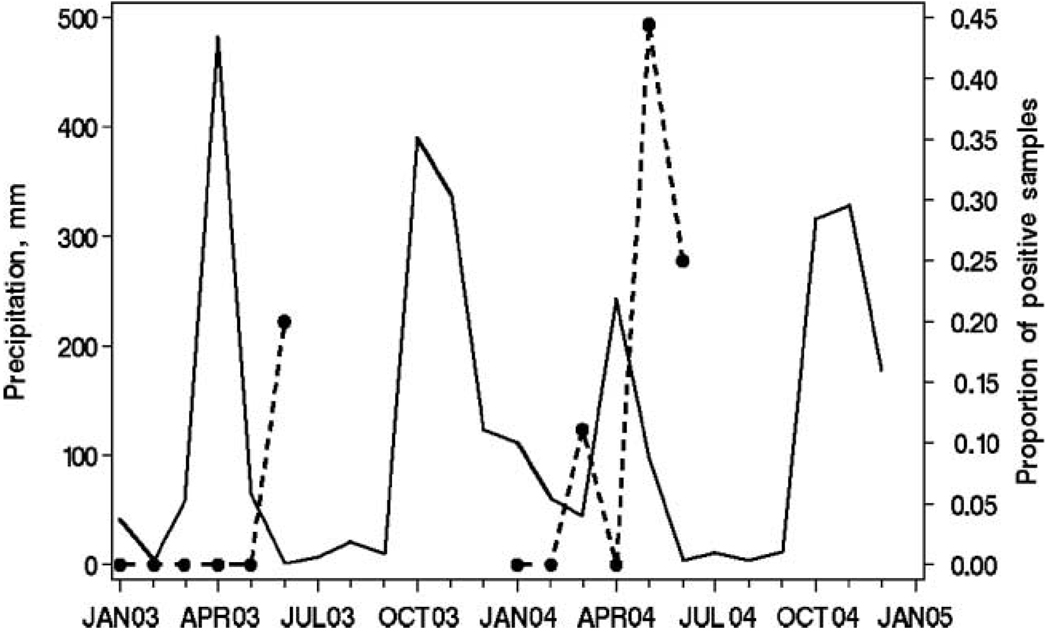
108 samples were acquired and 9 (8.3%) were PCR positive for oocysts (Table 1). Two of 60 (3.3%) samples obtained in 2003, versus 7 of 48 (14.6%) samples from 2004, were positive. The detection rate in 2004 was 4.375 fold higher than in 2003, which did not reach statistical significance (p = 0.075, 2-tailed Fisher’s exact test). Oocyst detection rates varied significantly by month (χ2 = 13.53, 5 df, p < 0.019). Table 2 displays detection rates before, during, and after the peak in precipitation which occurred in March and April of each year, together with monthly cumulative precipitation for each time period. The apparent seasonal period peak of detection at the end of the rainy season is significant (χ2 = 13.17, p < 0.0014). Figure 3 displays precipitation data recorded in Meru. Two peaks of precipitation usually occur each year. The years of 2003 and 2004 were atypical in that there was rainfall in the months separating the late 2003 and early 2004 precipitation peaks. The months when we detected oocysts corresponded to the nadirs immediately following seasonal peaks of precipitation (June 2003, March 2004, June 2004) or the descending right-hand slope of a period of decreasing precipitation after a peak (May 2004).
Table 1.
