Abstract
PURPOSE
To determine the existence of a relatively higher abundance of potential TFs in glaucomatous trabecular meshwork (TM) that may bind putative promoter regions and affect cochlin protein expression in glaucomatous compared to normal TM.
METHODS
Combinatorial bioinformatics and biochemical analyses, using human glaucomatous and normal donor tissue (n = 4 each). Biochemical analysis included electrophoretic mobility shift assays (EMSAs), filter binding assays (FBAs), coupled in vitro transcription–translation (TNT) assays and promoter mutation analysis.
RESULTS
Combinatorial bioinformatics and biochemical analyses revealed the existence of a higher abundance of TFs in glaucomatous than in normal TM nuclear extracts. The evidence of a relatively high abundance of TFs, leading to increased expression of cochlin predicted by bioinformatic and biochemical analyses (EMSA and FBA), was further supported by TNT and promoter mutation TNT assays.
CONCLUSIONS
These results support the finding that the observed increased cochlin expression in glaucomatous TM is due to relative elevated abundance of TFs. The results also demonstrate the utility of combinatorial bioinformatic and biochemical analyses for genes with uncharacterized promoter regions.
Glaucoma is a group of irreversible blinding eye diseases associated with optic neuropathy. Primary open-angle glaucoma (POAG) is often associated with elevated intraocular pressure (IOP), which is due to an imbalance between aqueous humor production and outflow in the anterior chamber of the eye.1 Aqueous humor is a clear liquid produced by the ciliary epithelium that exits through the trabecular meshwork (TM) after bathing the anterior segment structures, such as the cornea and lens, with nutrients. Aqueous outflow is believed to encounter increased resistance at the level of the TM in glaucoma. The mechanisms that impede aqueous outflow elevate IOP are poorly understood.
Cochlin, a secretory extracellular matrix (ECM) protein of unknown function, was identified by proteomic analyses to be differentially expressed in glaucomatous compared with normal TM.2 Cochlin is the product of the COCH gene3 located on human chromosome 14, region q12–13.3,4 The cochlin protein sequence is highly conserved, with 94% and 79% amino acid identity with human to mouse and chicken sequences, respectively. 5 Cochlin contains a short signal peptide, an N-terminal factor C homology, and two von Willebrand factor A-like domains. 2,4 In situ hybridization has shown that cochlin mRNA is expressed in the TM, suggesting that the protein is likely expressed and deposited locally.2
Elevated IOP is a significant risk factor for optic nerve damage. Changes in fluid dynamics and incremental fluctuations in IOP results in stress and stretch on TM cells and are thought to trigger early biochemical responses.6 Stress- and stretch-induced modulation of protein expression are mediated by transcription factors (TFs).6,7
Increased cochlin expression has been reported in the TM8 and in the inner ear4; however, the promoter region of cochlin and the details of cochlin gene expression remain to be characterized. Deciphering mechanisms that lead to transcriptional regulation of cochlin expression in the TM is critical for understanding the role cochlin may play in glaucoma’s pathogenesis. We used a combinatorial approach of bioinformatics and molecular and biochemical analyses to determine whether an increased abundance of transcription factors with the potential to bind and enhance transcription in the promoter region of cochlin was present in nuclear extracts of glaucomatous TM compared with the control.
MATERIAL AND METHODS
Tissue Procurement and Preparation of Nuclear Extracts
Glaucomatous and normal control eyes were obtained from the National Disease Research Institute (Philadelphia, PA) and the Lions Eye Bank (Miami, FL), respectively. The eyes had been enucleated within 10 hours of death and placed in a moisture chamber at 4°C and transported. They were dissected within 48 hours, and the TM was carefully excised for study. The available details of the donor were recorded. According to available information, all glaucomatous donor eyes had POAG (see Supplementary Table S1; all Supplementary Tables are online at http://www.iovs.org/cgi/content/full/50/7/3106/DC1)
Bioinformatic Analyses
The human cochlin upstream promoter gene region was analyzed up to 5000 bp upstream of the translational start site (ATG; accession number BC007230; National Center for Biotechnology Information [NCBI], Bethesda, MD). The cochlin DNA sequence was obtained from the UCSC genome browser (http://genome.ucsc.edu/ provided in the public domain by the University of California at Santa Cruz). Putative TF binding sites were identified with commercial software (MatInspector; Matrix Family Library Version 6.3; Genomatix, Munich, Germany). The analysis parameters used have been provided in respective tables. TFs related to the eye (see Supplementary Table S2) were further investigated for their correlation with glaucoma, the TM, the anterior chamber, or the eye, as reported in the literature and for its expression in the eye per the UniGene database (http://www.ncbi.nlm.nih.gov/UniGene; provided in the public domain by NCBI; see Supplementary Table S3). Tissue-specific associations of TFs in putative cochlin promoter regions for ear, brain, central nervous system (CNS), embryonic tissue and liver were also analyzed (MatInspector; Genomatix), and a list of tissue-specific TFs was generated (not shown). Only those TFs present in at least two search terms were investigated further. For the selected TFs, 5′ biotin end-labeled oligonucleotide sequences were generated for the relevant consensus binding sites as well as for their respective complementary sequences. These results were confirmed and/or cross-validated with other bioinformatic Web-based programs including the International HapMap Project (www.hapmap.org), a commercial bioinformatic database (Transfac; www.biobase-international.com; provided at a cost by Biobase Biological Databases, Wolfenbuettel Germany).
Nuclear Extract Preparation
TM nuclear protein extracts were obtained from glaucomatous and normal tissue (NE-PER Nuclear and Cytoplasmic Extraction Reagents kit, cat. no. 78833; Pierce Biotechnology, Rockford, IL) according to protocols recommended by the manufacturer. The proteins recovered were quantified spectrophotometrically using the Bradford protein assay and subsequently aliquoted for use or stored at −80°C for future analysis. All protein aliquots were either used immediately or subjected to only one freeze–thaw cycle.
