Abstract
GRASP-1 is a neuronally enriched protein that interacts with the AMPA-type glutamate receptor/GRIP complex. GRASP-1 can be cleaved by Caspase-3 in both normal and ischemic brains although the functional significance of this cleavage remains elusive. We investigated signal transduction pathways that might lie downstream of GRASP-1 and found that GRASP-1 potently activates JNK pathway signaling, with no effect on ERK signaling. Such JNK pathway-activating activity requires binding of GRASP-1 to both JNK and the upstream JNK pathway activator MEKK-1. Furthermore, mutations that prevent Caspase 3-cleavage of GRASP-1 dramatically inhibit the JNK pathway-activating activity of GRASP-1, suggesting a novel link between Caspase-3 activation and JNK pathway signaling. These results suggest that GRASP-1 serves as a scaffold protein to facilitate MEKK-1 activation of JNK signaling in neurons.
INTRODUCTION
GRASP-1 is a neuronally-enriched protein that was initially isolated due to its interaction with the PDZ domain containing adaptor protein GRIP1 (glutamate receptor interacting protein) [1]. GRIP1 is a neuronal scaffold protein [2] that organizes protein complexes containing AMPA-type glutamate receptors (AMPA-Rs), and GRASP-1 over-expression regulates AMPA-R trafficking in neurons [1]. The N-terminus of GRASP-1 is homologous to guanine nucleotide exchange factors (GEFs) for the small GTP-binding protein Ras and indeed GRASP-1 possesses RasGEF activity in vitro [1]. In normal brain, a fraction of GRASP-1 is cleaved by Caspase-3, separating the RasGEF domain from the GRIP-binding domain. Ischemia enhances the Caspase-cleavage of GRASP-1 [3]. However, the functional significance of Caspase-3 regulation of GRASP-1 remains unknown.
Ras triggers activation of several signaling pathways, most notably the protein kinase cascade leading to activation of the mitogen-activated protein kinase (MAPK) ERK [4]. Ras-ERK signaling is required for several important processes in neurons, including the regulation of dendritic morphology, AMPA receptor trafficking and long-term potentiation [5]. However, it is now clear that homologous MAPK pathways, leading to the activation of cJun N-terminal kinases (JNKs) and p38 MAPK can also regulate synaptic function [6–8]. While ERK activation in neurons, mediated via glutamate, growth factor receptors and calcium channels, is well documented [9,10], mechanisms that control neuronal JNK activity (which is basally very high in neurons) [11,12] are less clear.
JNKs bind, phosphorylate and activate the transcription factor c-jun [13]. Three JNK genes have been identified, of which JNK1 and 2 are expressed ubiquitously while JNK3 is enriched in brain, heart, and testis [13]. JNKs are phosphorylated and activated by the upstream kinases SEK1/MKK4 and MKK7. MKK4 and MKK7 are in turn phosphorylated and activated by a variety of upstream kinases, including MEK kinases (MEKKs; [14, 15]), mixed-lineage kinases (MLKs) and apoptosis signal-regulating kinase (ASKs; [4.13]). It is unclear which of these upstream kinases control neuronal JNK activation physiologically, though data from knockout mice demonstrate that MEKKs are essential for JNK activation in response to certain stimuli [16].
The functions of JNKs in non-neuronal tissues are diverse, ranging from T cell differentiation and apoptosis to tumorigenesis [13]. Within the brain JNKs are also implicated in a number of neurological disorders, including Parkinson’s disease, stroke and Alzheimer’s disease [17]. However, physiological roles for JNKs in neurons are also now being revealed, with JNKs implicated in the regulation of dendritic morphology, synaptic vesicle release and AMPA receptor trafficking [7,8,18].
These diverse roles for JNKs in neurons suggest that different pools of JNK may exist in different cellular locations, and both biochemical fractionation experiments (G.M.T., R.L.H. et al., in preparation) and the distinct phenotypes of different JNK knockout mice [7,18,19] support this hypothesis. The mechanisms used by cells to differentially target JNKs to different subcellular locations are not fully clear, but attractive candidate mediators are various multi-domain scaffold proteins that are known to bind both JNK signaling pathway components and other proteins.
Scaffold proteins often bind several signaling molecules to create multi-enzyme complexes and play important roles in defining the specificity and efficiency of signaling pathways. The first MAPK scaffold protein, Ste5p, was identified genetically in yeast and subsequent structure-function analysis demonstrated that distinct regions of Ste5p interact with the Map kinase kinase kinase (MKKK) Ste11p, the Map kinase kinase (MKK) Ste7p and the MAP kinase Fus3p/Kss1p [4] and that Ste5p facilitates signal transduction from Ste11p to Fus3p/Kss1p. More recently, MAPK scaffold proteins have been identified in mammalian cells. These mammalian proteins share little sequence homology with Ste5p but function in a similar manner. For example, JNK-interacting protein1 (JIP1) interacts with multiple components of the JNK signaling pathway, including the mixed-lineage protein kinase (MLK) family of MKKKs, MKK7 and JNK [20], and brings upstream and downstream kinases in these cascades into close proximity through protein-protein interactions [20, 21].
Here we report that GRASP-1 functions as a JNK pathway scaffold protein, binding both JNK and the upstream kinase MEKK1 in neurons, and that these interactions facilitate JNK signaling. We also show that the scaffolding function of GRASP-1 requires Caspase-3 cleavage, which releases the JNK/MEKK1 binding regions of GRASP-1 from the RasGEF domain.