Temporal pattern of Cryptosporidium detection in Meru, Kenya waters, using a calcium chloride precipitation technique (CCF) or filtration, followed by immunomagnetic bead separation and PCR
| 2003 testing using CCF | 2004 testing using filtration | Totals based on both years | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Month | Positive | Negative | Total 2003 | Positive | Negative | Total 2004 | Positive | Negative | Total |
| January | 0 | 10 | 10 | 0 | 4 | 4 | 0 | 14 | 14 |
| February | 0 | 10 | 10 | 0 | 9 | 9 | 0 | 19 | 19 |
| March | 0 | 10 | 10 | 1 | 8 | 9 | 1 | 18 | 19 |
| April | 0 | 10 | 10 | 0 | 9 | 9 | 0 | 19 | 19 |
| May | 0 | 10 | 10 | 4 | 5 | 9 | 4 | 15 | 19 |
| June | 2 | 8 | 10 | 2 | 6 | 8 | 4 | 14 | 18 |
| Total | 2 | 58 | 58 | 7 | 41 | 48 | 9 | 99 | 108 |
Table 2.
Cryptosporidium water sample testing by seasonal period
| Water sample testing by seasonal period | |||
|---|---|---|---|
| Seasonal period, cumulative precipitation in mm water (Mean ± SD) | Positive | Negative | Total |
| Before peak rainfall (January–February), 54 ± 44 mm |
0 | 33 | 33 |
| Peak rainfall (March–April), 207 ± 204 mm |
1 | 37 | 38 |
| After peak rainfall and start of traditional dry season (May–June), 41 ± 48 mm |
8 | 29 | 37 |
| Totals | 9 | 99 | 108 |
Figure 3.
Actual monthly precipitation in millimetres in Meru, Kenya by month of 2003 and 2004 and proportion of positive Meru Region surface water samples by month of 2003 and 2004. The solid black line represents precipitation in mm (left axis scale), and the dotted black line represents the proportion of positive surface water samples (right axis scale), by month (x axis, horizontal).
Genotypes of Cryptosporidium detected in water samples
C. parvum was detected six times. It was isolated three times from the sewage waste stream from Meru Town, from Meru Town’s water intake site, from a site downstream of Meru Town, and from a pool of water frequently used by elephants (Table 3). C. hominis was not detected from any site. C. andersoni was detected 3 times (Table 3). This species has been linked to cattle and does not appear to be pathogenic to humans. All 3 C. andersoni isolates were detected from the Kiina River, where pastoralists water their herds of cattle.
Table 3.
Genotypes of detected Cryptosporidium by month and year, and by site
| Genotypes of Detected Cryptosporidium by time and site | |||
|---|---|---|---|
| Year | Month | Genotype | Site |
| 2003 | June | C. parvum (formerly called bovine genotype Type 2) | Site D. Karinda (sewage waste stream from Meru Town) on the Kathita River |
| 2003 | June | C. andersoni | Site G. Kiina River |
| 2004 | March | C. andersoni | Site G. Kiina River |
| 2004 | May | C. parvum | Site F. Luuria (standing pool in elephant path in forest) |
| 2004 | May | C. parvum | Site C. (Intake for Meru Town water supply), Kathita River |
| 2004 | May | C. parvum | Site D. Karinda |
| 2004 | May | C. andersoni | Site G. Kiina River |
| 2004 | June | C. parvum | Site E. Tharaka (Marimanti) on the Kathita River |
| 2004 | June | C. parvum | Site D. Karinda |
DISCUSSION
In this pilot study, we detected Cryptosporidium surface water contamination in the Meru, Kenya region over the first six months of two sequential years (2003 and 2004). The period at the end of the sampled rainy season, and the beginning of the following drier, cool season (May and June) was significantly associated with oocyst detection in surface waters. This corresponded to the nadirs of precipitation between precipitation peaks, or to periods of tapering rainfall (Figure 3).
Cryptosporidium oocysts are believed to be flushed by rain into surface waters, and so one might have expected peak detection during the onset of the rainy months. During March and April, the volume of water which falls is so great that it is possible that the concentration of oocysts in surface waters is relatively low, because of dilution. Alternatively, there may be little Cryptosporidium transmission during March and April in Meru, and thus there may be fewer fecally excreted parasites to be swept into water bodies. Our surface water data does not allow us to speculate further on the merits of these and other possible explanations regarding transmission density, as human and animal cryptosporidiosis incidence data is not yet available from Meru.
