Abstract
Background and Aims
Laeliinae are a neotropical orchid subtribe with approx. 1500 species in 50 genera. In this study, an attempt is made to assess generic alliances based on molecular phylogenetic analysis of DNA sequence data.
Methods
Six DNA datasets were gathered: plastid trnL intron, trnL-F spacer, matK gene and trnK introns upstream and dowstream from matK and nuclear ITS rDNA. Data were analysed with maximum parsimony (MP) and Bayesian analysis with mixed models (BA).
Key Results
Although relationships between Laeliinae and outgroups are well supported, within the subtribe sequence variation is low considering the broad taxonomic range covered. Localized incongruence between the ITS and plastid trees was found. A combined tree followed the ITS trees more closely, but the levels of support obtained with MP were low. The Bayesian analysis recovered more well-supported nodes. The trees from combined MP and BA allowed eight generic alliances to be recognized within Laeliinae, all of which show trends in morphological characters but lack unambiguous synapomorphies.
Conclusions
By using combined plastid and nuclear DNA data in conjunction with mixed-models Bayesian inference, it is possible to delimit smaller groups within Laeliinae and discuss general patterns of pollination and hybridization compatibility. Furthermore, these small groups can now be used for further detailed studies to explain morphological evolution and diversification patterns within the subtribe.
Key words: Laeliinae, Orchidaceae, ITS, trnL intron, trnL-F spacer, matK
INTRODUCTION
Many phylogenetic studies based on molecular data have been carried out in different groups of Orchidaceae. With the advent of DNA sequencing, it was possible to begin disentangling the relationships between orchids and other families of monocotyledons and the internal structure of Orchidaceae (Cameron et al., 1999; Freudenstein et al., 2000, 2004). An analysis sampling 171 orchid taxa for rbcL gave a good idea of the relationships among subfamilies of orchids (Cameron et al., 1999), although the level of variation was not enough to assist in delimitation of tribes and subtribes.
Laeliinae are strictly neotropical and comprise about 50 genera and 1500 species (Dressler, 1981, 1993), being the third largest subtribe in the family after Pleurothallidinae and Oncidiinae. Some genera such as Cattleya, Guarianthe and Rhyncholaelia are of outstanding horticultural value, and others such as Encyclia, Epidendrum and Prosthechea are common floristic elements in the neotropics. Morphological diversity is extremely high, probably due to specialization for particular pollinators coupled with adaptation to a broad range of habitats (van der Pijl and Dodson, 1969). Chromosome numbers are nearly constant within the subtribe (Tanaka and Kamemoto, 1984). The production of artificial hybrids for horticulture is possible for nearly any combination of genera, and many natural intergeneric and interspecific hybrids also occur (Adams and Anderson, 1958; Pabst and Dungs, 1975, 1977; Borba and Semir, 1998; Azevedo et al., 2006).
Subtribe Laeliinae was established by Bentham (1881). Pfitzer (1889) divided the subtribe into two series: Ponereae and Cattleyeae, based on the presence of a column foot in the former. This concept was followed by Schlechter (1926) and most subsequent systems until Dressler removed Meiracyllium (Dressler, 1960) and Arpophyllum (Dressler, 1990) to their own monogeneric subtribes, based on pollinarium structure. More recent treatments included Baker (1972), based on leaf anatomic data, Brieger (1976), who divided the subtribe into four alliances (as ‘Gattungsreihen’), mostly based on column-foot presence and habit, Dressler (1981), who proposed six alliances based on Baker (1972), presence of column-foot and habit; and Szlachetko (1995) who split the genera into three subtribes: Laeliinae, Epidendrinae and Ponerinae (based on column structure, pollinium number and habit). These systems differ in which genera are placed in alliances and subtribes because of the different intuitive emphasis on morphological characters by each author; all of them have some aspects that appear to be highly artificial (e.g. emphasis on pollinium number, column-foot, and reed-stem habit).
Baker (1972) used leaf anatomy data for inferring relationships within Laeliinae and between Laeliinae and related subtribes. However, he did not carry out an explicit phylogenetic analysis, and many characters were polymorphic even within genera. He suggested alliances based on his results as a reticulate diagram. His results did not show Meiracyllium as distinct from Laeliinae. However, he found distinct features in Arpophyllum. His diagram was adapted and used by Dressler (1981) to propose generic alliances. The broad DNA studies published to date on orchids (Neyland and Urbatsch, 1996; Cameron et al., 1999; Freudenstein et al., 2000, 2004) did not have enough sampling to address most questions regarding circumscription of Laeliinae in relation to Epidendreae or to provide a clear picture of whether Arpophyllum and Meiracyllium were sister or embedded in Laeliinae. However, both Cameron et al. (1999) and Freudenstein et al. (2000) have shown Dilomilis to be sister to Pleurothallidinae rather than Laeliinae. A study centred on Epidendroideae and Epidendreae (van den Berg et al., 2005) circumscribed Epidendreae as an exclusively neotropical tribe, and also showed that Laeliinae should should include Arpophyllum and Meiracyllium. It also showed that Helleriella, Isochilus, Ponera and Nemaconia should be a separate subtribe (nomenclatural changes published by Soto-Arenas et al., 2007) and confirmed that Dilomilis and Neocogniauxia are part of Pleurothallidinae as shown by Pridgeon et al. (2001).
The only broad phylogenetic analysis within Laeliinae was performed using internal transcribed spacer (ITS) data for 295 taxa (van den Berg et al., 2000). They found little resolution and support along the spine of the tree, but relationships were clear enough to show that some groups were polyphyletic, which led to the transfer of many species from Laelia to Sophronitis (van den Berg and Chase, 2000, 2001, 2005). The other study available emphasized Encyclia and relatives (Higgins et al., 2003) and showed that there are distinctions between Encyclia and many genera previously included there, such as Artorima, Dinema, Prosthechea and Psychilis, as well and provided support for re-establishing Microepidendrum and describing Oestlundia. A detailed chronology of the taxonomic history and changing generic circumscriptions within Laeliinae can be found in van den Berg and Chase (2004).
In this study, a broad analysis of Laeliinae and putative outgroups is performed, based on six DNA regions: plastid trnL intron and trnL-F spacer (Taberlet et al., 1991), matK gene, trnK introns up and downstream from matK (Johnson and Soltis, 1994, 1995) and the ITS data of van den Berg et al. (2000). In this paper, the main goals were to clarify the internal topology within Laeliinae, to assess how reliable previous ITS topologies are and to increase resolution in order to establish generic alliances for further investigation better.
MATERIALS AND METHODS
Plant material and voucher information for this analysis are given in the Appendix, for which a dataset of DNA sequences was assembled from 125 terminals. Distant outgroups Earina valida and Polystachya galeata were chosen based on the analysis of Epidendroideae (van den Berg et al., 2005) and Cameron et al. (1999). Representatives of all other main clades of Epidendreae as defined by van den Berg et al. (2000, 2005) were included in the ingroup. Within Laeliinae, the aim to include all genera that have been listed in recent systems (Brieger, 1976; Dressler, 1981, 1993; Szlachetko, 1995), most infrageneric subgroups from the taxonomic literature and those that emerged from the ITS analysis of van den Berg et al. (2000). It was not possible to obtain material of Pygmaeorchis and Pinelianthe sensu stricto (this genus is now included in Homalopetalum by Soto-Arenas et al. 2007), and did not include Basiphyllaea due to technical difficulties in sequencing all five regions. However, Goldman et al. (2001) and van den Berg et al. (2005) clearly showed that Basyphyllaea is related to Bletiinae rather than Laeliinae.