Western Blot Analyses
Approximately 10 µg of nuclear protein extract was fractionated on 4% to 20% tris-glycine polyacrylamide gradient gels (Invitrogen, Carlsbad, CA), transferred onto polyvinylidene fluoride (PVDF) membranes and incubated overnight at 4°C with the appropriate antibodies (~5 µg/mL). Before antibody incubation, blots were blocked with 5% milk (cat no. 170–6404; Bio-Rad Laboratories, Hercules, CA). Rabbit polyclonal antibodies against Nrf2, FoxC1, FoxQ1 (cat. nos. ART38754_T100, ARP32300_T100, ARP39754_T100; Aviva Systems Biology, San Diego, CA, respectively), and Brn3a (cat. no. AB5945; Chemicon Inc., Temecula, CA), as well as a goat polyclonal antibody against Brn3b (cat. no. sc-6026; Santa Cruz Biotechnology Inc., Santa Cruz, CA) were used. The Pax6 protein was detected using previously published rabbit antisera against a 17-residue C-terminal mouse Pax6 peptide.9,10 All primary antibodies were detected with the appropriate horseradish peroxidase–conjugated secondary antibodies and ECL (cat. no. 32106; Pierce Biotechnology).
Gel Mobility Shift Experiments
The nonradioactive light shift chemiluminescent electrophoretic mobility shift assay (EMSA) kit (cat. no. 20148; Pierce Biotechnology) and 5′ biotin end-labeled oligonucleotides were used according to the manufacturer’s recommended protocol to detect DNA-TF interactions. All identified TF consensus sites were used to generate oligonucleotides for further analysis, except Pax6, for which only a subset of oligonucleotides were generated and used (Table 1). The oligonucleotide sequences were annealed to the respective complementary oligonucleotides. Nuclear extracts (3.5 µg) were incubated with 0.25 picomoles of double-stranded, biotin-labeled oligonucleotide, 1 × binding buffer, 1 M KCl, 100 mM MgCl2, 200 mM EDTA, 50% glycerol, 50 ng poly-dI-dC, and 1% NP-40, in a total volume of 20 µL for 20 minutes at room temperature. To determine binding specificity, supershift analysis was performed with the addition of 1 µg of antibody to select samples after 20 minutes of binding reaction and incubated for an additional 30 minutes at room temperature. Antibodies used were anti-Nrf2, anti-FoxC1, anti-FoxQ1 (Aviva Systems Biology, San Diego, CA) anti-Pax6,10,11 and anti-Brn3b (Santa Cruz Biotechnology). Specific and nonspecific competitions were performed by using nonbiotiny-lated oligonucleotide of specific sequences and poly-dI-dC, respectively (data not shown). The samples were then run on a 6% DNA retardation gel (cat. no. EC6252BOX; Invitrogen, Carlsbad, CA) at 100 V for 90 minutes. The gels were electrophoretically transferred at a constant current of 380 mA for 1 hour on ice to a positively charged nylon membrane (cat. no. 77016; Pierce Biotechnology) and the membrane was immediately UV cross-linked for 60 seconds at 120 mJ/cm2 in a UV transilluminator equipped with 254-nm bulbs. Streptavidin-horseradish peroxidase conjugate and the light shift chemiluminescent substrate were used to detect the biotin end-labeled DNA. Nylon membranes were then exposed to X-ray film for detection.
TABLE 1.
Relative Abundance of Select Transcription Factors According to the Filter Binding Assay for Consensus Binding Sites in the Cochlin Upstream Region
| Transcription Factor | Oligonucleotide Sequences (Consensus Binding Sites) |
Reported Association with | |||||
|---|---|---|---|---|---|---|---|
| Filter Binding Assay | |||||||
| Glaucoma | TM | Ant. Chamber |
Eye | Normal | Glaucomatous | ||
| Fork head–related activator-3 (FoxC1) | ataaaGTAAaaaaagac | + | + | + | + | 25.4 ± 0.7 | 17.6 ± 2.1 |
| aaaaaGTAAaaaatgag | + | + | + | + | 22.0 ± 1.0 | 18.3 ± 0.5 | |
| gccatGAAAataaacat | + | + | + | + | 23.7 ± 0.6 | 17.3 ± 0.5 | |
| Paired domain and homeodomain (Pax6)* | cggcgacttCCAGctccgc | + | + | + | + | 29.2 ± 2.8 | 29.5 ± 2.3 |
| cccgctgctCCAGgccagc | + | + | + | + | 25.8 ± 0.7 | 24.7 ± 0.6 | |
| POU-IV protein (Brn3) | ctatgatagATTAtagagc | + | + | 26.8 ± 1.7 | 27.8 ± 2.0 | ||
| ataatagTAATtaataaca | + | + | 25.4 ± 0.7 | 27.1 ± 1.0 | |||
| ttgttatTAATtactatta | + | + | 25.4 ± 0.7 | 32.2 ± 2.9 | |||
| ttcatctTAATtattttgt | + | + | 24.7 ± 0.6 | 30.9 ± 1.8 | |||
| HNF-3/Fkh homolog 1 (FoxQ1) | cctataTAAActaagag | + | + | 22.7 ± 2.1 | 28.8 ± 1.7 | ||
| acaataTAAActttttc | + | + | 26.8 ± 1.7 | 31.9 ± 1.8 | |||
| Nuclear factor (erythroid-derived 2)-like 2, (Nrf2) |
gcatagttTTGActctgccaaatca | + | + | 23.4 ± 1.5 | 27.8 ± 0.7 | ||
| ttcagtgaGTGAtttggcagagtca | + | + | 26.8 ± 1.7 | 30.2 ± 1.3 | |||
| Homeobox TAAT motif-binding transcription factor (Barx2) |
aataTAATtggtctggg | + | + | 25.8 ± 1.7 | 31.2 ± 2.9 | ||
| ttatTAATtactattat | + | + | 25.8 ± 0.7 | 28.8 ± 0.7 | |||
| caaaTAATgaggccggg | + | + | 24.8 ± 1.6 | 30.5 ± 2.3 | |||
| atctTAATtattttgtt | + | + | 24.1 ± 1.1 | 29.2 ± 1.2 | |||
| aaaaTAATtaagatgaa | + | + | 26.1 ± 1.2 | 29.1 ± 1.0 | |||
| ttttTAATggttacaga | + | + | 24.7 ± 0.6 | 29.5 ± 0.8 | |||
| tataTAATtaggaagag | + | + | 28.5 ± 2.3 | 30.9 ± 3.4 | |||
Association of a TF in the literature with glaucoma. TM, anterior chamber or eye is represented by plus sign.