However, data from other East African sites may be informative. In a recent survey of 4,899 pediatric fecal samples from laboratories near Nairobi, the rainy months of April and May had the lowest infection prevalence (1.5%), suggesting that transmission is less common during this period (Gatei et al. 2006). Infection rates were highest in children during the November to February period, which usually has little precipitation. Studies from Rwanda, Malawai, and Kenya (Bogaerts et al. 1987; Peng et al. 2003; Gatei et al. 2006), respectively, report that the peak incidence of cryptosporidiosis is at the end of rainy seasons and beginning of the drier months, as has a study by Moodley et al. (1991) in Durban, South Africa. In contrast, in Guinea-Bissau, West Africa, the peak prevalence of clinical cryptosporidiosis in children is just before, or at the onset of, the rainy season from May to July (Molbak et al. 1990, 1993; Perch et al. 2001). Similar results were reported from Zambia (Nchito et al. 1998). The East African human incidence pattern is congruent with our findings regarding oocysts in surface waters. It is possible that the apparent seasonality of surface water contamination and of human disease, is reflective of different transmission pathways, hosts, and/or Cryptosporidium species in East Africa when compared to other locations. We note that as climate change occurs, transmission patterns of many waterborne diseases may shift, and studies in locations with unusual seasonality patterns may help inform our understanding of what climate change may bring (Rose et al. 2002; Hunter 2003; Jagai et al. 2007; Naumova et al. 2007).
A limitation of our study is that we did not directly compare our two oocyst isolation methods in a side-by-side evaluation during the same year. We substituted the filtration method for the CCF method after unexpectedly low rates of isolation in 2003. While the direct field filtration of 30–40 L of water led to higher PCR detection rates than did the CCF method with 10 L samples, this did not reach statistical significance. The differences in detection that we encountered might be due to the natural variation between the years tested; inhibition of PCR by environmental contaminants (Sluter et al. 1997; Loge et al. 2002); or additional losses encumbered in the “double purification” via CCF and then IMS. Egorov et al. (2002) and Karanis et al. (2006) found CCF useful when oocysts were detected via immunofluorescent identification. Although Monis & Saint (2001) reported that the precipitation method is inhibitory for PCR detection, preliminary data (Kato et al. 2003) had suggested that our initial approach would be successful. In order to address this issue properly, side by side comparisons of the two methods would be needed which would quantify overall recovery efficiency, as we may easily have suffered from falsely negative results in 2003. We used PCR methods for detection, rather than visual ones, because of our interest in the genotypes of the Cryptosporidium that might be found in this ecologically unusual region.
C. parvum, but not C. hominis, was thrice detected from human sewage waste waters from Meru Town, as well as from the intake site for the town water supply and downstream of Meru Town. Studies from Kenya and Uganda (Tumwine et al. 2005; Gatei et al. 2006) show that C. hominis is the dominant species infecting children in the urban centers of Nairobi and Kampala. C. parvum is adapted to multiple host species, including wildlife, humans and cattle (Fayer 2004). C. hominis has only been reliably detected in primate hosts, although two studies report its detection in dugongs (Morgan et al. 2000) and cattle (Smith et al. 2005). Cattle herding is uncommon in the intensely farmed Meru Town region, while pastoralist herding is more common northeast of Meru Town. We detected C. andersoni thrice from the Kiina River, where pastoralists water their cattle. C. andersoni is not thought to be pathogenic for humans, and is dominantly a parasite of older cattle. C. parvum infects HIV infected and uninfected children, while C. meleagridis and C. muris are more frequently found in HIV-infected persons (Caccio 2005). In the rural regions of Kenya such as Meru, the spectrum of Cryptosporidium species infecting humans may differ from that in urban areas given different land use patterns (Naumova et al. 2005) and the use of the same water by wildlife as well as by people and herded animals. Our data is consistent with, but not proof of, endemic waterborne Cryptosporidium transmission in Meru, including potential transmission between wildlife, domestic animals, and humans. Further environmental, epidemiological, and molecular investigations are needed to confirm or refute this thesis. In particular, molecular analysis of Cryptosporidium isolates at the sub-type level from potential animal reservoirs, surface waters and humans may be helpful in this regard (Ajjampur et al. 2007). We find it intriguing that C. parvum was detected from a pool after use by migrating elephants at Luuria, as according to the local population these waters cause diarrhea in people after the elephants have passed through.