DNA was extracted from fresh leaves, fresh flowers and silica gel-dried leaves and flowers (Chase and Hills, 1991), using in most cases the 2× CTAB protocol of Doyle and Doyle (1987). For samples that presented difficulties due to the presence of polysaccharides, DNA was extracted using the Nucleon Phytopure Kit (Amersham Plc., Little Chalfont, Bucks, UK). Total DNAs were purified either by caesium chloride/ethidium bromide gradient or on silica columns (QIAGEN, Ltd) and sometimes by a combination of both methods. The ITS was amplified following van den Berg et al. (2000). For the trnL-F region, the four universal primers (c, d, e and f) of Taberlet et al. (1991) were used and a programme consisting of 28–30 cycles of 94 °C denaturation for 1 min, 50 °C annealing for 30 s and 72 °C of extension for 1 min. Most species were amplified with primers c and f, but difficult samples had to be amplified in two halves with the consequent insertion of missing characters in the area corresponding to primers d and e, which are reverse complements. The trnK/matK region was amplified by using the primers –19F (Molvray et al., 2000) and trnK-2R (Johnson and Soltis, 1994). PCR conditions were a hot start with 2 min of initial denaturation at 94 °C, followed by 28–30 cycles of 94 °C denaturation, 52 °C annealing for 45 s and 72 °C for an initial time of 2·5 min with auto-extension of 8 s per cycle. Purification of PCR products was performed with QIAquick (QIAGEN Ltd) and Concert (Gibco BRL Ltd) silica columns. For ITS only, an extra wash with 35 % guanidinium chloride solution was added to help to remove primer dimers. PCR products were sequenced in both directions using the Big Dye system and an ABI 377 automated sequencer following the manufacturer protocols (PE Applied Biosystems Inc., Warrington, Cheshire, UK). The following additional primers were employed for sequencing the matK gene: matK163F (Molvray et al., 2000), matK458F (Molvray et al., 2000), matK556R (Molvray et al., 2000), matK731F (Pridgeon et al., 2001), matK881R (Pridgeon et al., 2001), matK877F (Molvray et al., 2000), matK1155F (5′ TTC ACT TTT GGT YTC ACC CT 3′) and matK1592R (Goldman et al., 2001). Electropherograms were assembled and edited using Sequencher 3·0 and 3·1 (Genecodes Inc., Ann Arbor, MI, USA). All sequences were aligned by eye using a coloured font in PAUP 4.0 (Swofford, 1998). Gaps were treated as missing characters, but were translated into a manually coded binary gap-matrix (presence/absence) with all non-autapomorphic, unambiguous indels in the trnL-F, ITS and matK gene datasets. Gaps in the trnK intron were not coded, because this part of the intron was not sequenced by some collaborators, thus precluding sensible gap coding.
Maximum parsimony (MP) analyses were performed using PAUP 4.0, with Fitch parsimony (equal weights, unordered; Fitch, 1971). Four separate searches were performed: the first with plastid data only, the second with ITS data only, the third with all data combined and the fourth with the combined data but deleting four ITS sequences suspected of being paralogues. These were all Cattleya species (C. lawrenceana, C. lueddemanniana, C. maxima and C. wallisii). Each search consisted of 1000 random taxa-addition replicates, with the tree-bisection-reconnection (TBR) swapping limited to 15 trees per replicate to eliminate extensive swapping on a single replicate. The resulting trees were then used as starting trees for TBR swapping with an upper limit of 50 000 trees. Internal support for groups was evaluated using 1000 replicates of character bootstrapping (Felsenstein, 1985), with simple taxon addition and TBR, saving 15 trees per replicate. The separate analyses were compared to assess phylogenetic incongruence, by comparing disagreement in moderate to well-supported clades among analyses. For bootstrap support, bootstrap percentages (BP) of 50–70 were considered as weak, 71–85 as moderate and >85 as strong (Kress et al., 2002).
As an alternative phylogenetic reconstruction method, a Bayesian analysis of the combined dataset under mixed models was performed by using MRBAYES 3·1·2 (Ronquist and Huelsenbeck, 2003). Models for individual regions were selected by using hierachical likelihood ratio tests in MRMODELTEST 2 (Nylander, 2004) for four data partitions: ITS, trnL-F, matK gene and the two trnK introns. A fifth data partition was composed of indel characters, which were treated under the restriction site (binary) model assuming that characters that are constant in all taxa cannot be observed, as recommended by Ronquist et al. (2005). The values for all parameters in the data partitions were unlinked in MRBAYES, to allow more independent evolutionary models among DNA regions. For analysis, two simultaneous runs of four chains (one ‘cold’ and three ‘heated’) each were carried out with the MCMC algorithm, for 3000 000 generations, sampling one tree for each 100, until the average standard deviation of split ranges was smaller than 0·01, as recommended in the manual. Likelihoods of the trees produced were analysed graphically and, after discarding the initial 450 trees of each chain as burn-in, a majority-rule consensus was generated for the remaining trees in PAUP 4.0 to assess topology and clade posteriors.
RESULTS
General features of the datasets
General characteristics of the DNA datasets in relation to the combined trees are given in Table 1. A region of 480 bp in the trnL intron was of ambiguous alignment and was therefore excluded from the analyses. The non-coding portions of the trnK intron (upstream and downstream of the matK gene) were considered together. The most variable dataset was ITS (as a whole), followed by the trnK introns. The trnL-F (intron + exon + spacer) and matK gene had similar levels of variation. In terms of informativeness as measured by the retention index (RI) of each dataset, the matK gene and the trnL-F (intron + exon + spacer) performed similarly and slightly better than ITS. The indel matrix was composed of 26 indels from trnL-F, 23 from ITS and only three from matK. This matrix was used only in the combined parsimony and Bayesian analyses.
Table 1.
Features of DNA datasets used in this study
| DNA region | Aligned length | No. variable sites | No. potentially parsimony informative | No. of changes/variable site | Fitch tree length | CI | RI | ts:tv |
|---|---|---|---|---|---|---|---|---|
| ITS region | 789 | 461 (58·43 %) | 339 (42·97 %) | 5·05 | 2326 | 0·35 | 0·52 | 2·20 |
| ITS1 | 306 | 227 (74·18 %) | 169 (55·23 %) | 5·23 | 1188 | 0·35 | 0·51 | 2·15 |
| 5·8S | 158 | 23 (14·56 %) | 10 (6·33 %) | 1·87 | 43 | 0·65 | 0·58 | 2·31 |
| ITS2 | 325 | 211 (64·92 %) | 160 (49·23 %) | 5·19 | 1095 | 0·34 | 0·53 | 2·26 |
| trnL-F region | 1350 | 495 (36·66 %) | 223 (16·5 %) | 1·97 | 974 | 0·63 | 0·64 | 0·95 |
| trnL-F intron | 723 | 251 (34·72 %) | 104 (14·38 %) | 1·97 | 495 | 0·62 | 0·65 | 1·09 |
| trnL-F exon | 50 | 9 (18 %) | 2 (4 %) | 2·33 | 21 | 0·52 | 0·29 | 0·17 |
| trnL-F interg. spacer | 596 | 250 (41·95 %) | 124 (20·8 %) | 2·01 | 502 | 0·64 | 0·61 | 0·74 |
| trnK introns | 600 | 297 (49·5 %) | 118 (19·67 %) | 1·92 | 571 | 0·68 | 0·56 | 0·85 |
| matK gene | 1347 | 551 (40·91 %) | 259 (19·23 %) | 2·12 | 1167 | 0·58 | 0·64 | 1·03 |
| matK (1st positions) | 331 (28·36 %) | 0·67 | 0·63 | |||||
| matK (2nd positions) | 357 (30·59 %) | 0·59 | 0·69 | |||||
| matK (3rd positions) | 479 (41·04 %) | 0·52 | 0·59 | |||||
| All plastid data (except excluded bases) | 2739 | 0·63 | 0·64 | |||||
| All data (except excluded bases) | 5154 | 0·49 | 0·58 |
CI, consistency index; RI, retention index; ts:tv, transition/transversion ratio.
Individual analyses
Many possible trees were found for the plastid and ITS analyses (limited to 50 000 in the search). Because the trees do not show any major differences, and are largely unresolved along the spine of the tree, they are not shown here – only the overall patterns and discrepancies found are mentioned. In the plastid analysis, relationships are well resolved only in the outgroup and some of the terminal clades within the subtribe. Branch lengths along the spine of the tree are short. Many nodes collapse in the strict consensus, and BP are generally low in the spine of the tree but increase towards the terminal nodes. Within Laeliinae there are few groups having 50 BP or more. The relationships between the main clades within the subtribe do not appear consistently in all trees, and the strict consensus is largely unresolved. In the ITS analysis, several subtribes of Epidendreae are monophyletic, but their relationships are not clear, with most branches collapsing in the strict consensus. The separation of Ponerinae from Laeliinae (94 BP) and Dilomilis and Neocogniauxia from Pleurothallidinae (93 BP) supports the results of van den Berg et al. (2000, 2005). The only differences noteworthy between the ITS and plastid analyses are the position of some species within the Cattleya alliance. In the plastid trees, Cattleya maxima and C. araguaiensis form a clade that is sister to the whole of the alliance. In the ITS tree, C. maxima is sister to Cattleya section Cattleyodes and C. araguaiensis is sister to Guarianthe. Also in plastid trees, C. lueddemanniana, C. percivaliana and C. wallisii are collectively sister to the other species of unifoliate Cattleya, but in the ITS analysis they are sister to Cattleya section Hadrolaelia and C. section Cattleyodes. For this reason two combined analyses were run for MP: the first with a complete dataset and the second excluding the ITS sequences of C. lueddemanniana, C. maxima, C. percivaliana and C. wallisii.