MATERIALS AND METHODS
ERK and JNK assays on immunoprecipitates
HEK293T cells were transfected with HA-tagged ERK1 or JNK1 plus other DNA constructs of interest, using a Ca2+-phosphate method. Drug treatment and kinase assays were performed 40 hours after transfection. For BDNF treatment, cells were incubated in medium containing 50 ng/ml purified BDNF for 10 minutes. For nocodazole treatment, cells were incubated with 0.25 μg/ml nocodazole for 1 hour. After drug treatment, cells were washed once with ice-cold PBS and lysed in ice-cold lysis buffer (20 mM HEPES pH 7.4, 10 mM EGTA, 40 mM beta-glycerophosphate, 1% Triton-X100, 2.5 mM MgCl2, 1mM DTT, 2 mM sodium orthovanadate, 0.1 mM PMSF and 20 units/ml Trasylol). ERK1 or JNK1 were immunoprecipitated from lysates with anti-HA antibody (12CA5, Roche Molecular Biochemicals, IN) at 4°C. Immunoprecipitates were washed 3 times with TBS containing 1% Triton X-100 and 2 mM sodium orthovanadate, once with 100 mM Tris-HCl, pH 7.4 containing 0.5 M LiCl, and once with kinase reaction buffer (25 mM HEPES, pH 7.4, with 10 mM Mg acetate and 1 mM DTT). Reaction mix (50 μl) was added to the immunoprecipitates, which were then incubated at 30°C for 20 minutes. For the ERK assay, reaction mix contained 1 μCi 32P-γ-ATP, 20 μM ATP and 0.5 mg/ml MBP in kinase reaction buffer. For the JNK assay, reaction mix contained 5 μCi 32P-γ-ATP, 20 μM ATP and 2 μg GST-cJun (1–169) in reaction buffer. Reactions were stopped by the addition of 10 μl of 0.5 M EDTA. 32P radioactivity incorporated into substrates was analyzed either by spotting reaction mix onto P81 filter paper followed by extensive washing and Cerenkov counting, or by SDS-PAGE and subsequent autoradiography. A small fraction of each sample was used for SDS-PAGE and subsequent immunoblotting to determine the relative amount of HA-ERK1 and HA-JNK1 in the immunoprecipitates.
Cell transfection and Co-immunoprecipitation
HEK293 cells were transfected with HA-tagged JNK1 or MEKK1 together with various Myc-tagged GRASP-1 constructs. A region of rat MEKK1 (between E1206 and the stop codon [22]) that is slighter larger than the kinase domain was tagged with a HA-tag at the N-terminus and used in co-immunoprecipitation experiments to examine the GRASP1-MEKK1 interaction. Mouse MEKK1 full length with an N-terminal HA-tag was used in all other experiments. Forty-eight hours later, cells were lysed in immunoprecipitation buffer (IPB: 1xPBS, 1% Triton X-100, PMSF and aprotinin) and centrifuged at 12,000 g to pellet insoluble material. The soluble fraction was incubated with anti-HA antibody and Protein A-sepharose (1:1 slurry) for 2 hours at 4 °C. The mixture was then washed once with IPB, twice with IPB plus 500 mM NaCl, and three times with IPB without Triton X-100. Proteins were eluted with Laemmli sample buffer and loaded onto SDS-polyacrylamide gels. Western analysis was performed with anti-Myc antibody.
Detection of ERK and JNK activation using phospho-specific antibodies
HEK293T cells transfected with various GRASP-1 or MEKK1 constructs were washed with ice-cold 1X PBS and lysed in lysis buffer for ERK and JNK assays on immunoprecipitates. After centrifugation at 12,000 g for 15 minutes, supernatants were denatured with Laemmli sample buffer, separated by 10% SDS-PAGE and transferred to PVDF membrane. The membrane was blocked with blocking buffer (1X PBS, 1% BSA and 0.1% Tween-20) and then incubated with anti-phospho-ERK1/2 (Cell Signaling Technology) or anti-phospho-JNK (Cell Signaling Technology) diluted with blocking buffer. After three washes with blocking buffer, the membrane was incubated with peroxidase-linked anti-mouse IgG or anti-rabbit IgG secondary antibodies and subsequently visualized using enhanced chemiluminescence. To normalize phosphorylation levels to JNK expression levels, we stripped anti-phospho-JNK antibody from the Western blot and probed again with an antibody against JNK (Cell Signaling Technology). The intensity of phospho-JNK (Ip) and total JNK (It) signals were quantified with the software ImageJ (National Institutes of Health, Bethesda, Maryland). Because the anti-JNK antibody recognizes JNK2/3 (54 kD) much better than JNK1 (46 kD), the quantification was performed on JNK2/3. In all the experiments described in this paper, phosphorylation of JNK1 always parallels to that of JNK2/3. The equation Ip X 100/It was used to calculate the normalized level of JNK phosphorylation.