CONCLUSION
The Meru region of Kenya is an appropriate ecological zone for studying potential transmission through water between wildlife, humans and domesticated animals. We document C. parvum in the Kathita River, which serves all these groups, as well as Cryptosporidium from waters which serve humans, cattle and wildlife, such as elephants. Oocysts were significantly more commonly detected in surface waters at the end of rainy seasons and at the beginning of dry seasons, congruent with studies which have shown peak human disease during the same period in East Africa. This seasonality differs from that noted in West Africa and northern temperature regions. Continued environmental, epidemiological, and molecular studies will be needed to address questions regarding the actors in, and timing of, transmission in this region.
ACKNOWLEDGEMENTS
We thank Denise Castronovo (Mapping Sustainability, www.mappingsustainability.com) for her help with Figure 1, and Siobhan Mor for her comments on cryptosporidiosis seasonality in Africa. We thank the Meru Weather Station, Government of Kenya for the precipitation data. This study was supported by a grant to JKG, MM, ENN and JE from the US National Institutes of Health’s Fogarty International Center (R23 TW05821, “An Ecological Analysis of Cryptosporidium in Kenya”), and R01 AI43415 to JKG and ENN from the National Institute of Allergy and Infectious Diseases, entitled “Waterborne Emerging Diarrheal Diseases Study”.
Contributor Information
John M. Muchiri, Kenya Methodist University, PO Box 267-60200, Meru, Kenya
Luke Ascolillo, Department of Public Health and Family Medicine, Tufts University School of Medicine, 136 Harrison Avenue, Boston, Massachusetts 02111, USA.
Mutuma Mugambi, Kenya Methodist University, PO Box 267-60200, Meru, Kenya.
Titus Mutwiri, Kenya Methodist University, PO Box 267-60200, Meru, Kenya.
Honorine D. Ward, Department of Public Health and Family Medicine, Tufts University School of Medicine, 136 Harrison Avenue, Boston, Massachusetts 02111, USA
Elena N. Naumova, Department of Public Health and Family Medicine, Tufts University School of Medicine, 136 Harrison Avenue, Boston, Massachusetts 02111, USA
Andrey I. Egorov, Department of Public Health and Family Medicine, Tufts University School of Medicine, 136 Harrison Avenue, Boston, Massachusetts 02111, USA
Seth Cohen, Department of Public Health and Family Medicine, Tufts University School of Medicine, 136 Harrison Avenue, Boston, Massachusetts 02111, USA.