Combined parsimony analyses
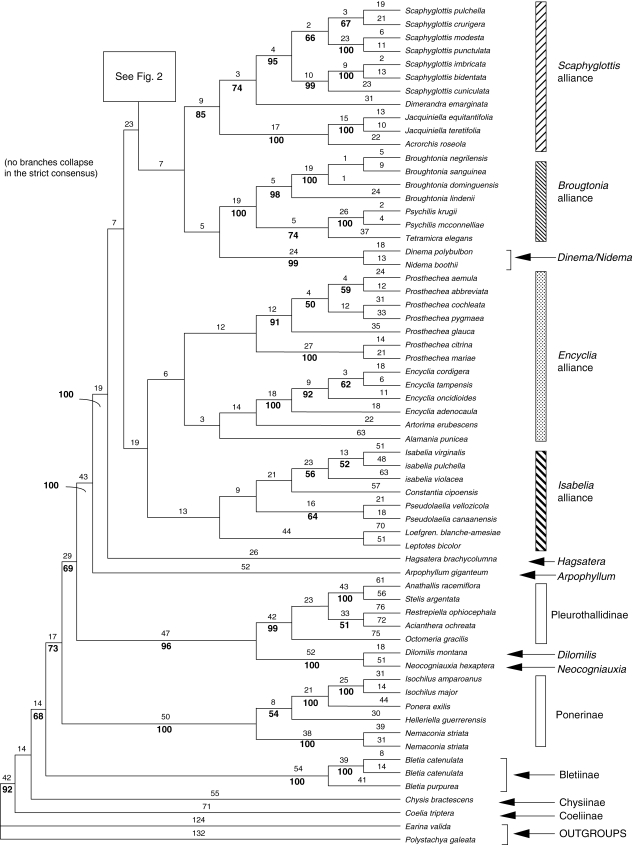
The analysis with all sequences recovered 360 trees (Figs 1 and 2) with tree length of 5154, consistency index (CI) = 0·49 and RI = 0·58. The strict consensus is much more resolved than the individual plastid or ITS analyses.
Fig. 1.
First part of one of the 360 most-parsimonious trees for the combined analysis of six DNA regions in Laeliinae. L = 5154, CI = 0·49, RI = 0·58. Numbers above the branches are Fitch lengths and numbers below the branches are bootstrap percentages (branches without numbers received <50 % bootstrap support).
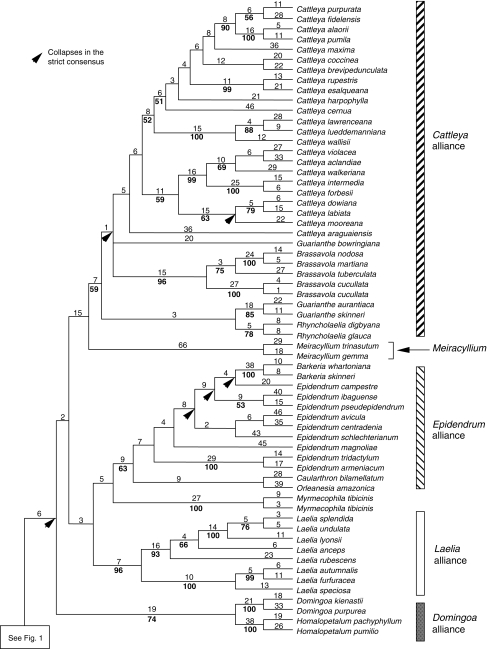
Fig. 2.
Second part of the tree in Fig. 1. Numbers above the branches are Fitch lengths and numbers below the branches are bootstrap percentages (branches without numbers received <50 % bootstrap support).
The outgroup relationships were nearly the same as in the plastid trees, with Arpophyllum sister to the rest of Laeliinae (100 BP). The immediate sister group of Laeliinae was Pleurothallidinae (69 BP), followed successively by Ponerinae (73 BP), Bletiinae (68 BP), Chysiinae (<50 BP) and finally Coeliinae (92 BP). Even though these relationships did not have high support, the monophyly of each subtribe did have high support: Bletiinae (100 BP), Ponerinae (100 BP), Pleurothallidinae including Dilomilis and Neocogniauxia (96 BP) and Laeliinae (including Arpophyllum and Meiracyllium; 100 BP).
Within Laeliinae most nodes of the spine are resolved in the strict consensus tree. The only branch with some (weak) internal support is the one leading to the Cattleya alliance (59 BP; Fig. 2). Hagsatera is placed between Arpophyllum and the rest of Laeliinae with <50 BP. The main groups with internal support above 50 BP in the combined trees were: Dinema/Nidema (99 BP), the Scaphyglottis alliance (85 BP), Domingoa with Homalopetalum (74 BP), Laelia sensu stricto and Schomburgkia (96 BP), the Epidendrum alliance (63 BP), Encyclia (100 BP), Prosthechea (91 BP), the Broughtonia alliance (100 BP), Brassavola (96 BP), a subclade of Cattleya including the type (C. labiata, 59 BP) and a group including three unifoliate Cattleya species (C. lawrenceana, C. lueddemanniana and C. wallisii) and the species formerly attributed to Laelia and Sophronitis (52 BP). It should be noted, however, that many of the groups follow previous taxonomic categories based on morphology, both at the generic and infrageneric levels. These previously recognized suites of characters increase our confidence in the tree, despite the low BPs.
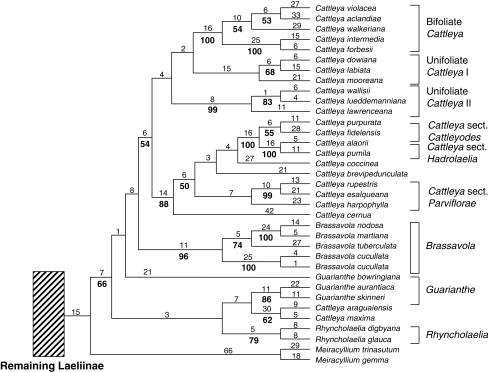
The second analysis, excluding the ITS sequences of four Cattleya species, was identical to the complete dataset in the topology of all outgroup and major clades within the subtribe, except for relationships within the Cattleya alliance. Therefore, only this portion of the tree is shown (in Fig. 3). With the exclusion of the ITS sequences of C. lawrenceana, C. lueddemaniana and C. wallisii (our ‘unifoliate Cattleya II’, Fig. 3), this group is no longer sister to the species previously attributed to Sophronitis (Cattleya sections Cattleyodes and Hadrolaelia in Fig. 3). However, it is not sister to our ‘unifoliate Cattleya I’ as would be expected (these species were once considered subspecies of C. labiata). Rather it is sister to the remaining members of Cattleya (with less than 50 BP). When the ITS sequence of C. maxima (which we expected to cluster with other unifoliate species of Cattleya) is excluded, it moves to be sister to C. araguaiensis, and these two are sister to Guarianthe (<50 BP).
Fig. 3.
Cattleya alliance portion of the combined analysis of six DNA regions in Laeliinae, excluding ITS sequences of four Cattleya species with incongruent placement in individual analyses. Numbers above the branches are Fitch lengths and numbers below the branches are bootstrap percentages (branches without values received <50 % bootstrap support).