Several binding sites were identified as being present in the cochlin promoter region (Supplementary Table S3); however, the two oligonucleotide sequences used were arbitrarily selected for filter binding or gel mobility shift assays.
Filter Binding Assay
Filter binding assays for relative quantification of protein-DNA complexes were performed using grade 1 filters (Whatman, Florham Park, NJ) and cut precisely in the shape of 8-mm discs by using a punch. The filters were then soaked with a 0.5 × TBE buffer for at least 30 minutes, each filter disc was placed in an 8-mm column, and vacuum was applied with a vacuum station (Qiagen, Valencia, CA). The vacuum was adjusted so that the filtering rate was slow enough not to dry the filters. The samples were prepared with nuclear extracts from glaucomatous and normal TM (3.5 µg) incubated with 0.25 picomoles of double-stranded, biotin-labeled oligonucleotide, 1 × binding buffer, 1 M KCl, 100 mM MgCl2, 200 mM EDTA, 50% glycerol, 50 ng poly-dI-dC, and 1% NP-40, in a total volume of 20 µL for 20 minutes at room temperature. In the control experiments, nuclear extracts were omitted from the binding reactions. The samples were then added to the column and vacuum was applied. After the samples had passed through, the filters were washed once with the same sample volume of 0.5× TBE and air dried. Next, the filter discs were cross-linked at 120 mJ/cm2 for 60 seconds in a UV transilluminator. The detection of biotin-labeled DNA by chemiluminescence was performed according to the manufacturer’s instructions (cat. no. 20148; Pierce Biotechnology). The filters were then exposed to X-ray film from 20 seconds to 2 hours for detection.
The images obtained from gel mobility shift assays and filter binding assays on films were subjected to densitometric scan on a commercial imaging system (Alpha Innotech, San Leandro, CA) and relative quantification of densitograms were performed with the system-associated software (Alpha Ease FC; Alpha Innotech). For relative quantification, all relative calculations were performed on the same film and a relative ratio of total area was determined. For semiquantitative estimates, known band area values in the same film were used for comparison with unknowns. Some bound filters were also subjected to luminometric counting on a scintillation counter. A linear correlation was found between luminometric and film-based measurements performed for filter binding assays (data not shown).
Coupled Transcription-Translation (TNT)
A quick coupled transcription-translation system (TNT T7, cat no. L1170; Promega Corp., Madison, WI) was used for these experiments after the manufacturer’s recommendation. Cochlin BAC clone (2 µg of circular plasmid DNA; cat no. RP11-48L1 containing the region from 30444864 to 30560848 of 14q12 on chromosome 14) from Children’s Hospital Oakland Research Institute (Oakland, CA) was added to the TNT quick master mix and incubated for 90 minutes at 30°C. Glaucomatous and normal nuclear extracts depleted of polyadenylated RNA, DNA, and cochlin by incubation with oligo-dT, hydroxyapatite and anti-cochlin-bound beads, respectively, were added to the mix. For the control experiments, only the BAC clone was added with nuclear extract. Additional controls, with plasmid not related to cochlin or no template, were also added to the TNT reaction. After incubation, synthesized proteins were immediately analyzed by direct ELISA. For this purpose, a 96-well flexible PVC plate (cat no. 353912; BD Biosciences, San Jose, CA) was incubated with 100 µL of 1:1 dilution in 1× PBS of the sample at 37°C for 1 hour. For the control, equal amounts of the TNT mixture or depleted nuclear extracts were used. The plate was washed three times with 1× PBS and then blocked with 0.2% BSA at 37°C for 1 hour. Custom peptide antibody against cochlin peptide (KR LKK TPE KKT GNK DC) from cochlin coding region 147–162 designated as hCochlin12 and an alkaline phosphatase–coupled secondary antibody and a plate reader were used for detection.
