Summary
DeconMSn accurately determines the monoisotopic mass and charge state of parent ions from high resolution tandem mass spectrometry data, offering significant improvement for LTQ_FT and LTQ_Orbitrap instruments over the commercially delivered Thermo Fisher Scientific’s extract_msn tool. Optimal parent ion mass tolerance values can be determined using accurate mass information, thus improving peptide identifications for high mass measurement accuracy experiments. For low-resolution data from LCQ and LTQ instruments, DeconMSn incorporates a support-vector-machine-based charge detection algorithm that identifies the most likely charge of a parent species through peak characteristics of its fragmentation pattern.
1 INTRODUCTION
Peptide identification through tandem mass spectrometry has become a widely used technique in proteomics-based research. Common methods for analyzing LC-MS/MS fragmentation results from Thermo Fisher mass spectrometers involve the use of extract_msn (part of the SEQUEST™ analysis software package) to create “.dta” file representations of spectra, that contain the parent mass, charge and observed fragmentation pattern in the form of peak lists. Search tools such as SEQUEST, MASCOT or X!Tandem are then used to analyze these spectrum files to associate each parent with a corresponding peptide sequence.
Extract_msn determines the parent mass and charge for each spectrum file based on instrument-supplied information. The acquisition software detects accurate precursor masses for subsequent fragmentation from high-resolution data (LTQ_FT/LTQ_Orbitrap) only if the instrument is set to recognize monoisotopic precursor masses. A drawback of this setting is that it leads to fewer peptide identifications (see supplementary information). If the instrument is set to fragment all parent species to ensure higher coverage, then extract_msn records only the mass of the fragmented parent which on many occasions, is offset from the correct monoisotopic mass of the parent by one or more isotopes. As a result, a significant number of peptides will not be identified unless, contrary to the nature of high-mass accuracy experiments, a wide-mass tolerance search is performed.
Our DeconMSn tool determines more accurately, the parent charge state and monoisotopic mass for high resolution data by “deisotoping” the high mass measurement accuracy precursor spectrum, regardless of which mass was selected for fragmentation and independent of instrument settings, thus enabling improvements in peptide identifications through tools such as SEQUEST, MASCOT or X!Tandem. Additionally, a support-vector-machine-based charge detection algorithm is implemented for determining parent mass for low-resolution data (LCQ/LTQ) that offers advantages in terms of reduced search time.
2 METHODS
Using the same algorithmic core as Decon2LS (Jaitly et al., 2006), DeconMSn uses a modified approach to THRASH (Horn et al., 2000), to accurately deisotope the high-resolution parent ion distribution. This step is accomplished by first calculating the charge using autocorrelation-based peak fitting methods (Senko et al., 1995), following which the monoisotopic mass is calculated by overlapping a calculated theoretical profile (Senko et al., 1995b; Rockwood et al., 1995) with the observed profile. Corresponding fit scores are determined in this way for each distribution within a window of the parent distribution to ensure proper selection of the monoisotopic mass. The fit score is Euclidean based and measures the distance between the calculated theoretical distribution and the observed distribution. The monoisotopic mass and charge that corresponds to the theoretical distribution closest to the observed distribution and which contains the parent peak is returned. Then a spectrum file is created for that MS/MS spectrum by using the determined charge and protonated monoisotopic mass. If THRASH fails for a particular distribution (for e.g. due to a bad fit score), then the above step is repeated again except that the parent isotopic distribution is now summed across a window of scans (for details on the empirically chosen parameter values for deisotoping, see supplementary information). By summing across the retention time dimension, the parent distribution quality improves, thus maximizing the chances of finding the correct monoisotopic mass for low-quality data and low-abundant species. The summing of parent spectra across the retention time dimension is not performed by default so as to avoid the risk of introducing errors that occur when noise peaks get accidentally grouped with isotopic distributions.
Summing is also not conducive for overlapping peptide. Also, summing is performed simply across retention time and not across the peak of the parent elution profile as the detection and characterization of such profiles are computationally complex. The neutral mass is also calculated separately, by identifying the monoisotopic peak through peak-finding algorithms that identify peak existence by stepping a distance of 1.003/(charge state) from the parent. If both methods, i.e. THRASH and neutral mass determination trough peak-finding, return the same charge of the precursor then the THRASH value of the monoisotopic mass is used to create the spectrum file. This is performed even if the two methods return different monoisotopic mass values for the precursor, as given a spectrum (single or summed) THRASH has higher likelihood of arriving at the correct value for monoisotopic mass than through peak-finding. If, on the other hand, both methods return conflicting charge states then separate spectrum files are created corresponding to each charge state. Finally, if all steps fail, then the spectrum files are generated with default charge states of +2 and +3.