Bayesian analysis
The models selected by successive hierachical likelihood ratio tests were GTR + G for all three plastid non-coding data partitions (trnL-F, trnK introns up and downstream from matK), and GTR + I for ITS and the coding region of matK. The combined tree obtained from a majority-rule consensus of 59 100 trees produced by the two runs of MCMC in the mixed model context is presented in Fig. 4. Most relationships were similar to the combined MP tree with all sequences (Figs. 1 and 2). Because posterior probabilities (PP) from Bayesian analysis (BA) can be considered inflated in relation to the conservative values obtained for parsimony BP (Erixon et al., 2003) those below 95 PP are considered as weakly supported. Even under this criterion, a much greater number of nodes in the BA tree attained high support compared with MP (often 100 PP). The outgroup relationships were essentially the same, with most relationships being strongly supported, except for Pleurothallidinae as sister to Laeliinae (86 PP). The position of Arpophyllum was identical (100 PP), as was that of Hagsatera (88 PP). There is a large polytomy within Laeliinae, which is equivalent to the many clades with <50 BP in MP. Some novel well-supported relationships that did not appear in the parsimony tree were: (a) the Scaphyglottis alliance including Dinema and Nidema (98 PP); (b) the Epidendrum and Laelia alliances as sister (96 PP); (c) the Broughtonia, Cattleya, Epidendrum and Laelia alliances forming a clade (95 PP). Relationships of the Cattleya alliance did not differ from the parsimony results, but many with <50 BP in the MP trees were well supported in BA, such as Cattleya s.l. (98 PP) and Guarianthe/Rhyncholaelia (96 PP). However, relationships among the Brassavola, Cattleya and Guarianthe/Rhyncholaelia clades remained unresolved.
Fig. 4.
Bayesian tree of 59 100 trees obtained from two runs 3000 000 chains of MCMC. Numbers above branches represent the posterior probabilities (PP). Nodes with PPs below 50 % have been collapsed.
DISCUSSION
Molecular evolution
The variation of the ITS dataset was similar to that in van den Berg et al. (2000). However, the performance (in terms of RI) of this region was worse than the plastid datasets. This could be explained by the fact that the ITS dataset has a higher number of changes per variable position than the plastid loci, and is therefore more likely to be affected by taxon-sampling (incomplete taxon sampling could preclude the reconstruction of multiple changes at a given position). In general, the levels of variation found in different regions in the present study were lower than those found by van den Berg et al. (2005), whereas CI and RI were higher, as would be expected when dealing with more closely related taxa. This effect is less obvious for matK, reinforcing the fact that this gene is often useful at all taxonomic levels.
Outgroup relationships
Arpophyllum is always sister to a strongly supported clade of the remaining members of Laeliinae (ITS and plastids, 98 BP; combined MP and BA, 98 BP and 98 PP). The sister group of Laeliinae is probably Pleurothallidinae (including Dilomilis and Neocogniauxia), which had already been seen in the previous ITS analysis (van den Berg et al., 2000; <50 BP). This sister relationship is not supported in the plastid consensus tree but has weak (69 BP, 86 PP) support in the MP and BA combined. The relationship between Bletiinae and Ponerinae also remains ambiguous. In the plastid trees there is a polytomy among Pleurothallidinae, Ponerinae and Bletiinae, and in the combined analysis they are successive sister groups as in the ITS tree of van den Berg et al. (2000). However, in an analysis of all Epidendreae based on the same DNA regions as in this paper (van den Berg et al., 2005), Bletiinae and Ponerinae were sister to each other with 90 BP and strongly supported in the BA tree. This pattern is different here due to the less extensive outgroup sampling in the present study. The position of Meiracyllium, deeply embedded in Laeliinae, appears in all analyses (and also in Cameron et al., 1999; van den Berg et al., 2000, 2005; Goldman et al., 2001). Finally, the position of Chysis and Coelia is the same in all analyses, which agrees with ITS alone (van den Berg et al., 2000). One important point to mention is the embedded placement of Meiracyllium in Laeliinae. Plastid analysis places it as sister to the Cattleya alliance (<50 BP), whereas in the ITS analysis of van den Berg et al. (2000) it was sister to some species of Prosthechea (P. mariae and P. citrina; 61 BP). The plastid placement reappeared in the combined analysis (but with <50 BP). The long branch-length leading to this genus, although correlated with the striking morphological peculiarities, could indicate long-branch attraction. This relationship was also found with BA (Fig. 4), but this method is also not immune against long-branch attraction artefacts.
Generic alliances within Laeliinae
When considering the topologies obtained within Laeliinae in this study one first important detail is that, although there is no conflict between our ITS trees and the larger analysis (295 taxa) used by van den Berg et al. (2000), the latter had many fewer branches collapsing in the strict consensus and stronger bootstrap support for relationships. This is probably the effect of taxon sampling since the alignment of both matrices was the same (the ITS dataset used here was produced by deleting taxa from the larger one without alteration of gaps). In this study, weak incongruence was found between the topologies resulting from the plastid and ITS analyses, but none of these relationships has BP >50, suggesting that most of the incongruence could be due to character sampling error. Results of the combined analyses are in agreement with the ITS data alone, at least for the few areas where ITS had internal support in van den Berg et al. (2000). To a much more limited extent there is a similarity between the DNA trees in the present study and the alliances proposed by Dressler (1981) based on the leaf anatomical characters of Baker (1972). The main weaknesses of his alliances were the inability to detect polyphyletic genera such as Laelia and Cattleya and also that the genera of Ponerinae (Helleriella, Isochilus, Nemaconia, Ponera), Dilomilis and Neocogniauxia did not belong to Laeliinae. All alliances proposed by Dressler (1981) each appear to include a few unrelated genera, and a system of generic alliances based on the results of the present study would need a larger number of smaller alliances, although this may be reduced as more data are collected, and the relationships among larger clades within Laeliinae are resolved. The generic alliances presented in this discussion are based on highly supported clades in BA (Fig. 4), which do not contradict any well-supported clades in the combined parsimony analysis.
Isabelia alliance
This is a small group (five genera and approx. 28 species) mainly from south-eastern Brazil, a few species extending north to Bahia State (Brazil) and south-west to Paraguay and northern Argentina. Many species grow exclusively as epiphytes on Vellozia (Velloziaceae) or lithophytes. They are generally small- (<5 cm) to medium-flowered (5–10 cm) for the subtribe, and have a short column in relation to the lip and a stigma whch is much wider than long, adnation between the base of the column and the lip, and lateral lobes reduced to spreading auricles. These flower characters resemble Hagsatera (unresolved within the subtribe) and Dilomilis (sister to Pleurothallidinae) and might represent an ancestral suite of characters in this group.
Domingoa alliance
This is a clade with approx. 12 species and only Domingoa and Homalopetalum. These are small plants (<10 cm), mostly Caribbean, which often have a well-developed column-foot. Based on van den Berg et al. (2000) and preliminary plastid data, Soto-Arenas et al. (2007) synonymized Nageliella with Domingoa. In the same work, Pinelianthe was lumped in Homalopetalum, but no material of Pinelianthe has been obtained for DNA sequencing so far, and therefore their decision was based solely on similarities in flower structure and small habit, pending confirmation by molecular data.
Encyclia alliance
This alliance comprises some 250 species in four genera, and has been the most difficult group to circumscribe in the current study, with weak support in both the combined MP and BA. The main genera here are Encyclia and Prosthechea (the latter a segregate of the former; Higgins, 1997), which are only weakly supported as distinct; this pattern, however, was confirmed by the leaf anatomical data of Pires et al. (2003) despite the incomplete taxon sampling of the latter. Most genera and species have ovoid to clavate heteroblastic pseudobulbs and partial fusion between the column and lip, with only rare exceptions. Relationships between them were not included in this paper, but have been extensively discussed by Higgins et al. (2003).
Scaphyglottis alliance
This group includes four genera and approx. 50 species, most of which have a conspicuous column-foot (not as well developed in Jacquiniella). Despite the vegetative resemblance to Epidendrum, Dimerandra fits here, which is supported by floral morphology. Although unresolved in the present ITS trees, Dimerandra was related to Dinema and Nidema in the Encyclia clade in van den Berg et al. (2000). The internal topology of the Scaphyglottis alliance was studied in detail by Dressler et al. (2004), who synonymized Hexadesmia, Hexisea, Reichenbachanthus and Platyglottis with Scaphyglottis. The highly supported association of Dinema and Nidema with this clade in the present study is useful for understanding this group and is likely to indicate a Caribbean origin for the whole alliance. The Scaphyglottis alliance with Jacquiniella was present in the ITS results of van den Berg et al. (2000) with BP <50. The plastid and combined datasets show it as a moderately to well-supported clade (81 BP and 85, 98 PP, respectively)
Broughtonia alliance
This is a small group (approx. 30 species) of exclusively Caribbean genera including Broughtonia, Psychilis and Tetramicra. Despite the fact that species of Psychilis were considered members of Encyclia for a long time, there are many floral similarities between the latter and Tetramicra. Although Quisqueya was not sampled in the current study, it was shown to be closely related to Tetramicra by the combined molecular and plastid analyses of Higgins et al. (2003).