RESULTS
Overexpression of GRASP-1 activates the JNK pathway
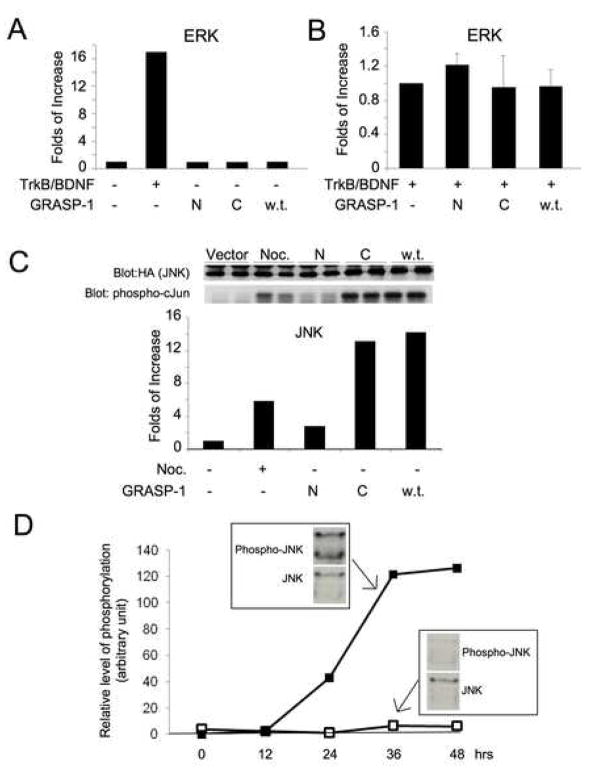
GRASP-1 acts as a guanine nucleotide exchange factor for the small GTP binding protein Ras in vitro [1] and Ras signaling is known to mediate ERK activation [4]. We therefore investigated whether expression of GRASP-1 in heterologous cells triggered increases in ERK activity. HA-tagged ERK1 was co-transfected into HEK 293T cells plus either empty vector, an N-terminal fragment of GRASP-1 encoding the region upstream of the Caspase-cleavage site (GRASP1-N), a C-terminal fragment of GRASP-1 encoding the region downstream of the Caspase-cleavage site (GRASP1-C) or full length GRASP-1 containing a wild-type Caspase-cleavage site (w.t.). ERK activity was then examined by specific immunoprecipitation and subsequent in vitro kinase assays. Although GRASP-1 acts as a Ras guanine nucleotide exchange factor (Ras-GEF) in vitro, none of the GRASP-1 constructs activated ERK in HEK-293 cells (Figure 1A). ERK was dramatically activated by BDNF in cells transfected with the receptor tyrosine kinase TrkB, confirming the fidelity of our ERK assay. Moreover, GRASP-1 constructs had no effect on BDNF-induced ERK activation (Figure 1B).
Figure 1.
GRASP-1 activates the JNK signaling pathway. (A) GRASP-1 does not activate ERK signaling. The histogram shows the quantification of results from ERK assay on immunoprecipitates of HA-ERK1. HEK293T cells were transfected with HA-tagged ERK1 together with empty vector pRK5, a plasmid encoding the receptor tyrosine kinase TrkB, the region of GRASP-1 upstream of the caspase-cleavage site (N), the region of GRASP-1 downstream of the caspase-cleavage site (C) or GRASP-1 wild-type full-length (w.t.). TrkB-transfected cells were treated with 50 ng/ml purified BDNF for 10 minutes prior to the assay. Kinase activity towards the ERK substrate MBP was analyzed following immunoprecipitation with HA antibody and is plotted relative to control (empty vector transfected) cells. Western blotting analysis with anti-HA antibody confirmed similar amounts of HA-ERK1 in immunoprecipitates (data not shown). (B) GRASP-1 does not enhance TrkB-mediated ERK activation. HEK293T cells transfected with HA-tagged ERK1 and TrkB cDNAs together with empty vector pRK5, or GRASP-1-N, GRASP-1-C or GRASP-1 wild-type full-length (w.t.) cDNAs. Every dish of transfected cells was treated with 50 ng/ml purified BDNF for 10 minutes prior to lysis and assay as in (A). (C) GRASP-1 activates JNK signaling. HA immunoprecipitation and subsequent kinase assay using GST-cJun as substrate was performed using lysates from HEK293T cells co-transfected with HA-JNK1 plus the GRASP-1 constructs described in (A). One dish of cells transfected with empty vector was treated with 0.25 μg/ml nocodazole for 1 hour and used as positive control for JNK activation. (D) Time course of JNK phosphorylation at Thr183/Tyr185 promoted by GRASP-1 C-terminal fragment (GRASP1-C). HEK293T cells were transfected with either empty plasmid vector or GRASP1-C plasmids. Cell lysates were collected at different time points following transfection as indicated. Representative Western blot is shown for 36 hrs after transfection. The plot shows the levels of JNK phosphorylation that are normalized to the levels of JNK protein.
Surprisingly, when similar assays were performed to assess activation of JNK, a MAP kinase homologous to ERK, we found that transfected HA-JNK1 was activated more than 10-fold in GRASP-1 transfected cells compared to cells transfected with empty vector. Furthermore, the GRASP1-C fragment was sufficient for this activation (Figure 1C), while expression of the N-terminal fragment of GRASP-1 (GRASP1-N, which contains the RasGEF domain) did not lead to JNK activation.
To explore the mechanism of GRASP-1-induced JNK pathway activation, we next examined whether the phosphorylation of JNK at threonine 183 (Thr183) and tyrosine 185 (Tyr185) was enhanced by GRASP-1 expression. Phosphoryation of Thr183 and Tyr185 by SEK1/MKK4, which is in turn phosphorylated and activated by several kinases including MEKK1, is known to activate JNK1/2/3 [4, 15]. We found that expression of GRASP1-C robustly increased JNK phosphorylation (Figure 1D). The GRASP-1- induced increase in JNK phosphorylation was detectable 12 hours after transfection and reached a plateau about 36 hrs after transfection (Figure 1D).
These results suggest that the region of GRASP-1 downstream of the Caspase-3-cleavage site is capable of activating the JNK signaling pathway by enhancing the phosphorylation of JNK.