James G. Else, Yerkes National Primate Research Center, Emory University, 954 Gatewood Road, Atlanta, Georgia 30329, USA
Jeffrey K. Griffiths, Department of Public Health and Family Medicine, Tufts University School of Medicine, 136 Harrison Avenue, Boston, Massachusetts 02111, USA
REFERENCES
- Abrahamsen MS, Templeton TJ, Enomoto S, Abrahante JE, Zhu G, Lancto CA, Deng M, Liu C, Widmer G, Tzipori S, Buck GA, Xu P, Bankier AT, Dear PH, Konfortov BA, Spriggs HF, Iyer L, Anantharaman V, Aravind L, Kapur V. Complete genome sequence of the apicomplexan, Cryptosporidium parvum. Science. 2004;304:441–445. doi: 10.1126/science.1094786. [DOI] [PubMed] [Google Scholar]
- Abubakar I, Aliyu SH, Arumugam C, Hunter PR, Usman UK. Prevention and treatment of cryptosporidiosis in immunocompromised patients. Cochrane Database Syst. Rev. 2007;1 doi: 10.1002/14651858.CD004932.pub2. CD004932. [DOI] [PubMed] [Google Scholar]
- Ajjampur SS, Gladstone BP, Selvapandian D, Muliyil JP, Ward H, Kang G. Molecular and spatial epidemiology of cryptosporidiosis in children in a semiurban community in South India. J. Clin. Microbiol. 2007;45:915–920. doi: 10.1128/JCM.01590-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akiyoshi DE, Feng X, Buckholt MA, Widmer G, Tzipori S. Genetic analysis of a Cryptosporidium parvum human genotype 1 isolate passaged through different host species. Infect. Immun. 2002;70:5670–5675. doi: 10.1128/IAI.70.10.5670-5675.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagley ST, Auer MT, Stern DA, Babiera MJ. Sources and fate of Giardia cysts and Cryptosporidium oocysts in surface water. J. Lake Res. Manage. 1998;14:379–392. [Google Scholar]
- Bogaerts J, Lepage P, Rouvroy D, van Goethem C, Nsengumuremye F, Mohamed O, Habyalimana JB, Vandepitte J. Cryptosproridiosis in Rwanda. Clinical and epidemiological features. Ann. Soc. Belge. Méd. Trop. 1987;67:157–165. [PubMed] [Google Scholar]
- Caccio SM. Molecular epidemiology of human cryptosporidiosis (Review) Parassitologia. 2005;47(2):185–192. [PubMed] [Google Scholar]
- Egorov A, Paulauskis J, Petrova L, Tereschenko A, Drizhd N, Ford T. Contamination of water supplies with Cryptosporidium parvum and Giardia lamblia and diarrheal illness in selected Russian cities. Int. J. Hyg. Environ. Health. 2002;205:281–289. doi: 10.1078/1438-4639-00153. [DOI] [PubMed] [Google Scholar]
- Fayer R. Cryptosporidium: a water-borne zoonotic parasite. Vet. Parasitol. 2004;126:37–56. doi: 10.1016/j.vetpar.2004.09.004. [DOI] [PubMed] [Google Scholar]
- Gatei W, Wamae CN, Mbae C, Waruru A, Mulinge E, Waithera T, Gatika SM, Kamwati SK, Revathi G, Hart CA. Cryptosporidiosis: prevalence, genotype analysis, and symptoms associated with infections in children in Kenya. Am. J. Trop. Med. Hyg. 2006;75:78–82. [PubMed] [Google Scholar]
- Gendrel D, Treluyer JM, Richard-Lenoble D. Parasitic diarrhea in normal and malnourished children (Review) Fundam. Clin. Pharmacol. 2003;17(2):189–197. doi: 10.1046/j.1472-8206.2003.00169.x. [DOI] [PubMed] [Google Scholar]
- Griffiths JK. Human cryptosporidiosis: epidemiology, transmission, clinical disease and diagnosis. In: Saul T, editor. Emerging Human Enteric Protozoan Infections. Vol. 40. London, UK: Academic Press; 1998. pp. 37–85. Adv. Parasitol. [DOI] [PubMed] [Google Scholar]
- Hunter PR. Climate change and waterborne and vector-borne diseases. J. Appl. Microbiol. 2003;94:37S–46S. doi: 10.1046/j.1365-2672.94.s1.5.x. [DOI] [PubMed] [Google Scholar]
- Hunter P, Syed Q, Naumova EN. Possible undetected outbreaks of cryptosporidiosis in areas of the north west of England supplies by an unfiltered surface water source. Commun. Dis. Public Health. 2001;4:136–138. [PubMed] [Google Scholar]
- IDEXX. [Accessed 28 January, 2008];Filta-Max Operator’s Guide. Protocol for Use with the Manual Wash Station. 2002 http://www.idexx.com/water/refs/060458201MAN.pdf.