TF Binding Site Mutation Analysis
A plasmid containing the cochlin coding sequence and cochlin upstream region up to 2000 bp (cochlin composite construct) was generated by DNA cloning using standard molecular biology protocols. Cochlin structural gene was subcloned from a pCDNA 3.0–based clone readily available in our laboratory, and the promoter region was cloned from the cochlin BAC clone (RP11-48L1). The PCR products of the cochlin gene and the promoter region were ligated in a final vector pQE1 (Qiagen). Briefly, the following primer pairs were used for amplification of the upstream (~2000 bp) promoter region and the cochlin structural gene: ATTACGTCAGATATCTCAAAACAAAATAATTAAG( forward); ATATTAAAGATCTGGTGACTGATAGGCT (reverse); TACATTAGATCTATGTCCGCAGCCTG (forward); and AGATTCATAGAGCTCTTATTTGCTGCATCATG (reverse). The amplification products were digested with BglII and ligated, and the resulting insert was digested with EcoRV, SacI, before its final ligation in the pQE1 vector digested with same enzymes. The presence of cochlin structural gene enabled detection and quantitation of the translation product using antibodies against cochlin. Point mutations to select TF DNA-binding sequences located in the upstream region of the cochlin gene were generated by PCR-mediated mutagenesis with a site-directed mutagenesis kit (QuickChange XL; cat. no. 200516; Stratagene, La Jolla, CA). The wild-type promoter region served as a control. A nonspecific sequence, not corresponding to any TF DNA-binding sequence, was also mutated and used as a control. All mutagenic primer pairs were HPLC-purified and contained the desired mutation to allow annealing to the same sequence on opposite strands of the plasmid. The primers were between 25 and 45 bases long, with a melting temperature of ≥78°C and a minimum GC content of 40%. The desired mutation was located in the middle of the primer and 10 to 15 bases of the original sequence were kept on both sides. The effect of the inserted mutations on cochlin expression was analyzed subsequently with coupled TNT (TNT T7 System; Promega Corp.) with equal amounts of normal TM nuclear extract– derived protein. The product of TNT was subjected to ELISA analysis using the cochlin antibody. The following primer pairs were used for mutagenesis of the indicated TF binding sites (the wild-type sequence is shown in parentheses): Brn3-Forward primer: CTTAAATGTTTCAAAACAAccccccccAGATGAATCTCAGTAAAC; Brn3-reverse primer: GTTTACTGAGATTCATCTGGGGGGGGTTGTT TTGAAACATTTAAG (CTTAAATGTTTCAAAACAAAATAATTAAGATGAATCTCAGTAAAC); Nrf2-forward primer: GAAATAGCTGCATAGTTccccCTCTGCCAAATCACTCACTG; Nrf2-reverse primer: CAGTGAGTGATTTGGCAGAGggggAACTATGCAGCTATTTC (GAAATA GCTGCATAGTTTTGACTCTGCCAAATCACTCACTG); and FOXQ1-forward primer: GGCTATTTGAAAAAGccccccTTGTACCAGAAA GGTTAGC; FOXQ1-reverse primer: GCTAACCTTTCTGGTACAAggggggCTTTTTCAAATAGCC (GGCTATTTGAAAAAGTTTATATTGTACCAGAAAGGTTAGC).
A nonspecific (NS) sequence, not known to bind any known mammalian TF, was also subjected to mutagenesis and served as a control. The mutated TF binding sequences were designated as m-brn3, m-Nrf2 and m-FOXQ1, respectively. NS-forward primer: CCTGTGGGGGCTCTGccccGCATCTACCCCTTTG; and NS-reverse primer: CAAAGGGGTAGATGCggggCAGAGCCCCCACAGG (CCTGTGGGGGCTCTGAATAGCATCTACCCCTTTG).
RESULTS
Analyses of the putative cochlin promoter region for potential TF binding sites in the 5′ upstream region from the start codon (500–5000 bp; MatInspector; Genomatix), specifying association with the eye, revealed sites for different TFs (Supplementary Table S2). The TFs found were then subjected to a search in the literature and the UniGene database for correlation with TM, glaucoma, and eye, which generated a shorter list (Supplementary Table S3). For most TFs, all identified binding sites (except Pax6) were used to design 5′ biotin end-labeled oligo- nucleotides for TF binding assays (Table 1 and Supplementary Table S2) and for EMSA.
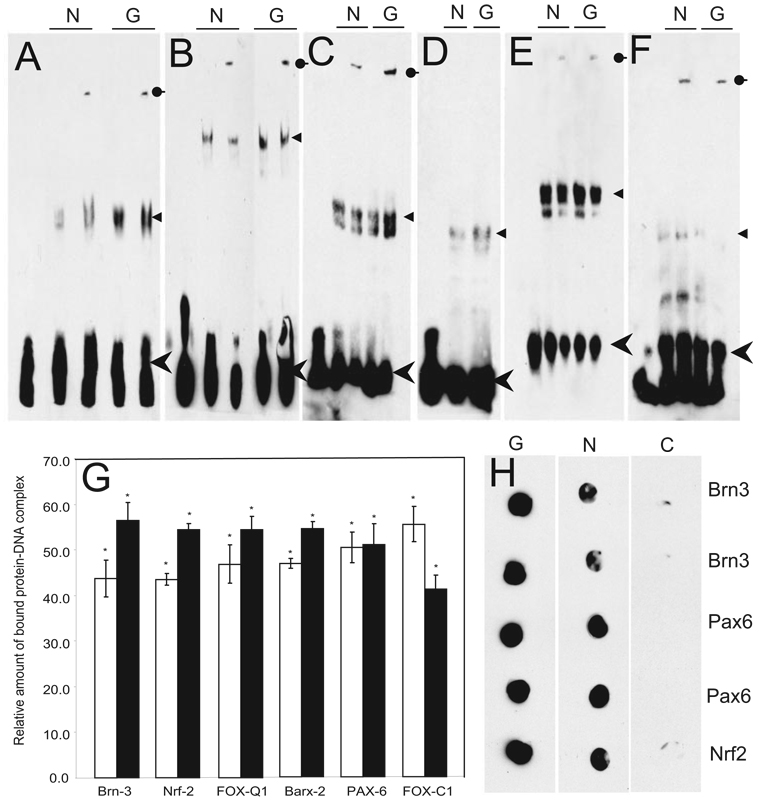
Equal amounts of nuclear protein extract (see Supplementary Table S1 for donor information) showed increased intensity of the shifted oligonucleotide band for Brn3, Nrf2, FoxQ1, and Barx2 (Fig. 1A–D), equal intensity for Pax6 (Fig. 1E) and decreased intensity for FoxC1 (Fig. 1F) in glaucomatous compared with normal TM. The specificity of this DNA-protein complex was confirmed by detecting antibody-mediated supershifts (Figs. 1A–C). The amount of bandshift was quantified from three independent EMSAs, by densitometry (Fig. 1G). In addition, filter binding assays (Fig. 1H) were performed as an independent method of obtaining statistically significant quantitative estimates (Table 1) consistent with the EMSA results.
FIGURE 1.