GRASP-1 interacts with MEKK1 and JNK1
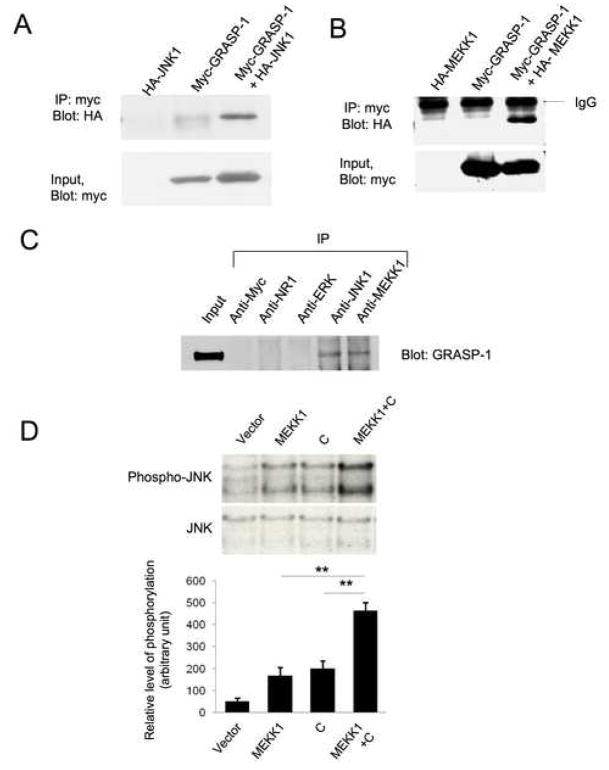
Scaffold proteins, such as JIP-1, activate the JNK pathway by bringing upstream and downstream kinases in these cascades into close proximity through protein-protein interactions [20, 21]. We wondered whether the ability of GRASP-1 to activate JNK signaling might be due to its ability to act as a scaffold of this type and therefore examined whether JNK directly interacts with GRASP-1. Indeed, JNK1 was clearly detected in myc immunoprecipitates from HEK293T cells co-expressing myc-GRASP-1 and HA-JNK1, but not from cells expressing HA-JNK1 alone (Figure 2A). Since JIP1-like scaffolds also bind upstream JNK pathway kinases [20] we also examined the ability of GRASP-1 to interact with MEKK1, a well-characterized JNK pathway ‘upstream’ kinase [4, 15]. HEK293T cells were therefore transfected with HA-tagged MEKK1, Myc-tagged GRASP1-C, or these two constructs together, and immunoprecipitations were performed from cell lysates using anti-HA antibody. GRASP1-C was detected in HA immunoprecipitates only in lysates from cells co-transfected with HA-MEKK1 and GRASP1-C (Figure 2B). These experiments demonstrated that GRASP-1 interacts with two key kinases in the JNK pathway. To examine whether GRASP-1 interacts with MEKK1 and JNK1 in neurons, co-immunoprecipitation experiments were performed with detergent-solubilized extracts from cultured cortical neurons (Figure 2C). Both anti-JNK1 and anti-MEKK1 antibodies immunoprecipitated GRASP-1 from neuronal lysates while antibodies against Myc, against the NR1 subunit of NMDA receptors or against ERK1 did not immunoprecipitate GRASP-1. These results suggest that GRASP-1 interacts with MEKK1 and JNK1 in neurons.
Figure 2.
GRASP-1 interacts with MEKK1 and JNK1. (A) JNK1 co-immunoprecipitates with GRASP-1 from transfected HEK293T cells. HEK293T cells were co-transfected with Myc-tagged GRASP-1 and HA-tagged JNK1 or Myc-GRASP-1 alone or HA-MEKK1 alone. Lysates were immunoprecipitated with anti-myc antibody and the immunoprecipitates were analyzed by immunoblotting with anti-HA antibody. A fraction of lysate from each transfection (Input) was examined for expression of Myc-GRASP-1. (B) MEKK1 co-immunoprecipitates with GRASP-1 from transfected HEK293T cells. HEK293T cells were co-transfected with Myc-tagged GRASP-1 and HA-tagged kinase domain of MEKK1 or Myc-GRASP-1 alone or HA-MEKK1 alone. Lysates were immunoprecipitated with anti-myc antibody and the immunoprecipitates were analyzed by immunoblotting with anti-HA antibody. A fraction of lysate from each transfection (Input) was examined for expression of Myc-GRASP-1. (C) GRASP-1 is present in MEKK1 and JNK1 immunoprecipitates from cortical neuronal cultures. Immunoprecipitation from cortical neuronal lysates was performed with the indicated antibodies and the immunoprecipitates were examined for the presence of GRASP-1. (D) GRASP1-C enhances MEKK1-induced JNK phosphorylation. The upper panel shows a representative Western blot. The histogram shows the quantification of results from 3 independent experiments. **: p<0.01 (t-test).
GRASP-1 is a scaffold protein for the JNK signaling pathway
While the results above suggested that GRASP-1 can bind JNK1 and MEKK1 in both heterologous cells and neurons, they did not address whether GRASP-1 acts as a JNK pathway scaffold and facilitates activation of JNK signaling. To begin to explore this possibility, we examined whether co-expression of GRASP-1 augments JNK activation by MEKK1 (which when transfected alone is known to activate JNK signaling [4, 15]). Indeed, HEK293T cells transfected with the C-terminal fragment of GRASP-1 together with MEKK1 displayed a greater degree of JNK activation than cells transfected with either construct alone (Figure 2D), consistent with the hypothesis that GRASP-1 acts as a scaffold protein for JNK pathway kinases.