Autocorrelation methods of charge state determination typically fail for low-resolution data due to lack of peak-profile information. To overcome this shortcoming, DeconMSn uses a support-vector-machine (SVM) based approach, modified from the one described elsewhere (Klammer et al., 2005), to determine the most likely charge of the parent ion. A set of 19 features are calculated from a given MSn spectrum (see supplementary information). The features from different datasets are used to train the SVM in MATLAB (Canu et al., 2005). DeconMSn then uses the trained SVM to assign a +1, +2, +3 or +4 charge state to each spectrum. Ambiguous spectra are assigned both charge states +2 and +3.
RESULTS
DeconMSn is a command-line executable program. Input can be in the form of Thermo .RAW file format or the .mzXML format. Options include setting of mass range, scan range, minimum ion count for spectra consideration and the ability to force specific charges on spectra. The resulting .dta or .MGF file can be submitted directly to SEQUEST/MASCOT/X!Tandem.
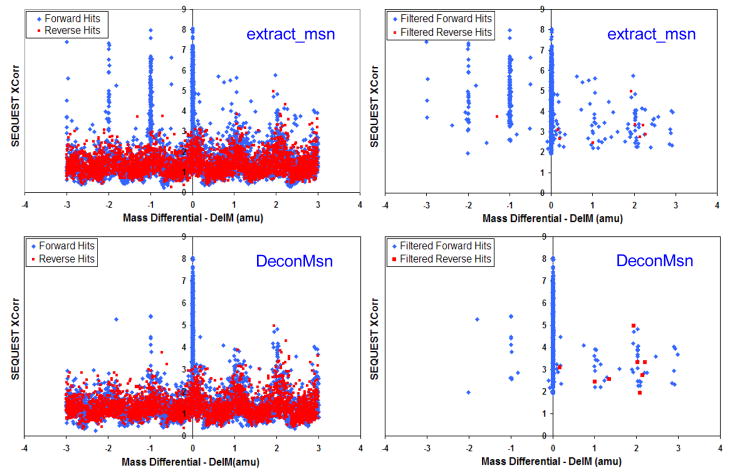
Figure 1 illustrates the effect of improvement in mass accuracy. The difference between calculated and actual peptide hits (Mass Differential or DelM) as a function of the SEQUEST identification score (Xcorr) are depicted for both original and filtered hits from a Shewanella oneidensis dataset run on a LTQ_Orbitrap instrument and processed using both software tools. The forward hits (blue) conform to hits to a forward sequence database, whereas the reverse hits (red) indicate hits to a reverse sequence database, which is used to estimate false discovery rates. Extract_msn detects the wrong parent monoisotopic masses by recording masses that are isotopes of the actual monoisotopic mass as evidenced by the streaks on the non-zero values of DelM. In contrast, DeconMSn correctly identified almost all peptides on either side of 0 DelM to be vicinal of 0 DelM (see supplementary information for more details).
Figure 1.
Mass Differential (DelM) vs. SEQUEST Xcorr for all hits [left] and filtered hits [right] for a single LTQ_Orbitrap LC-MS/MS dataset.
Filters used – DelCn2 >=0.1, Xcorr >= 1.9 for 1+ peptides, Xcorr >=2.2 for 2+ peptides, Xcorr >=3.2 for >= 3+ peptides.