- Jagai JS, Monchak J, McEntee JC, Castranovo D, Naumova EN. The Use of Remote Sensing to Assess Global Trends in Seasonality of Cryptosporidiosis; Proceedings of the 32nd International Symposium on Remote Sensing of Environment; June 25–29 2007; San Jose, Costa Rica. 2007. [Google Scholar]
- Kaplan JE, Hu DJ, Holmes KK, Jaffe HW, Masure H, De Cock KM. Preventing opportunistic infections in human immunodeficiency virus-infected persons: implications for the developing world. Am. J. Trop. Med. Hyg. 1996;55:1–11. [PubMed] [Google Scholar]
- Karanis P, Sotiriadou I, Kartashev V, Kourenti C, Tsvetkova N, Stojanova K. Occurrence of Giardia and Cryptosporidium in water supplies of Russia and Bulgaria. Environ. Res. 2006;102:260–271. doi: 10.1016/j.envres.2006.05.005. [DOI] [PubMed] [Google Scholar]
- Kato S, Ascolillo L, Egas J, Elson L, Gostyla K, Naples L, Else J, Sempertegui F, Naumova E, Egorov A, Ojeda F, Griffiths JK. Waterborne Cryptosporidium oocyst identification and genotyping: use of GIS for Ecosystem Studies in Kenya and Ecuador. J. Eukaryotic. Microbiol. 2003;50 Suppl:548–549. doi: 10.1111/j.1550-7408.2003.tb00624.x. [DOI] [PubMed] [Google Scholar]
- Loge FJ, Thompson DE, Call DR. PCR detection of specific pathogens in water: a risk-based analysis. Environ. Sci. Technol. 2002;36(12):2754–2759. doi: 10.1021/es015777m. [DOI] [PubMed] [Google Scholar]
- Moodley D, Jackson TF, Gathiram V, van den Ende J. Cryptosporidium infections in children in Durban. Seasonal variation, age distribution and disease status. S. Afr. Med. J. 1991;79:295–297. [PubMed] [Google Scholar]
- Molbak K, Hojlyng N, Ingholt L, Da Silva AP, Jepsen S, Aaby P. An epidemic outbreak of cryptosporidiosis: a prospective community study from Guinnea-Bissau. Pediatr. Infect. Dis. J. 1990;9:566–570. doi: 10.1097/00006454-199008000-00008. [DOI] [PubMed] [Google Scholar]
- Molbak K, Hojlyng N, Gottschau A, Sa JC, Ingholt L, da Silva AP, Aaby P. Cryptosporidiosis in infancy and childhood mortality in Guinea Bissau, west Africa. Br. Med. J. 1993;307:417–420. doi: 10.1136/bmj.307.6901.417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monis PT, Saint CP. Development of a nested-PCR assay for the detection of Cryptosporidium parvum in finished water. Water Res. 2001;35:1641–1648. doi: 10.1016/s0043-1354(00)00426-7. [DOI] [PubMed] [Google Scholar]
- Morgan UM, Xiao L, Hill BD, O’Donoghue P, Limor J, Lal A, Thompson RC. Detection of the Cryptosporidium parvum “human” genotype in a dugong (Dugong dugon) J. Parasitol. 2000;86:1352–1354. doi: 10.1645/0022-3395(2000)086[1352:DOTCPH]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- Naumova EN, Christodouleas J, Hunter PR, Syed Q. Temporal and spatial variability in cryptosporidiosis recorded by the surveillance system in North West England in 1990–1999. J. Water Health. 2005;3(2):185–196. [PubMed] [Google Scholar]