Electrophoretic mobility shift assay (EMSA) for select TFs in glaucomatous (G) and normal (N) TM nuclear extracts. (A) Brn3, (B) Nrf2, (C) FoxQ1, (D) Barx2, (E) Pax6, and (F) FoxC1. EMSA was performed with 5′-biotin end-labeled oligonucleotides and 3.5 µg TM nuclear extract. Arrowhead, arrow, round-end arrow: free oligonucleotides, DNA-protein complex (shift), and DNA-protein-antibody complex (supershift), respectively. In all EMSA experiments (except Barx2), specific antibody to the TF was used for supershift analysis. (G) Densitometric scan of EMSA results (A–F) converted into relative units. Each bar represents the mean ± SD of three independent experimental readings; results were found significantly different from 0.0 by the one-sample t-test: *P < 0.05. (H) Representative filter binding assay for select TFs in glaucomatous and normal TM nuclear extracts. Eight-millimeter filter discs were soaked on 0.5× TBE buffer and placed in 8-mm columns. TM nuclear extract (3.5 µg) and 5′- biotin end-labeled oligonucleotides in a 20-µL binding reaction were added to the columns. Vacuum was applied. For the control, nuclear extracts were omitted from the binding reactions. The filter binding for glaucomatous (G), normal (N), and control (C) conditions are as indicated. The TFs for which an oligonucleotide sequence was used for filter binding are as indicated.
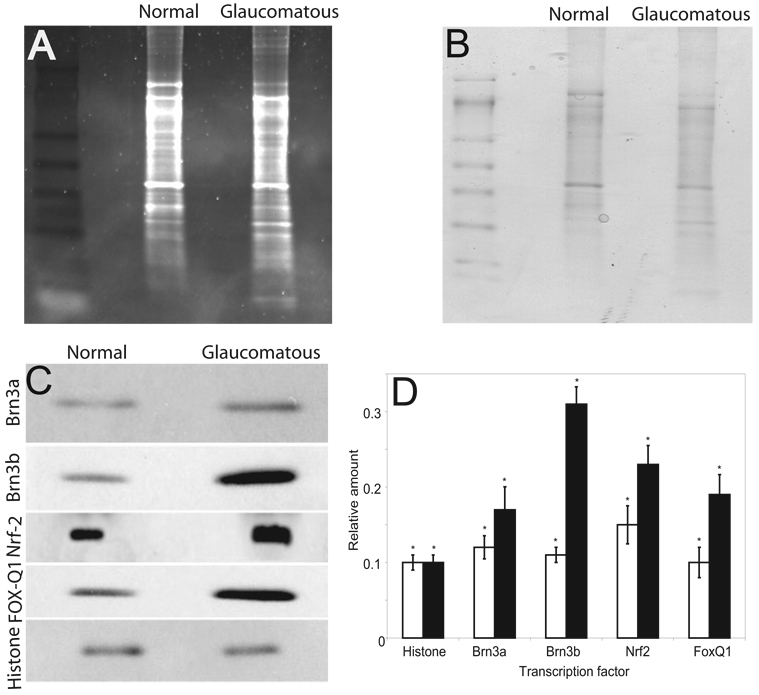
Western blot analyses were performed with total nuclear extract to determine the expression levels of different TFs. Spectro-photometry of total protein and densitometric quantification of dye-stained gels were performed to ensure equal protein loading (Figs. 2A, 2B). Elevated levels of Nrf2, Brn3a, Brn3b, and FoxQ1; similar Pax6; and decreased FoxC1 in glaucomatous nuclear extract compared with normal TM (Fig 2C) were observed, further corroborating EMSA results. Histone H3 immunoreactivity served as a protein-loading control.
FIGURE 2.
Representative Western blot analysis for TFs in glaucomatous and normal TM nuclear extracts. Protein (10 µg) was fractionated with 4% to 20% SDS-PAGE and transferred on a PVDF membrane and analyzed. (A) A representative corresponding gel after partial transfer stained with Sypro Ruby fluorescent red. (B) The corresponding gel after partial transfer stained with Coomassie blue dye. (C) The membrane probed with antibodies to TFs as indicated. Histone H3 was used as a loading control. (D) Relative densitometric quantification of Western blot results. Each bar represents the mean ± SD from three independent experiments; the results were found significantly different from 0.0 by the one-sample t-test: *P < 0.05.