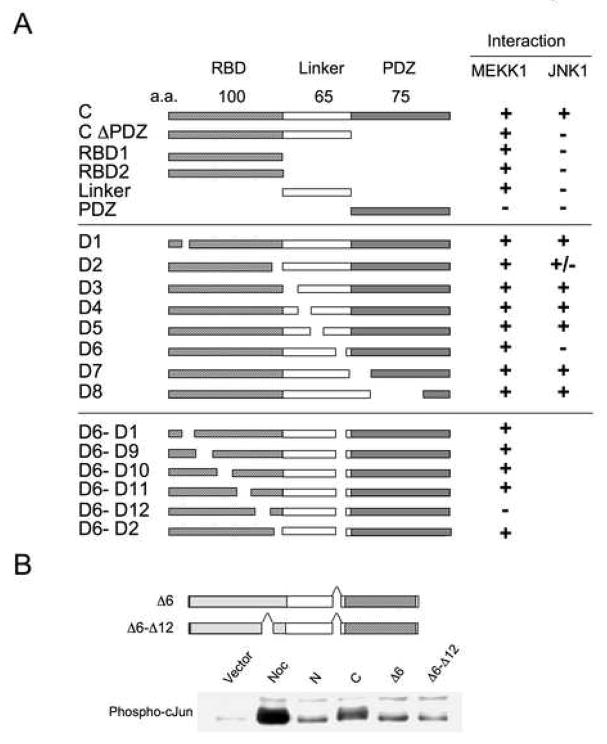
An alternative interpretation of this result, however, is that GRASP-1 and MEKK1 activate JNK via separate parallel pathways. To distinguish between these possibilities, we mapped the regions of GRASP-1 that are required for MEKK1 and JNK1 binding. These MEKK1- and JNK-interaction sites were mapped by co-immunoprecipitation using serial truncation and internal deletion mutants of GRASP-1 (Figure 3A). Deletion of a 16 amino acid sequence within the linker region of GRASP1 (D6) abolished the interaction with JNK1. MEKK-1 required two sites in the C-terminal fragment of GRASP-1, one within the RBD and second in the D6 linker region, and the interaction was maintained until both D6 and a 13 amino acid sequence (D12) were deleted (Fig 3A).
Figure 3.
GRASP-1 serves as a scaffold protein for the JNK signaling pathway. (A) Mapping of the MEKK1- and JNK1-binding sites in GRASP-1. MEKK1- and JNK-interaction sites were mapped by coimmunoprecipitation studies using HEK293T cells co-transfected with either HA-MEKK1 or HA-JNK1 plus the indicated truncation or internal deletion mutants of GRASP-1. The D6 deletion abolished the interaction with JNK1. MEKK-1 appeared to interact with two sites in the C-terminal fragment and the interaction existed until both D6 and D12 were deleted. (B) GRASP-1 mutants deficient in JNK (Δ6) and/or MEKK1 binding (Δ6–Δ12) do not activate JNK. Western analysis was performed with anti-phospho-Jun antibody on lysates from HEK293T cells transfected with empty vector pRK5, GRASP1-N, GRASP1-C, GRASP1-CΔ6 or GRASP1-CΔ6–Δ12. Nocodazole-treatment on cells transfected with pRK5 was used as a positive control.
The GRASP-1 deletion mutants were then tested for their ability to activate JNK. Interestingly, both D6 and D6–D12 mutants of GRASP-1 caused greatly reduced activation of JNK compared to that caused by the C-terminal fragment (Figure 3B), suggesting that JNK pathway activation by GRASP-1 requires interactions of GRASP-1 with JNK and MEKK1. These results support the hypothesis that GRASP-1 serves as a scaffold protein that can lead to the activation of the JNK signaling pathway
Caspase-3-cleavage is required for GRASP-1-mediated JNK pathway activation
GRASP-1 is one of the few neuron-specific substrates of Caspase-3. The cleavage of GRASP-1 by Caspase-3 releases the 30 kDa C-terminal fragment (GRASP1-C) from the rest of the protein [1,3]. This cleavage event occurs in both normal brain and ischemic brain [3]. Western blot analysis with antibodies against the C-terminal region of GRASP-1 suggests that a significant amount of the GRASP1-C fragment is present in normal brain [1] as well as in HEK293T cells (data not shown). However, the functional significance of this cleavage is unknown. Since the JNK pathway-activating activity of GRASP-1 is unique to the GRASP1-C fragment, we wondered whether Caspase-3-cleavage of GRASP-1 played a role in regulating this activity.
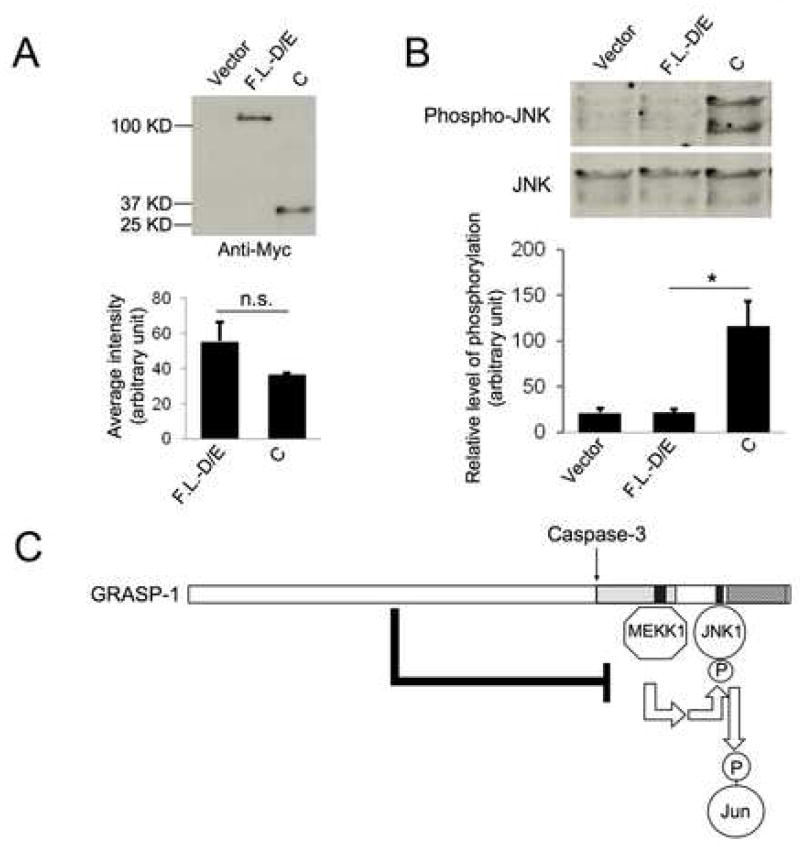
To examine this possibility, we compared the level of JNK phosphorylation in cells expressing GRASP1-C and those expressing a full length GRASP-1 that is resistant to Caspase-3 cleavage due to mutations in the cleavage site (F.L.-D/E) [1]. The amount of Myc-tagged GRASP1-C and GRASP-F.L.-D/E cDNA that was transfected was adjusted so these two proteins were expressed at comparable levels (Figure 4A). Strikingly, while GRASP1-C dramatically enhanced JNK phorphorylation, GRASP1-F.L-D/E did not significantly alter JNK phosphorylation (Figure 4B).