- Naumova EN, Jagai J, Matyas B, DeMaria A, MacNeill IB, Griffiths JK. Seasonality in six enterically transmitted diseases and ambient temperature. Epidemiol. Infect. 2007;135(2):281–292. doi: 10.1017/S0950268806006698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nchito M, Kelly P, Sianongo S, Luo NP, Feldman R, Farthing M, Baboo KS. Cryptosporidiosis in urban Zambian children: an analysis of risk factors. Am. J. Trop. Med. Hyg. 1998;59:435–437. doi: 10.4269/ajtmh.1998.59.435. [DOI] [PubMed] [Google Scholar]
- Peng MM, Meshnick SR, Cunliffe NA, Thindwa BDM, Hart CA, Broadhead RL, Xiao L. Molecular epidemiology of cryptosporidiosis in children in Malawi. J. Eukaryotic Microbiol. 2003;50 Suppl:557–559. doi: 10.1111/j.1550-7408.2003.tb00628.x. [DOI] [PubMed] [Google Scholar]
- Perch M, Sodemann M, Jakobsen MS, Valentiner-Branth P, Steinsland H, Fischer TK, Lopes DD, Aaby P, Molbak K. Seven years’ experience with Cryptosporidium parvum in Guinea-Bissau, West Africa. Ann. Trop. Paediatr. 2001;21:313–318. doi: 10.1080/07430170120093490. [DOI] [PubMed] [Google Scholar]
- Rose JB, Huffman DE, Gennaccaro A. Risk and control of waterborne cryptosporidiosis. FEMS Microbiol. Rev. 2002;26:113–123. doi: 10.1111/j.1574-6976.2002.tb00604.x. [DOI] [PubMed] [Google Scholar]
- Sluter SD, Tzipori S, Widmer G. Parameters affecting polymerase chain reaction detection of waterborne Cryptosporidium parvum oocysts. Appl. Microbiol. Biotechnol. 1997;48(3):325–330. doi: 10.1007/s002530051057. [DOI] [PubMed] [Google Scholar]
- Smith HV, Nichols RAB, Mallon M, MacLeod A, Tait A, Reilly WJ, Browning LM, Gray D, Reid SWJ, Wastling JM. Natural Cryptosporidium hominis infections in Scottish cattle. Vet. Rec. 2005;156:710–711. doi: 10.1136/vr.156.22.710. [DOI] [PubMed] [Google Scholar]
- Tumwine JK, Kekitiinwa A, Bakeera-Kitaka S, Ndeezi G, Downing R, Feng X, Akiyoshi DE, Tzipori S. Cryptosporidiosis and microsporidiosis in Ugandan children with persistent diarrhea with and without concurrent infection with the human immunodeficiency virus. Am. J. Trop. Med. Hyg. 2005;73:921–925. [PubMed] [Google Scholar]
- Tzipori S, Ward H. Cryptosporidiosis: biology, pathogenesis and disease. Microbes Infect. 2002;4:1047–1058. doi: 10.1016/s1286-4579(02)01629-5. [DOI] [PubMed] [Google Scholar]
- United States Environmental Protection Agency 2005 Method 1623. Cryptosporidium and Giardia in Water by Filtration/IMS/FA. 2005 December; Office of Water, EPA-815-R-05-002. Available online at http://epa.gov/nerlcwww/1623de05.pdf.
- Vesey G, Slade JS, Byrne M, Shepherd K, Fricker CR. A new method for the concentration of Cryptosporidium oocysts from water. J. Appl. Bacteriol. 1993;75:82–86. doi: 10.1111/j.1365-2672.1993.tb03412.x. [DOI] [PubMed] [Google Scholar]
- Xiao L, Escalante L, Yang C, Sulaiman I, Escalante AA, Montali RJ, Lal AA. Phylogenetic analysis of Cryptosporidium parasites based on the small-subunit rRNA gene locus. Appl. Environ. Microbiol. 1999;65(4):1578–1583. doi: 10.1128/aem.65.4.1578-1583.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]