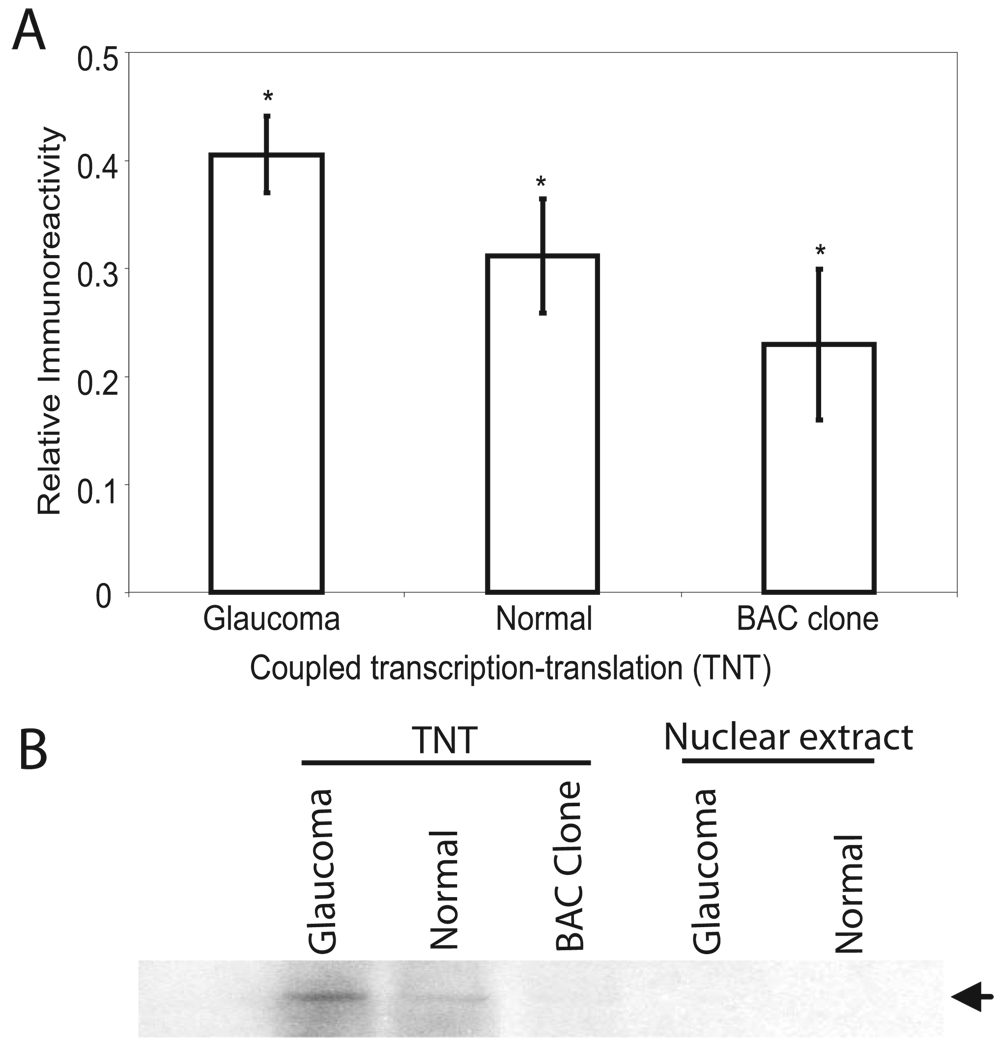
To determine whether the presence of elevated levels of TFs results in increased protein expression, cochlin BAC clone (RP11-48L1, containing the region from 30444864 to 30560848 of 14q12) was used as a template for coupled TNT with equal amounts of glaucomatous or normal TM nuclear extract–derived protein (Fig. 3). The addition of equal amounts of nuclear extract (DNA, mRNA, and cochlin depleted) from glaucomatous but not control TM or BAC clone alone in the TNT system resulted in increased formation of cochlin (Figs. 3A, 3B), supporting the EMSA, filter-binding, and Western blot results (Fig. 1, Fig 2).
FIGURE 3.
Representative coupled TNT analyses. (A) ELISA (SD from six experiments) of the cochlin BAC clone using nuclear extract from glaucomatous, normal, or clone alone as indicated. Readings were found significantly different from 0.0 by the one-sample t-test: *P < 0.05. (B) Representative Western analyses of TNT assay products as above and, as indicated, control nuclear extract.
Our results demonstrate elevated TF-DNA complex formation and protein levels of Brn3a, Brn3b, and stretch/stress-induced TFs Nrf2, Barx2, and FoxQ1 (Figs. 1A–G, Table 1); similar levels of Pax6 (Fig. 2C); and decreased levels of FoxC1 (Fig. 2C, Fig 1G; Table 1) in glaucomatous compared with normal TM nuclear extracts.
Although it demonstrated an elevation of some TFs (for example, Brn3a, Nrf2, Fox-Q1, and Barx-2) in glaucomatous compared to control TM, our analyses did not show a dramatic change in the levels of TFs (Fig. 1, Fig 2). At the protein level, there was a clear difference in the expression of cochlin between normal and glaucomatous TM. Cochlin was not detectable in the normal TM by Western blot analysis8; however, in situ hybridization suggests the presence of cochlin mRNA expression in normal TM.2 Very low levels of cochlin expression, below the threshold of detection by Western blot analysis, may exist in normal TM.
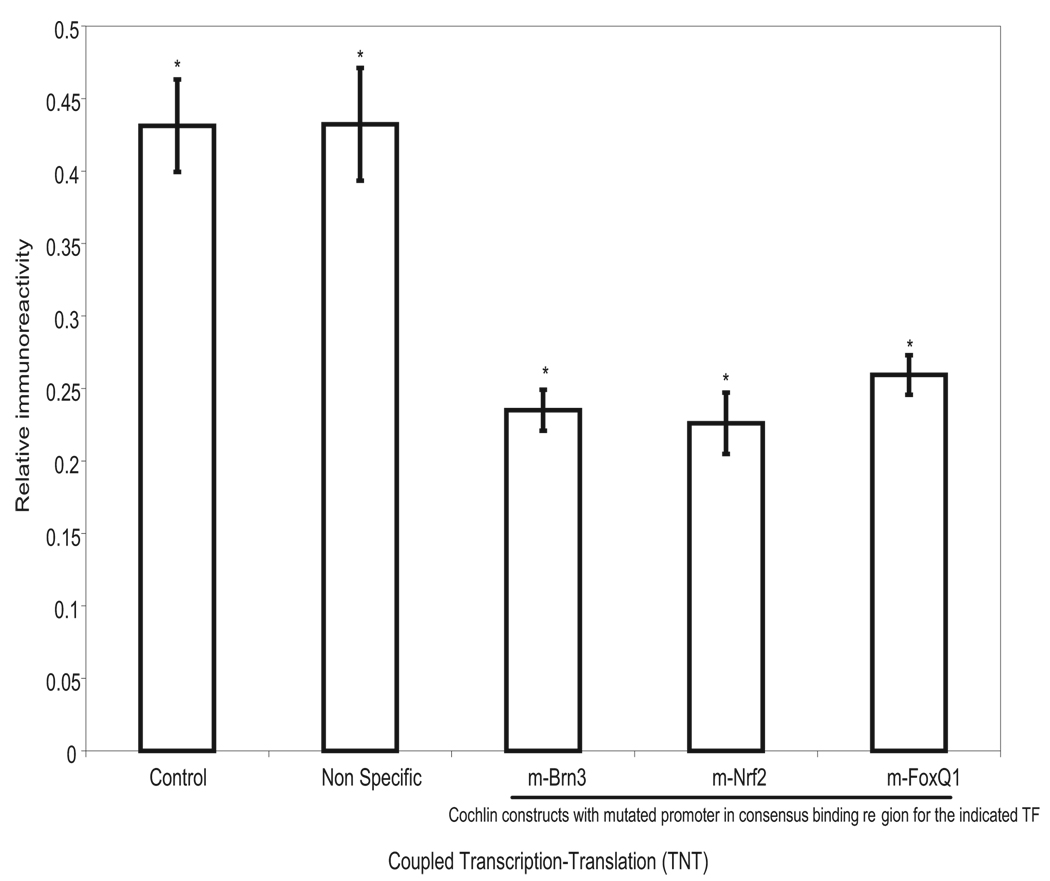
These observations are consistent with increased cochlin expression in glaucomatous TM, but not with an absolute change in the protein levels between normal and glaucomatous samples, as observed in human TM.8 Nuclear extracts alone did not show cochlin-positive immunoreactivity (Fig. 3). To further determine whether potential TF binding modulates cochlin expression in glaucomatous TM, we prepared a DNA construct with the promoter region and the structural cochlin gene. Several TF binding sites and a control promoter region (not known to bind any identified mammalian TF) were mutated. The constructs were subjected to a coupled TNT experiment and ELISA analysis that revealed that the mutation in the TF binding sequence for Brn3, Nrf2, and Fox-Q1 showed lower cochlin expression than that for wild-type or NS mutant control (Fig. 4), suggesting that these TFs regulate cochlin expression.