Laelia and Epidendrum alliances
The close relationship of these two alliances is an important result of this study. The Epidendrum alliance is the largest in number of species (Epidendrum alone has >1500 species) and was weakly supported in van den Berg et al. (2000) and the Laelia alliance (approx. 20 species) was mixed with the Domingoa alliance (<50 BP for both relationships). The Laelia alliance does seems to be characterized by plants with heteroblastic pseudobulbs, eight pollinia and large gullet-flowers which are not very different from those on the Cattleya alliance, which explains their placement previous to the first DNA analyses. The strongly supported paraphyly of Laelia in relation to Schomburgkia led to the synonymization of these genera by van den Berg and Chase (2005) and Soto-Arenas (2005). The Epidendrum alliance is composed largely of Epidendrum species, and can, in general terms, be characterized by a lip united with the column, four pollinia and a reed-stem habit. The free lip of Caularthron appears to be a plesiomophic character state in this clade, but within Epidendrum there are many reversals in character states, resulting in free lips, different pollinia numbers and Cattleya/Encyclia-like habit. A detailed analysis of Epidendrum was presented by Hágsater and Soto-Arenas (2005), which led to the inclusion of Amblostoma, Lanium, Nanodes and Oerstedella and 33 other genera in Epidendrum. The placement of Myrmecophila in this alliance is also noteworthy. ITS data placed this genus sister to the rest of the Cattleya alliance in van den Berg et al. (2000) and sister to Guarianthe aurantiaca in the ITS analysis of this study. However, plastid data place it in an unresolved clade above Meiracyllium. The MP combined analysis here places this genus in the Epidendrum clade (BP <50), but in BA it received 95 PP. This new placement is closer to Caularthron, and both genera are alone in Laeliinae in having hollow pseudobulbs that hold ant nests. It is also reasonably close to Laelia (including Schomburgkia, in which Myrmecophila was included). This could imply an ancestor with long stems and similar flower morphology.
Cattleya alliance
This alliance includes approx. 130 species in four genera. Relationships within this alliance remain confused due to several problems. Four Cattleya species together occupied an unexpected position (based on previous classifications and similar morphology to C. labiata) in the ITS analysis of this study and in van den Berg et al. (2000), but fell in a more intuitive position in the plastid trees. However, in the combined MP analysis, three of them were still grouped together, and C. maxima was sister to C. araguaiensis. These patterns could suggest reticulation events involving some members of this group. Due to the overall level of variation, the ITS data seem to override plastid patterns in the combined analyses. In fact, the plastid analysis produced a topology that is more in agreement with our understanding of the group from a morphological viewpoint: in the plastid analysis, the Cattleya species that were formerly Sophronitis sensu stricto and Brassavola form monophyletic groups, and Cattleya harpophylla clusters with two species of section Parviflorae, in agreement with the system of Withner (1990). The plastid analysis has fewer groups with internal support due to the lower levels of variation. The combined analysis followed more closely the ITS data. The four troublesome Cattleya species (C. maxima, C. lawrenceana, C. lueddemanniana and C. wallisii) still occupied the same positions as in the ITS dataset, but the topology for C. cernua, C. coccinea and C. brevipedunculata is remarkably different. The dominance of the ITS dataset is still clear even after the four troublesome Cattleya sequences are removed (Fig. 3). Although the position of the four species changed, the rest of the tree remained nearly the same as in the ITS trees. Because of the discrepancies between the ITS and plastid trees, the adequacy of ITS for resolving the overall phylogeny of the Cattleya alliance is questionable, and probably the best strategy would be to collect more plastid data or look for an appropriate single-copy nuclear gene to strengthen support for the plastid topologies. On the other hand, contrasting alternative topologies of the plastid and ITS results emphasize the need for detailed studies of hybridization in this group. Several natural interspecific and intergeneric natural hybrids have been reported (Adams and Anderson, 1958), and for this reason hybridization could have played a significant role in the evolution of the Cattleya alliance before their early diversification. Adding plastid data to the original ITS dataset was a great improvement in the bootstrap support within Cattleya. Also, Brassavola was monophyletic as in the plastid analysis with good internal support. The paraphyly of Brassavola in relation to Cattleya in the ITS trees of van den Berg et al. (2000) might have been due to character sampling effects. An increase in characters solved this problem, as in Sheahan and Chase (2000).
Inferences about the evolution of Laeliinae
Hybridization in relation to phylogeny
The ability to produce artificial interspecific and intergeneric crosses of Laeliinae and outgroups and within the subtribe is often a reflection of the phylogenetic relationships. Although there are thousands of hybrids in Cattleya (including species formerly placed in Sophronitis and Laelia; International Orchid Register at http://www.rhs.org.uk/plants/registration_orchids.asp), and across the subtribe with genera from different alliances (e.g. Sophronitis × Constantia and Scaphyglottis × Epidendrum), there are no hybrids between Arpophyllum and other Laeliinae. Genera previously considered to be members of Laeliinae and found in Pridgeon et al. (2001) and van den Berg et al. (2005) and this study to be part of other subtribes (Dilomilis, Helleriella, Isochilus, Neocogniauxia and Ponera) have not produced any registered hybrids. It could be argued there might have been no attempt to produce such hybrids because these genera are not showy. However, such attempts have probably been made at least with Arpophyllum and Isochilus, which are common in cultivation, and Neocogniauxia, which is showy. Even within generic alliances, the degree of fertility seems to be reduced (e.g. many hybrids of Cattleya with Brassavola and Rhyncholaelia have low seed viability). At the same time F1 hybrids between Cattleya and Epidendrum are generally sterile, despite the fact most species have the same chromosome number (2n = 40). In light of the newly clarified phylogenetic relationships, there is a framework in which new artificial crosses for evaluating hybridization potential should be attempted and recorded systematically.
Pollination systems
Bee pollination is the plesiomorphic state in Laeliinae, as stated by Borba and Braga (2003). It occurs in all alliances, despite the small number and lack of detail of early studies (e.g. Dodson and Frymire, 1961; Dodson, 1965). The most common bees are Bombus spp. reported in Cattleya and Pseudolaelia (Borba and Braga, 2003; Smidt et al., 2006) and Xylocopa spp. in Constantia (Matias et al., 1996). A critical taxon for which pollination data are still needed is Arpophyllum, although flower colour, fragrance and appearance would suggest small bees. In smaller subclades within the alliances, such as the rupicoulous species of Cattleya section Parviflorae, shifts to smaller bees associated with polyploidy may be the driving force of radiation (Blumenschein, 1961; Verola, 2008). All other types of specialized pollination occur in well-defined subclades within alliances, such as bird pollination in some Cattleya and Alamania and butterfly and moth polination in Epidendrum, Rhyncholaelia and Brassavola (van der Pijl and Dodson, 1969), which might be key innovations that led to speciation. This is the most likely explanation for the huge number (>1000) of species in Epidendrum.
The main factors of diversification within Laeliinae remain a rich area for research involving pollination mechanisms, habitat preferences and biogeographical patterns, due to the great variation of morphological features and the large number of species and generic and infrageneric groupings. The results of this study improve our understanding of the overall phylogeny within the subtribe and should help in delimiting smaller sets of taxa for more detailed studies which can isolate the different mechanisms responsible for the the richness of species in the subtribe.
ACKNOWLEDGEMENTS
We would like to thank A. Ando, F. Barros, S. Bell, E. L. Borba, D. Hayes, N. B. Machado, W. Foster and S. Beckendorf for plant material and A. de Bruijn, R. Cowan, J. Joseph, C. A. Oliveira and M. Powell for help in the laboratory. This work was supported by the American Orchid Society (USA), Conselho Nacional de Desenvolvimento Científico e Tecnológico (Brazil) and the Royal Botanic Gardens, Kew (UK).