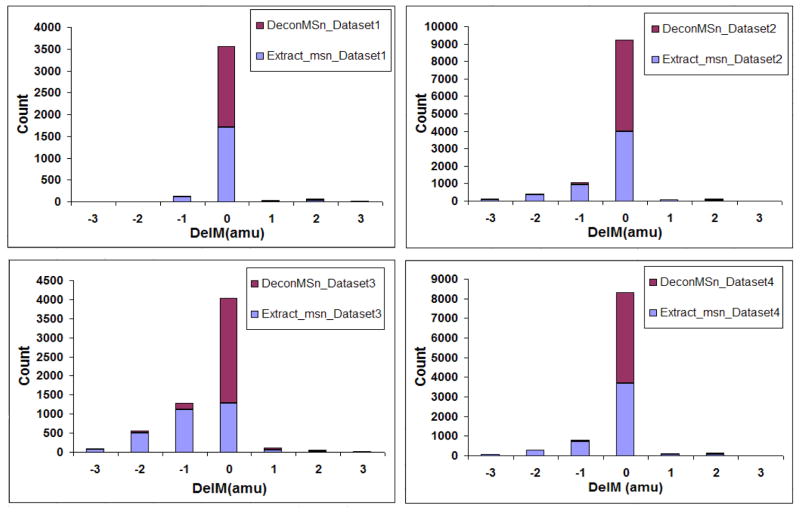
Figure 2 shows the DelM distributions, with a bin size of 1 amu, for filtered hits for four Shewanella oneidensis datasets including the one used in Figure 1. The figure clearly shows the increase in the number of hits in the region close to 0 DelM for DeconMSn with a simultaneous decrease in the −1 and −2 amu DelM region. Further, a narrow parent-mass tolerance search (e.g. 0.1 Da) performed using DeconMSn resulted in approximately 10% more peptide identifications than extract_msn. Please note that the remaining identifications that have offsets from the exact masses mostly correspond to low-quality data, low abundant species, overlapping peptides and even incorrect peptide identifications. For low-resolution data, DeconMSn detected 98% of all filtered peptides using the charge-state determination algorithm that extract_msn detected (see supplementary information). High mass accuracy experiments benefit from the use of DeconMSn in conjunction with search algorithms through a reduced peptide identification computational search space and concurrent reduction in false discovery rates.
Figure 2.
Histogram of DelM values for filtered hits across four Shewanella oneidensis LC-MS/MS datasets
All the datasets used for analysis are available for download from http://omics.pnl.gov. The DeconMSn installer, main source code, supporting source code such as the SVM MATLAB scripts and other tutorials are available for download from http://ncrr.pnl.gov.
Supplementary Material
Acknowledgments
This research was primarily supported by the NIH National Center for Research Resources (Grant RR018522) and the National Institute of Allergy and Infectious Diseases (NIH/DHHS through interagency agreement Y1-AI-4894-01). Experimental portions of this research were performed at the Environmental and Molecular Sciences Laboratory (EMSL), a U. S. Department of Energy (DOE) national scientific user facility located at Pacific Northwest National Laboratory in Richland, Washington, USA. PNNL is operated by Battelle Memorial Institute for the U.S. Department of Energy under contract DE-AC05-76RLO 1830.
Footnotes
Availability: http://ncrr.pnl.gov/software/ or http://www.proteomicsresource.org/
Contact: rds@pnl.gov
Supplementary Information: PowerPoint presentation/Poster on http://ncrr.pnl.gov/software/.
References
- Horn DM, et al. Automated reduction and interpretation of high resolution electrospray mass spectra of large molecules. J Am Soc Mass Spectrometry. 2000;11:320–332. doi: 10.1016/s1044-0305(99)00157-9. [DOI] [PubMed] [Google Scholar]
- Senko MW, et al. Automated assignment of charge states from resolved isotopic peaks for multiply charged ions. J Am Soc Mass Spectrometry. 1995;6:52–56. doi: 10.1016/1044-0305(94)00091-D. [DOI] [PubMed] [Google Scholar]
- Senko MW, et al. Determination of monoisotopic mass and ion populations for large biomolecules from resolved isotopic distributions. J Am Soc Mass Spectrometry. 1995b;6:52–56. doi: 10.1016/1044-0305(95)00017-8. [DOI] [PubMed] [Google Scholar]
- Jaitly N, et al. Open source tools for the Accurate Mass and Time (AMT) Tag proteomics pipeline. Proceedings of 54th ASMS Conference on Mass Spectrometry; Seattle, USA. 2006. [Google Scholar]
- Klammer AA, et al. Peptide charge state determination for low-resolution tandem mass spectra. Proceedings of the 2005 IEEE Computational Systems Bioinformatics Conference; Stanford, USA. 2005. [DOI] [PubMed] [Google Scholar]
- Canu S, et al. Perception Systemes et Information. INSA de Rouen; France: 2005. SVM and Kernel Methods MATLAB toolbox. [Google Scholar]
- Rockwoood AL, et al. Rapid calculation of isotope distributions. J Anal Chem. 1995;67:2699–2704. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.