Figure 4.
Caspase-3-cleavage is required for GRASP-1-mediated JNK pathway activation. (A–B) Full length GRASP-1 containing a mutated Caspase-3 cleavage site (F.L.-D/E) is a much less effective activator for JNK signaling. (A) Western blot and quantification show comparable expression levels of Myc-GRASP1-F.L.-D/E and Myc-GRASP1-C. n.s: not significant. N=3. (B) GRASP1-C but not GRASP1-F.L.-D/E enhances JNK phosphorylation. *: p<0.05 (t-test), n=3. (C) A model that summarizes our findings that GRASP-1 serves as a scaffold protein for JNK signaling pathway.
Taken together, these results suggest that full length GRASP-1 does not enhance JNK pathway activity, possibly due to the inhibitory effect of the N-terminal fragment on the C-terminal fragment. In contrast, Caspase-3 cleavage of GRASP-1releases the C-terminal fragment, which in turn activates JNK signaling by serving as a scaffold protein (Figure 4C).
DISCUSSION
In this study we have demonstrated that GRASP-1 serves as a neuronal scaffold protein for the JNK signaling pathway. We have also provided evidence that this function of GRASP-1 is regulated by Caspase-3.
Although GRASP-1 acts as a RasGEF in vitro [1], we did not observe GRASP-1-dependent activation of ERK (a well known ‘downstream’ target of Ras) in HEK293T cells. There are several possible explanations for this finding. Firstly, the Ras superfamily of small G proteins contains many members and the specific Ras that is regulated by GRASP-1 in vivo may not be expressed in HEK293T cells. Furthermore, a significant fraction of GRASP-1 localizes to specific intracellular vesicles [1] that are likely to be endosomes [23]. Different small G proteins localize to specific cellular membranes [24] and it is possible that in HEK293T cells the localization of GRASP-1 is distinct from that of Ras family proteins that GRASP-1 has the ability to regulate in vitro. A further possibility is that additional GRASP-1 binding partners (including JNK pathway kinases) might sterically obscure the Ras binding domain (RBD) of GRASP-1 in cells, thus limiting GRASP-1’s ability to function as a RasGEF. It will be interesting to investigate whether stimuli that induce changes in GRASP-1 localization and/or regulate GRASP-1’s interactions with its binding partners might differentially modulate the ability of GRASP-1 to modulate Ras, JNK and AMPA receptors.
Accumulating evidence suggests that glutamate receptors activate signal transduction through kinase cascades in neurons. For example, glutamate treatment [9] and membrane depolarization [10,25] activate ERK signaling through Ras in a calcium-dependent manner. In contrast, JNK activity in neurons is constitutively high [11] but specific pools of JNK can respond to certain stimuli [12]. Subcellular fractionation experiments reveal GRASP-1 immunoreactivity in both post-synaptic density and cytosolic fractions [1]. Whether GRASP-1/JNK pathway complexes contribute to synaptic or cytosolic JNK activity, or to both is an intriguing problem to solve in future.
We note that JNK plays many roles in neurons in different subcellular locations and is known to interact with a variety of scaffold proteins. It is therefore not surprising that only a small fraction of the total pool of JNK is complexed with GRASP-1 in neurons (Figure 2C). However, even if JNK-GRASP-1 complexes comprise only a minor fraction of total JNK scaffold complexes, JNK-GRASP-1 complexes might still play a critical role in certain subcellular locations, or in response to particular stimuli.
One set of stimuli that might regulate JNK-MEKK1-GRASP-1 complexes are those that induce Caspase activation. We have previously demonstrated that GRASP-1 is a direct Caspase-3 substrate [1, 3]. Here we demonstrate that the caspase-3 cleavage of GRASP-1 is necessary for the ability of GRASP-1 to enhance JNK pathway activity.
Interestingly, MEKK-1 is also a substrate of Caspase-3 and cleavage of MEKK-1 by Caspase-3 results in subsequent kinase activation [26]. Not only are both MEKK-1 and GRASP-1 Caspase-3 substrates, the cleavage sites of these two proteins are more homologous to each other than to any other Caspase-3 substrates, suggesting that Caspase-3 may bind GRASP-1 and MEKK-1 with similar affinity, and that their cleavage might be triggered under similar conditions. Cleavage of MEKK-1 by Caspase-3 is required for the apoptosis induced by loss of integrin-mediated contacts with the extracellular matrix (“anoikis”) in MDCK cells [27] and genotoxin-induced apoptosis in HEK293 cells [28]. Roles for MEKK-1 have also been reported in apoptosis of neuronally-derived cell lines [29, 30]. The functions of MEKK-1 can be either JNK-dependent or JNK-independent. For example, apoptosis mediated by MEKK-1 in rat PC12 pheochromocytoma cells is JNK-dependent, while MEKK-1-mediated apoptosis in Swiss 3T3 cells is JNK-independent [30, 31]. In neurons, prolonged activation of AMPA receptors leads to JNK pathway activation (B.Y. and R.L.H, unpublished results) and delayed neuronal cell death [32]. In addition, multiple forms of neuronal cell death including cerebral ischemia and kainate-induced excitotoxicity require JNK [17, 19]. Given the fact that GRASP-1 scaffolds JNK with MEKK1, and MEKK1 is an apoptosis-inducing kinase, it is conceivable that the JNK activation facilitated by GRASP-1 has apoptotic effects. It is an intriguing possibility that during neuronal apoptosis, JNK pathway activity may be enhanced not only by the activation of MEKK1 by Caspase-3 cleavage but also by the release of the GRASP-1 C-terminal fragment, which then serves as a scaffold protein for the JNK signaling pathway.