FIGURE 4.
Representative coupled TNT analyses. ELISA (SD from six experiments) of the cochlin promoter and structural region construct (cochlin composite construct) using nuclear extract from glaucomatous trabecular meshwork. Readings were found significantly different from 0.0 by the one-sample t-test: *P<0.02. The control used wild-type cochlin promoter. Nonspecific refers to a mutation in the promoter region of a sequence that has been identified as not binding to any known TF. m-Brn3, m-Nrf2, and m-FoxQ1 are the promoter region cochlin composite construct mutated for indicated TF. m, mutation in the consensus binding site.
DISCUSSION
These results demonstrate the utility of a combined bioinformatic and biochemical analysis approach for assessing transcriptional gene regulation. We assessed the expression of TFs in diseased compared with normal tissue, to explain enhanced transcription of a disease-associated protein. Promoter regions themselves might not undergo epigenetic or genetic changes in disease states in complex late-onset and progressive diseases; therefore, reporter assays alone may not provide entire information and additional experiments may provide complementary insight. Aberrant expression of many cytosolic proteins is capable of enhancing transcription of syn-expressed genes via regulatory feedback loops.12 The promoter region of cochlin is expected to be beyond 367 nucleotides (they are included in the cochlin transcript; NCBI accession number BC007230) from the start codon within the genomic sequence. We arbitrarily selected an ~5-kbp upstream region for analysis, which is higher than the usual length for the promoter region of most proteins. Questions concerning the real promoter region of the gene cannot be completely addressed by our study and require separate investigation. The cochlin homologues for which the promoter regions have been characterized—bone morphogenetic protein 2 and -4,13 and the von Willebrand factor14—are within the 2-kbp region upstream from the start codon.
Our results demonstrate elevated protein-DNA complex and protein levels for Brn3a, Brn3b, Nrf2, Barx2, and FoxQ1 and decreased levels for FoxC1 (Figs. 1A–G, Fig. 2; Table 1) in glaucomatous compared with normal TM nuclear extracts. The Brn3/POU domain TFs (Brn3a, Brn3b, and Brn3c) have similar consensus DNA-binding sites15 and were found in the cochlin promoter region. Nrf2 expression is increased in glaucoma,7 activating antioxidant genes,7,16 possibly in response to pathogenic accumulation of oxidative damage products,17 thereby supporting a role of Nrf2 in oxidative and mechanical stress.6 Barx2 has been implicated in Rieger syndrome and glaucoma.18 Pax6 and FoxC1 play critical roles in normal ocular development. Mutations in the Pax6 and Fox family of genes, including that in FoxC1, are involved in anterior segment dysgenesis and glaucoma.19,20 Experiments with human TM cells have shown that a reduction in FoxC1 expression leads to a decreased FoxO1A expression and increased apoptosis.21 A reduction in FoxC1 expression is likely to be a part of a negative regulating step.22
These results further corroborate and demonstrate the utility of the approach outlined herein. Additional transcriptional and posttranscriptional controls cannot be ruled out by the present analysis. Nevertheless, the findings in this study suggest that potential molecular and biochemical machinery exists for enhanced transcription of cochlin in the glaucomatous TM in comparison with the control. The approach outlined has the limitation of the possibility of missing some elements/TFs (because many TM-expressed genes have not yet been identified) resulting in their underidentification. The approach presented is applicable to a wide variety of tissues (for example, cochlin is expressed in the eye, ear ,and brain) to determine TF control of a specific protein expression in different states in a tissue.
Supplementary Material
Acknowledgments
The authors thank Alexis Garcia, Meera Ledwani, Jose Ruiz, Gabriel Gaidosh, and George Inana, MD, at BPEI, University of Miami for their technical assistance and critical reading of the manuscript, and Jim Lauderdale, PhD, at Cell Biology, University of Georgia for research gift of PAX6 antibody.