APPENDIX
Voucher information and Genbank accession numbers for the samples used in this study.
| Species name | Voucher | ITS | trnL-F | matK |
|---|---|---|---|---|
| Acianthera ochreata (Lindl.) Pridgeon & M.W.Chase | Harley 15636 (K spirit) | AF262858 | AY008446–7 | AY008458 |
| Acrorchis roseola Dressler | W.M. Whitten 399 (FLAS) | AY008521 | AY422389 | AY397086 |
| Alamania punicea La Llave & Lex. | van den Berg C184 (ESA) | AF260177 | AF267005 | AF263783 |
| Anathallis racemiflora (Lindl. ex Lodd.) Pridgeon & M.W.Chase | W.E. Higgins 140 (FLAS 198267) | AY008477 | AY422379 | AY396076 |
| Arpophyllum giganteum Hartw. ex Lindl. | Chase O-586 (K) | AF266742 | AF266975 | AF265485 |
| Artorima erubescens (Lindl.) Dressler & G.E.Pollard | Chase O-6412 (K) | AF260178 | AF267006–7 | AF263756 |
| Barkeria skinneri (Batem. ex Lindl.) Lindl. ex Paxton | van den Berg C250 (K spirit) | AF260171 | AF266066–7 | AF263750 |
| Barkeria whartoniana (C.Schweinf.) Soto Arenas | van den Berg C249 (K spirit) | AF260170 | AF266999 | AF263754 |
| Bletia catenulata Ruiz & Pav. | E.L. Borba 590 (UEC) | AY008462 | AF008449–50 | AY121720 |
| Bletia catenulata Ruiz & Pav. | W. Forster 10 (ESA) | AY008461 | AF219024–5 | AY121718 |
| Bletia purpurea DC. | van den Berg C342 (K spirit) | AY008463 | AY008451–2 | AF518022–3 |
| Brassavola cucullata (L.) R.Br. | W.E. Higgins 130 (FLAS 198290) | AY008589 | AF263819 | AY396097 |
| Brassavola martiana Lindl. | Unvouchered (Kew 1995-2685) | AF260220 | AF267060–1 | AF263821 |
| Brassavola nodosa (L.) Lindl. | Chase O-339 (K) | AF260219 | AF267059 | AF263820 |
| Brassavola tuberculata Hook. | Brieger Coll. 3497 (ESA) | AF260217 | AF267057 | AF263818 |
| Broughtonia dominguensis (Lindl.) Rolfe | W.E. Higgins 1039 (FLAS) | AF260187 | AF267016–7 | AF263791 |
| Broughtonia lindenii (Lindl.) Dressler | W.E. Higgins 251 (FLAS 198289) | AY008570 | AY422399 | AY396096 |
| Broughtonia negrilensis Fowlie | W.E. Higgins 152 (FLAS 198288) | AF008569 | AY422396 | AY396093 |
| Broughtonia sanguinea (Sw.) R.Br. | Brieger Coll. 14440 (ESA) | AF260186 | AF267015 | AF263790 |
| Cattleya aclandiae Lindl. | Brieger Coll. 32982 (ESA) | AF260207 | AF267040 | AF263810 |
| Cattleya alaorii Brieger & Bicalho | Brieger Coll. 19179 (ESA) | AF260195 | AF267026 | AF263799 |
| Cattleya araguaiensis Pabst | Unvouchered (Kew 1999-1443) | AF260215 | AF267054 | AF263817 |
| Cattleya brevipedunculata (Cogn.) Van den Berg | São Paulo B.G. s.n. IBDF (SP) | AF260202 | AF267034 | AF263805 |
| Cattleya cernua (Lindl.) Van den Berg | Brieger Coll. 15737 (ESA) | AF260200 | AF267032 | AF263803 |
| Cattleya coccinea (Lindl.) Rchb.f. | São Paulo B.G. 9577 (SP) | AF260201 | AF267033 | AF263804 |
| Cattleya dowiana Batem. | Chase O-282 (K) | AF260210 | AF267045 | AF263638 |
| Cattleya esalqueana (Blumensch. ex Pabst) Van den Berg | Brieger Coll. 4980 (ESA) | AF260198 | AF267029 | AF263751 |
| Cattleya fidelensis (Pabst) Van den Berg | C225-Machado s.n. (ESA) | AF260194 | AF267025 | AF263028 |
| Cattleya forbesii Lindl. | Brieger Coll. 2448 (ESA), W.E. Higgins 59 (FLAS 200709) | AY008617 (Brieger) | AY422405 (Higgins) | AY396102 (Higgins) |
| Cattleya harpophylla (Rchb.f.) Van den Berg | Brieger Coll. 6687 (ESA) | AF260199 | AF267030–1 | AF263802 |
| Cattleya intermedia Graham ex Hook. | Brieger Coll. 4095 (ESA) | AF260204 | AF267036 | AF263807 |
| Cattleya labiata Lindl. | Brieger Coll. 5487 (ESA) | AF008594 | AF267051 | AF263759 |
| Cattleya lawrenceana Rchb.f. | Brieger Coll. 3802 (ESA) | AF260208 | AF267041–2 | AF263811 |
| Cattleya lueddemanniana Rchb.f. | Brieger Coll. 3759 (ESA) | AF266744 | AF267052–3 | AF263816 |
| Cattleya maxima Lindl. | Unvouchered (Kew 1983-4362) | AY008631 | AY008456 | AY008460 |
| Cattleya mooreana Withner, D.Alison & Guenard | Unvouchered (Kew 1999-1599) | AF260216 | AF267055–6 | AF263760 |
| Cattleya pumila Hook. | Brieger Coll. 7794 (ESA) | AF260196 | AF267027 | AF263800 |
| Cattleya purpurata (Lindl. & Paxton) Van den Berg | Chase O-997 (K) | AY008641 | AF267024 | AF263797 |
| Cattleya rupestris (Lindl.) Van den Berg | van den Berg C33 (ESA) | AF260197 | AF267028 | AF263801 |
| Cattleya violacea (Kunth) Rolfe | Brieger Coll. 28495 (ESA) | AF260206 | AF267039 | AF263709 |
| Cattleya walkeriana Gardner | Brieger Coll. 1627 (ESA) | AF260205 | AF267037–8 | AF263808 |
| Cattleya wallisii (Linden ex Rchb.f.) Rchb.f. | Brieger Coll. 28787 (ESA) | AF260213 | AF267050 | AF263815 |
| Caularthron bilamellatum (Rchb.f.) R.E.Schultes | Brieger Coll. 3690 (ESA) | AF260173 | AF267001 | AF263780 |
| Chysis bractescens Lindl. | Chase O-436 (K) | AF260150 | AF266971 | AF263640 |
| Coelia triptera (Smith) G.Don ex Steud. | Chase O-324 (K) | AF260151 | AF266972 | AF263643 |
| Constantia cipoensis Pôrto & Brade | São Paulo B.G. s.n. (SP) | AF260193 | AF267023 | AF263796 |
| Dilomilis montana (Sw.) Summerh. | Chase O-206 (K) | AF260147 | AF266967 | AF263765 |
| Dimerandra emarginata (G.Mey.) Hoehne | Chase O-335 (K) | AF260179 | AF267008 | AF263784 |
| Dinema polybulbon (Sw.) Lindl. | Brieger Coll. 6052 (ESA) | AF260154 | AF266976–7 | AF263769 |
| Domingoa kienastii (Rchb.f.) Dressler | W.E. Higgins 225 (FLAS 198291) | AY008564 | AY422398 | AY396095 |
| Domingoa purpurea (Lindl.) Van den Berg & Soto Arenas | van den Berg C260 (K spirit) | AF266743 | AF266980 | AF263771 |
| Earina valida Rchb.f. | van den Berg C296 (Leiden 950080) | AF521077 | AY008448 | AY121741 |
| Encyclia adenocaula (La Llave & Lex.) Schltr. | W.E. Higgins 12 (FLAS 198274) | AY008526 | AY422414 | AY396111 |
| Encyclia cordigera (Kunth) Dressler | W.E. Higgins 24 (FLAS 198276) | AY008528 | AY422417 | AY396114 |
| Encyclia oncidioides (Lindl.) Schltr. | Brieger Coll. 5420 (ESA) | AF260184 | AF267013 | AF263788 |
| Encyclia tampensis (Lindl.) Small | W.E. Higgins 27 (FLAS 198277) | AY008529 | AY422418 | AY396115 |
| Epidendrum armeniacum (Lindl.) | Brieger Coll. 33081 (ESA) | AF260165 | AF266993 | AF263748 |
| Epidendrum schlechterianum Ames | Chase O-301 (K) | AF260172 | AF267000 | AF263779 |
| Epidendrum avicula (Lindl.) Dressler | Brieger Coll. 23319 (ESA) | AF260169 | AF266998 | AF263778 |
| Epidendrum campestre Lindl. | E.L. Borba 553 (UEC) | AF260174 | AF267002 | AF263781 |
| Epidendrum centradenia Rchb.f. | van den Berg C169 (K spirit) | AF260175 | AF267003 | AF263782 |