Acknowledgments
We thank Dr. David Ginty for insightful discussion and Elizabeth Hwang and Kate Searcy-Goldberg for technical assistance. This work was supported by funding from Howard Hughes Medical Institute and NINDS (5RO1NS036715-10, to R.L.H.)
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Ye B, Liao D, Zhang X, Zhang P, Dong H, Huganir RL. GRASP-1: a neuronal RasGEF associated with the AMPA receptor/GRIP complex. Neuron. 2000;26:603–617. doi: 10.1016/s0896-6273(00)81198-8. [DOI] [PubMed] [Google Scholar]
- 2.Dong H, O’Brien RJ, Fung ET, Lanahan AA, Worley PF, Huganir RL. GRIP: a synaptic PDZ domain-containing protein that interacts with AMPA receptors. Nature. 1997;386:279–284. doi: 10.1038/386279a0. [DOI] [PubMed] [Google Scholar]
- 3.Ye B, Sugo N, Hurn PD, Huganir RL. Physiological and pathological caspase cleavage of the neuronal RasGEF GRASP-1 as detected using a cleavage site-specific antibody. Neuroscience. 2002;114:217–227. doi: 10.1016/s0306-4522(02)00142-2. [DOI] [PubMed] [Google Scholar]
- 4.Pearson G, Robinson G, Beers Gibson T, Xu BE, Karandikar M, Berman K, Cobb MH. Mitogen-activated protein (MAP) kinase pathways: regulation and physiological functions. Endocr Rev. 2001;22:153–83. doi: 10.1210/edrv.22.2.0428. [DOI] [PubMed] [Google Scholar]
- 5.Thomas GM, Huganir RL. MAPK cascade signalling and synaptic plasticity. Nat Rev Neurosci. 2004;5:173–183. doi: 10.1038/nrn1346. [DOI] [PubMed] [Google Scholar]
- 6.Zhu JJ, Qin Y, Zhao M, Van Aelst L, Malinow R. Ras and Rap control AMPA receptor trafficking during synaptic plasticity. Cell. 2002;110:443–455. doi: 10.1016/s0092-8674(02)00897-8. [DOI] [PubMed] [Google Scholar]
- 7.Chen JT, Lu DH, Chia CP, Ruan DY, Sabapathy K, Xiao ZC. Impaired long-term potentiation in c-Jun N-terminal kinase 2-deficient mice. J Neurochem. 2005;93:463–473. doi: 10.1111/j.1471-4159.2005.03037.x. [DOI] [PubMed] [Google Scholar]
- 8.Zhu Y, Pak D, Qin Y, McCormack SG, Kim MJ, Baumgart JP, Velamoor V, Auberson YP, Osten P, van Aelst L, Sheng M, Zhu JJ. Rap2-JNK removes synaptic AMPA receptors during depotentiation. Neuron. 2005;46:905–916. doi: 10.1016/j.neuron.2005.04.037. [DOI] [PubMed] [Google Scholar]
- 9.Bading H, Greenberg ME. Stimulation of protein tyrosine phosphorylation by NMDA receptor activation. Science. 1991;253:912–914. doi: 10.1126/science.1715095. [DOI] [PubMed] [Google Scholar]
- 10.Wu GY, Deisseroth K, Tsien RW. Spaced stimuli stabilize MAPK pathway activation and its effects on dendritic morphology. Nat Neurosci. 2001;4:151–158. doi: 10.1038/83976. [DOI] [PubMed] [Google Scholar]
- 11.Xu X, Raber J, Yang D, Su B, Mucke L. Dynamic regulation of c-Jun N-terminal kinase activity in mouse brain by environmental stimuli. Proc Natl Acad Sci USA. 1997;94:12655–12660. doi: 10.1073/pnas.94.23.12655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Coffey ET, Smiciene G, Hongisto V, Cao J, Brecht S, Herdegen T, Courtney MJ. c-Jun N-terminal protein kinase (JNK) 2/3 is specifically activated by stress, mediating c-Jun activation, in the presence of constitutive JNK1 activity in cerebellar neurons. J Neurosci. 2002;22:4335–4345. doi: 10.1523/JNEUROSCI.22-11-04335.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Davis RJ. Signal transduction by the JNK group of MAP kinases. Cell. 2000;103:239–252. doi: 10.1016/s0092-8674(00)00116-1. [DOI] [PubMed] [Google Scholar]
- 14.Hagemann C, Blank JL. The ups and downs of MEK kinase interactions. Cell Signal. 2001;13:863–875. doi: 10.1016/s0898-6568(01)00220-0. [DOI] [PubMed] [Google Scholar]
- 15.Yan M, Dai TC, Deak JC, Kyriakis JM, Zon LI, Woodgett JR, Templteon DJ. Activation of stress-activated protein kinase by MEKK1 phosphorylation of its activator SEK1. Nature. 1994;372:798–800. doi: 10.1038/372798a0. [DOI] [PubMed] [Google Scholar]
- 16.Yujiri T, Ware M, Widmann C, Oyer R, Russell D, Chan E, Zaitsu Y, Clarke P, Tyler K, Oka Y, Fanger GR, Henson P, Johnson GL. MEK kinase 1 gene disruption alters cell migration and c-Jun NH2-terminal kinase regulation but does not cause a measurable defect in NF-kappa B activation. Proc Natl Acad Sci USA. 2000;97:7272–7277. doi: 10.1073/pnas.130176697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Waetzig V, Herdegen T. Neurodegenerative and physiological actions of c-Jun N-terminal kinases in the mammalian brain. Neurosci Lett. 2004;361:64–67. doi: 10.1016/j.neulet.2004.02.041. [DOI] [PubMed] [Google Scholar]