Footnotes
Disclosure: R.G. Picciani, None; A. Diaz, None; R.K. Lee, None; S.K. Bhattacharya, None
References
- 1.Morrison JC, Acott TS. Glaucoma: Science and Practice. New York: Thieme Medical Publishers, Inc.; 2003. pp. 34–41. [Google Scholar]
- 2.Picciani R, Desai K, Guduric-Fuchs J, Cogliati T, Morton CC, Bhattacharya SK. Cochlin in the eye: functional implications. Prog Retin Eye Res. 2007;26:453–469. doi: 10.1016/j.preteyeres.2007.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Robertson NG, Skvorak AB, Yin Y, et al. Mapping and characterization of a novel cochlear gene in human and in mouse: a positional candidate gene for a deafness disorder, DFNA9. Genomics. 1997;46:345–354. doi: 10.1006/geno.1997.5067. [DOI] [PubMed] [Google Scholar]
- 4.Robertson NG, Hamaker SA, Patriub V, Aster JC, Morton CC. Subcellular localisation, secretion, and post-translational processing of normal cochlin, and of mutants causing the sensorineural deafness and vestibular disorder, DFNA9. J Med Genet. 2003;40:479–486. doi: 10.1136/jmg.40.7.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Robertson NG, Lu L, Heller S, et al. Mutations in a novel cochlear gene cause DFNA9, a human nonsyndromic deafness with vestibular dysfunction. Nat Genet. 1998;20:299–303. doi: 10.1038/3118. [DOI] [PubMed] [Google Scholar]
- 6.Vittal V, Rose A, Gregory KE, Kelley MJ, Acott TS. Changes in gene expression by trabecular meshwork cells in response to mechanical stretching. Invest Ophthalmol Vis Sci. 2005;46:2857–2868. doi: 10.1167/iovs.05-0075. [DOI] [PubMed] [Google Scholar]
- 7.Malone PE, Hernandez MR. 4-Hydroxynonenal, a product of oxidative stress, leads to an antioxidant response in optic nerve head astrocytes. Exp Eye Res. 2007;84:444–454. doi: 10.1016/j.exer.2006.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bhattacharya SK, Rockwood EJ, Smith SD, et al. Proteomics reveal Cochlin deposits associated with glaucomatous trabecular meshwork. J Biol Chem. 2005;280:6080–6084. doi: 10.1074/jbc.M411233200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Andrews GL, Mastick GS. R-cadherin is a Pax6-regulated, growth-promoting cue for pioneer axons. J Neurosci. 2003;23:9873–9880. doi: 10.1523/JNEUROSCI.23-30-09873.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kim J, Lauderdale JD. Analysis of Pax6 expression using a BAC transgene reveals the presence of a paired-less isoform of Pax6 in the eye and olfactory bulb. Dev Biol. 2006;292:486–505. doi: 10.1016/j.ydbio.2005.12.041. [DOI] [PubMed] [Google Scholar]
- 11.Mastick GS, Davis NM, Andrew GL, Easter SS., Jr Pax-6 functions in boundary formation and axon guidance in the embryonic mouse forebrain. Development. 1997;124:1985–1997. doi: 10.1242/dev.124.10.1985. [DOI] [PubMed] [Google Scholar]
- 12.Qian J, Dolled-Filhart M, Lin J, Yu H, Gerstein M. Beyond synexpression relationships: local clustering of time-shifted and inverted gene expression profiles identifies new, biologically relevant interactions. J Mol Biol. 2001;314:1053–1066. doi: 10.1006/jmbi.2000.5219. [DOI] [PubMed] [Google Scholar]
- 13.Helvering LM, Sharp RL, Ou X, Geiser AG. Regulation of the promoters for the human bone morphogenetic protein 2 and 4 genes. Gene. 2000;256:123–138. doi: 10.1016/s0378-1119(00)00364-4. [DOI] [PubMed] [Google Scholar]
- 14.Guan J, Guillot PV, Aird WC. Characterization of the mouse von Willebrand factor promoter. Blood. 1999;94:3405–3412. [PubMed] [Google Scholar]
- 15.Mu X, Klein WH. A gene regulatory hierarchy for retinal ganglion cell specification and differentiation. Semin Cell Dev Biol. 2004;15:115–123. doi: 10.1016/j.semcdb.2003.09.009. [DOI] [PubMed] [Google Scholar]
- 16.Lee JM, Li J, Johnson DA, et al. Nrf2, a multi-organ protector? FASEB J. 2005;19:1061–1066. doi: 10.1096/fj.04-2591hyp. [DOI] [PubMed] [Google Scholar]
- 17.Govindarajan B, Laird J, Salomon RG, Bhattacharya SK. Isolevuglandin-modified proteins, including elevated levels of inactive calpain-1, accumulate in glaucomatous trabecular meshwork. Biochemistry. 2008;47:817–825. doi: 10.1021/bi701517m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hjalt TA, Murray JC. The human BARX2 gene: genomic structure, chromosomal localization, and single nucleotide polymorphisms. Genomics. 1999;62:456–459. doi: 10.1006/geno.1999.6037. [DOI] [PubMed] [Google Scholar]
- 19.van Heyningen V, Williamson KA. PAX6 in sensory development. Hum Mol Genet. 2002;11:1161–1167. doi: 10.1093/hmg/11.10.1161. [DOI] [PubMed] [Google Scholar]
- 20.Lehmann OJ, Ebenezer ND, Jordan T, et al. Chromosomal duplication involving the forkhead transcription factor gene FOXC1 causes iris hypoplasia and glaucoma. Am J Hum Genet. 2000;67:1129–1135. doi: 10.1016/s0002-9297(07)62943-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Berry FB, Skarie JM, Mirzayans F, et al. FOXC1 is required for cell viability and resistance to oxidative stress in the eye through the transcriptional regulation of FOXO1A. Hum Mol Genet. 2008;17:490–505. doi: 10.1093/hmg/ddm326. [DOI] [PubMed] [Google Scholar]
- 22.Berry FB, Lines MA, Oas JM, et al. Functional interactions between FOXC1 and PITX2 underlie the sensitivity to FOXC1 gene dose in Axenfeld-Rieger syndrome and anterior segment dysgenesis. Hum Mol Genet. 2006;15:905–919. doi: 10.1093/hmg/ddl008. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.