| Epidendrum ibaguense Lindl. | W.E. Higgins 60 (FLAS 198270) | AY008505 | AY422382 | AY396079 |
| Epidendrum magnoliae Muhl. | W.E. Higgins 244 (FLAS 198271) | AY008506 | AY422383 | AY396080 |
| Epidendrum pseudepidendrum Rchb.f. | van den Berg C4 (ESA) | AF260160 | AF266986 | AF263753 |
| Epidendrum tridactylum Lindl. | Brieger Coll. 15628 (ESA) | AF260164 | AF266692 | AF263775 |
| Guarianthe aurantiaca (Batem. ex Lindl.) Dressler & W.E.Higgins | Brieger Coll. 124 (ESA) | AF260209 | AF267043–4 | AF263812 |
| Guarianthe bowringiana (J.H.Veitch) Dressler & W.E.Higgins | Chase O-1174 (K) | AF260212 | AF267048–9 | AF263814 |
| Guarianthe skinneri (Batem.) Dressler & W.E.Higgins | Kew DNA bank MWC 6497* | AF260211 | AF267046–7 | AF263813 |
| Hagsatera brachycolumna (L.O.Williams) R.González | W.E. Higgins 229 (FLAS 198272) | AY008515 | AY422391 | AY396088 |
| Helleriella guerrerensis Dressler & Hágsater | van den Berg C172 (K spirit) | AF260142 | AF266961 | AF518029 |
| Homalopetalum pachyphyllum (L.O.Williams) Dressler | M. Soto 7640 (AMO) | AF260155 | AF266978–9 | AF263770 |
| Homalopetalum pumilio (Rchb.f.) Schltr. | W.E. Higgins 234 (FLAS 200730) | AY429389 | AY422392 | AY396089 |
| Isabelia pulchella (Kraenzl.) Van den Berg & M.W.Chase | Brieger Coll. 6367 (ESA) | AF260163 | AF266990–1 | AF263745 |
| Isabelia violacea (Lindl.) Schltr. | van den Berg C127 (ESA) | AF260168 | AF266997 | AF263777 |
| Isabelia virginalis Barb.Rodr. | Brieger Coll. 30243 (ESA) | AF260161 | AF266987 | AF263747 |
| Isochilus amparoanus Schltr. | Chase O-204 (K) | AF260143 | AF266962 | AF263762 |
| Isochilus major Cham. & Schltdl. | W.M. Whitten 91348 (FLAS) | AY008481 | AY422381 | AY396078 |
| Jacquiniella teretifolia Britton & P.Wilson | W.M. Whitten 97026 (FLAS) | AY008519 | AY422390 | AY396087 |
| Jaquiniella equitantifolia (Ames) Dressler | van den Berg C171 (K spirit) | AF260158 | AF266982–3 | AF263773 |
| Laelia anceps Lindl. | Chase O-998 (K) | AF260191 | AF267021 | AF263794 |
| Laelia autumnalis (La Llave & Lex.) Lindl. | Chase O-1314 (K) | AF260189 | AF267019 | AF263759 |
| Laelia furfuracea Lindl. | Chase 6410 (K) | AF260190 | AF267020 | AF263793 |
| Laelia lyonsii (Lindl.) L.O.Williams | Brieger Coll. 16846 (ESA) | AF260222 | AF267063 | AF263823 |
| Laelia rubescens Lindl. | Chase O-284 (K) | AY008575 | AY422401 | AY396098 |
| Laelia speciosa (Kunth) Schltr. | Chase O-6088 (K) | AF260188 | AF267018 | AF263792 |
| Laelia splendida (Schltr.) L.O.Williams | W.M. Whitten 93026 (FLAS) | AY008573 | AY422408 | AY396105 |
| Laelia undulata (Lindl.) L.O.Williams | Chase O-1251 (K) | AF260223 | AF267064–5 | AF263749 |
| Leptotes bicolor Lindl. | Brieger Coll. 1068 (ESA) | AF260185 | AF267014 | AF263789 |
| Loefgrenianthus blanche-amesiae (Loefgr.) Hoehne | São Paulo B.G. s.n. (SP) | AF260183 | AF267012 | AF263787 |
| Meiracyllium gemma Rchb.f. | M. Soto 8731 (AMO) | AF260153 | AF266974 | AF263767 |
| Meiracyllium trinasutum Rchb.f. | Chase O-202 (K) | AF260152 | AF266973 | AF263670 |
| Myrmecophila tibicinis (Batem.) Rolfe | Brieger Coll. 6128 (ESA) | AF260203 | AF267035 | AF263806 |
| Myrmecophila tibicinis (Batem.) Rolfe | Chase O-281 (K) | AF008581 | AY422402 | AY396099 |
| Nemaconia striata Lindl. | Chase 6178 (K) | AF260145 | AF266965 | AY121728 |
| Nemaconia striata Lindl. | W.E. Higgins 197 (FLAS 198268) | AY008484 | AY422380 | AY396077 |
| Neocogniauxia hexaptera (Cogn.) Schltr. | van den Berg C244 (K) | AF260148 | AF266968–9 | AF263766 |
| Nidema boothii (Lindl.) Schltr. | W.E. Higgins 192 (FLAS 198273) | AY008522 | AY422384 | AY396081 |
| Octomeria gracilis Lodd. ex Lindl. | Chase O-977 (K) | AF262911 | AF265526 | AF265484 |
| Orleanesia amazonica Barb.Rodr. | São Paulo B.G. 15936 (SP) | AF260176 | AF267004 | AF263755 |
| Polystachya galeata Rchb.f. | van den Berg C283 (K spirit) | AY008470 | AY008453 | AY008464 |
| Ponera exilis Dressler | M. Soto s.n. Paracho, Michoacán (AMO) | AF260144 | AF266963–4 | AF263763 |
| Prosthechea abbreviata (Schltr.) W.E.Higgins | Brieger Coll. 10092 (ESA) | AF260181 | AF267010 | AF263757 |
| Prosthechea aemula (Lindl.) W.E.Higgins | W.E. Higgins 17 (FLAS 198279) | AY008544 | AY422428 | AY396125 |
| Prosthechea citrina (La Llave & Lex.) W.E.Higgins | W.E. Higgins 54 (FLAS 198269) | AY008501 | AY422409 | AY396106 |
| Prosthechea cochleata (L.) W.E.Higgins | MBG 75-0658 (FLAS 198280) | AY008545 | AY422429 | AY396126 |
| Prosthechea glauca Knowles & Westc. | W.E. Higgins 176 (FLAS 200722) | AY429410 | AY422433 | AY396130 |
| Prosthechea mariae (Ames) W.E.Higgins | Chase O-158 (K) | AF260192 | AF267022 | AF263795 |
| Pseudolaelia canaanensis Ruschi | Brieger Coll. 16205 (ESA) | AF260167 | AF266995–6 | AF263746 |
| Pseudolaelia vellozicolla (Hoehne) Pôrto & Brade | São Paulo B.G. 13362 (SP) | AF260166 | AF266994 | AY121748 |
| Psychilis krugii (Bello) Sauleda | Chase O-1062 (K) | AF260157 | AF266891 | AF263772 |
| Psychilis macconnelliae Sauleda | W.E. Higgins 53 (FLAS 198287) | AY008568 | AY422394 | AY396091 |
| Restrepiella ophiocephala (Lindl.) Garay & Dunst. | Chase O-291 (K) | AF262909 | AF265523 | AF265482 |
| Rhyncholaelia digbyana (Lindl.) Schltr. | Chase O-331 (K) | AF260221 | AF267062 | AF263822 |
| Rhyncholaelia glauca (Lindl.) Schltr. | van den Berg C30 (ESA), W.E. Higgins 134 (FLAS) | AY008584 (van den Berg) | AY422404 (Higgins) | AY396101 (Higgins) |
| Scaphyglottis bidentata (Lindl.) Dressler | Brieger Coll. 1253 (ESA) | AF260162 | AF266988–9 | AF263774 |
| Scaphyglottis crurigera (Lindl.) Ames & Correll | Chase O-336 (K) | AF260180 | AF267009 | AF263785 |
| Scaphyglottis cuniculata (Schltr.) Dressler | W.M. Whitten 96051 (FLAS) | AY008551 | AY422387 | AY396084 |
| Scaphyglottis imbricata (Lindl.) Dressler | W.M. Whitten (FLAS 97039) | AY429388 | AY422386 | AY396083 |
| Scaphyglottis modesta (Rchb.f.) Schltr. | Brieger Coll. 2756 (ESA) | AF260159 | AF266984–5 | AF263752 |
| Scaphyglottis pulchella (Schltr.) L.O.Williams | W.M. Whitten 208 (FLAS) | AY174740 | AY422385 | AY396082 |
| Scaphyglottis punctulata (Rchb.f.) C.Schweinf. | Chase O-299 (K) | AF260182 | AF267011 | AF263786 |
| Stelis argentata Lindl. | Kew 1984-7410 (K spirit 60886) | AF262878 | AF265503 | AF265464 |
| Tetramicra elegans (Ham.) Cogn. | W.E. Higgins 160 (FLAS 198285) | AY008566 | AY422397 | AY396094 |
* This sample was originally taken from a plant in the living collection (Kew 1986-04870).