- 18.Bjorkblom B, Ostman N, Hongisto V, Komarovski V, Filen JJ, Nyman TA, Kallunki T, Courtney MJ, Coffey ET. Constitutively active cytoplasmic c-Jun N-terminal kinase 1 is a dominant regulator of dendritic architecture: role of microtubule-associated protein 2 as an effector. J Neurosci. 2005;25:6350–6361. doi: 10.1523/JNEUROSCI.1517-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang DD, Kuan CY, Whitmarsh AJ, Rincon M, Zheng TS, Davis RJ, Rakic P, Flavell RA. Absence of Excitotoxicity-induced Apoptosis in the Hippocampus of Mice Lacking the Jnk3 gene. Nature. 1997;389:865–870. doi: 10.1038/39899. [DOI] [PubMed] [Google Scholar]
- 20.Whitmarsh AJ, Cavanagh J, Tournier C, Yasuda Y, Davis RJ. A mammalian scaffold complex that selectively mediates MAP kinase activation. Science. 1998;281:1671–1674. doi: 10.1126/science.281.5383.1671. [DOI] [PubMed] [Google Scholar]
- 21.Dard N, Peter M. Scaffold proteins in MAP kinase signaling: more than simple passive activating platforms. Bioessays. 2006;28:146–156. doi: 10.1002/bies.20351. [DOI] [PubMed] [Google Scholar]
- 22.Xu S, Robbins DJ, Christerson LB, English JM, Vanderbilt CA, Cobb MH. Cloning of rat MEK kinase 1 cDNA reveals an endogenous membrane-associated 195-kDa protein with a large regulatory domain. Proc Natl Acad Sci USA. 1996;93:5291–5295. doi: 10.1073/pnas.93.11.5291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stinton LM, Selak S, Fritzler MJ. Identification of GRASP-1 as a novel 97 kDa autoantigen localized to endosomes. Clin Immunol. 2005;116:108–117. doi: 10.1016/j.clim.2005.03.021. [DOI] [PubMed] [Google Scholar]
- 24.Chiu VK, Bivona T, Hach A, Sajous JB, Silletti J, Wiener H, Johnson RL, 2nd, Cox AD, Philips MR. Ras signalling on the endoplasmic reticulum and the Golgi. Nat Cell Biol. 2002;4:343–350. doi: 10.1038/ncb783. [DOI] [PubMed] [Google Scholar]
- 25.Rosen LB, Ginty DD, Weber MJ, Greenberg ME. Membrane Depolarization and Calcium Influx Stimulate MEK and MAP Kinase via Activation of Ras. Neuron. 1994;12:1207–1221. doi: 10.1016/0896-6273(94)90438-3. [DOI] [PubMed] [Google Scholar]
- 26.Widmann C, Gerwins P, Johnson NL, Jarpe MB, Johnson GL. MEK kinase 1, a substrate for DEVD-directed caspases, is involved in genotoxin-induced apoptosis. Mol Cell Biol. 1998;18:2416–2429. doi: 10.1128/mcb.18.4.2416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cardone MH, Salvesen GS, Widmann C, Johnson G, Frisch SM. The regulation of anoikis: MEKK-1 activation requires cleavage by caspases. Cell. 1997;90:315–323. doi: 10.1016/s0092-8674(00)80339-6. [DOI] [PubMed] [Google Scholar]
- 28.Kim KH, Min YK, Baik JH, Lau LF, Chaqour B, Chung KC. Expression of angiogenic factor Cyr61 during neuronal cell death via the activation of c-Jun N-terminal kinase and serum response factor. J Biol Chem. 2003;278:13847–13854. doi: 10.1074/jbc.M210128200. [DOI] [PubMed] [Google Scholar]
- 29.Wood JL, Russo AF. Autoregulation of cell-specific MAP kinase control of the tryptophan hydroxylase promoter. J Biol Chem. 2001;276:21262–21271. doi: 10.1074/jbc.M007520200. [DOI] [PubMed] [Google Scholar]
- 30.Xia Z, Dickens M, Raingeaud J, Davis RJ, Greenberg ME. Opposing effects of ERK and JNK-p38 MAP kinases on apoptosis. Science. 1995;270:1326–1331. doi: 10.1126/science.270.5240.1326. [DOI] [PubMed] [Google Scholar]
- 31.Johnson NL, Gardner AM, Diener KM, Lange-Carter CA, Gleavy J, Jarpe MB, Minden A, Karin M, Zon LI, Johnson GL. Signal transduction pathways regulated by mitogen-activated/extracellular response kinase kinase kinase induce cell death. J Biol Chem. 1996;271:3229–3237. doi: 10.1074/jbc.271.6.3229. [DOI] [PubMed] [Google Scholar]
- 32.Koh JY, Goldberg MP, Hartley DM, Choi DW. Non-NMDA receptor-mediated neurotoxicity in cortical culture. J Neurosci. 1990;10:693–705. doi: 10.1523/JNEUROSCI.10-02-00693.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]