LITERATURE CITED
- Adams H, Anderson E. A conspectus of hybridization in the Orchidaceae. Evolution. 1958;12:512–518. [Google Scholar]
- Azevedo CO, Borba EL, van den Berg C. Evidence of natural hybridization and introgression in Bulbophyllum involutum Borba, Semir & Barros and B. weddellii (Lindl.) Rchb.f. (Orchidaceae) im the Chapada Diamantina, Brazil, by using allozyme markers. Revista Brasileira de Botânica. 2006;29:415–421. [Google Scholar]
- Baker RK. Foliar anatomy of the Laeliinae (Orchidaceae) Washington University: St Louis; 1972. PhD Thesis. [Google Scholar]
- Bentham G. Notes on Orchideae. Journal of the Linnaean Society. 1881;18:281–360. [Google Scholar]
- van den Berg C, Chase MW. Nomenclatural notes on Laeliinae-I. Lindleyana. 2000;15:115–119. [Google Scholar]
- van den Berg C, Chase MW. Nomenclatural notes on Laeliinae. II. Additional combinations and notes. Lindleyana. 2001;16:109–112. [Google Scholar]
- van den Berg C, Chase MW. A chronological view of Laeliinae taxonomical history. Orchid Digest. 2004;68:226–254. [Google Scholar]
- van den Berg C, Chase MW. Nomenclatural notes on Laeliinae. IV. New combinations in Laelia and Sophronitis. Kew Bulletin. 2005;59:565–567. [Google Scholar]
- van den Berg C, Higgins WE, Dressler RL, et al. A phylogenetic analysis of Laeliinae (Orchidaceae) based on sequence data from internal transcribed spacers (ITS) of nuclear ribosomal DNA. Lindleyana. 2000;15:96–114. [Google Scholar]
- van den Berg C, Goldman DH, Freudenstein JV, Pridgeon AM, Cameron KM, Chase MW. An overview of the phylogenetic relationships within Epidendroideae (Orchidaceae) inferred from multiple DNA regions and recircumscription of Epidendreae and Arethuseae (Orchidaceae) American Journal of Botany. 2005;92:613–624. doi: 10.3732/ajb.92.4.613. [DOI] [PubMed] [Google Scholar]
- Blumenschein A. Estudos citológicos nas orquídeas. In: Brieger FG, editor. Atas do Primeiro Congresso Sul-Americano de Genética. Piracicaba: Cadeira de Citologia e Genética Gerald a Escola Superior de Agricultura ‘Luiz de Queiroz’; 1961. pp. 161–163. [Google Scholar]
- Borba EL, Braga PIS. Biologia reprodutiva de Pseudolaelia corcovadensis (Orchidaceae): melitofilia e autoimcompatibilidade em uma Laeliinae basal. Revista Brasileira de Botânica. 2003;26:541–549. [Google Scholar]
- Borba EL, Semir J. Bulbophyllum × cipoense (Orchidaceae), a new natural hybrid from the Brazilian ‘campos rupestres’: description and biology. Lindleyana. 1998;13:113–120. [Google Scholar]
- Brieger FG. Subtribus Epidendrinae. In: Brieger FG, Maatsch R, Senghas K, editors. Schlechter's Die Orchideen. 3rd edn. Berlin: Paul Parey; 1976. pp. 460–635. [Google Scholar]
- Cameron KM, Chase MW, Whitten WM, et al. A phylogenetic analysis of the Orchidaceae: evidence from rbcL nucleotide sequences. American Journal of Botany. 1999;86:208–224. [PubMed] [Google Scholar]
- Chase MW, Hills HG. Silica gel: an ideal material for field preservation of leaf samples for DNA studies. Taxon. 1991;40:215–220. [Google Scholar]
- Dodson CH. Agentes de polinización e su influencia sobre la evolución en la família Orchidaceae. Iquitos: Universidad Nacional de la Amazonia Peruana, Instituto General de Investigaciones; 1965. [Google Scholar]
- Dodson CH, Frymire GP. Natural pollination of orchids. Missouri Botanical Garden Bulletin. 1961;49:133–152. [Google Scholar]
- Doyle JJ, Doyle JL. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochemical Bulletin of the Botanical Society of America. 1987;19:11–15. [Google Scholar]
- Dressler RL. Nomenclatural notes on Orchidaceae I. Taxon. 1960;9:213–214. [Google Scholar]
- Dressler RL. The orchids: natural history and classification. Cambridge, MA: Harvard University Press; 1981. [Google Scholar]
- Dressler RL. The major clades of the Orchidaceae-Epidendroideae. Lindleyana. 1990;5:117–125. [Google Scholar]
- Dressler RL. Phylogeny and classification of the orchid family. Portland, OR: Dioscorides Press; 1993. [Google Scholar]
- Dressler RL, Whitten WM, Williams NH. Phylogenetic relationships of Scaphyglottis and related genera (Laeliinae: Orchidaceae) based on nrDNA ITS sequence data. Brittonia. 2004;56:58–66. [Google Scholar]
- Erixon P, Svennblad B, Britton T, Oxelman B. Reliability of Bayesian posterior probabilities and bootstrap frequencies in phylogenetics. Systematic Biology. 2003;52:665–673. doi: 10.1080/10635150390235485. [DOI] [PubMed] [Google Scholar]
- Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- Fitch WM. Toward defining the course of evolution: minimum change for a specific tree topology. Systematic Zoology. 1971;20:406–416. [Google Scholar]
- Freudenstein JV, Senyo DM, Chase MW. Mitochondrial DNA and relationships in the Orchidaceae. In: Wilson KL, Morrison DA, editors. Monocots: systematics and evolution. Melbourne: CSIRO Publishing; 2000. pp. 421–429. [Google Scholar]
- Freudenstein JV, van den Berg C, Goldman DH, Kores PJ, Molvray M, Chase MW. An expanded plastid DNA phylogeny of the Orchidaceae and analysis of jackknife branch support strategy. American Journal of Botany. 2004;91:149–157. doi: 10.3732/ajb.91.1.149. [DOI] [PubMed] [Google Scholar]
- Goldman DH, Freudenstein JV, Kores PJ, et al. Phylogenetics of Arethuseae (Orchidaceae) based on plastid matK and rbcL sequences. Systematic Botany. 2001;26:670–695. [Google Scholar]
- Hágsater E, Soto-Arena MA. In: Genera orchidacearum. Pridgeon AM, Cribb PJ, Chase MW, Rasmussen FN, editors. Vol. 4. Oxford: Oxford University Press; 2005. pp. 236–251. Epidendrum. [Google Scholar]
- Higgins WE. A reconsideration of the genus Prosthechea (Orchidaceae) Phytologia. 1997;82:370–383. [Google Scholar]
- Higgins WE, van den Berg C, Whitten WM. A combined molecular phylogeny of Encyclia (Orchidaceae) and relationships within Laeliinae. Selbyana. 2003;24:165–179. [Google Scholar]
- Johnson LA, Soltis DE. matK DNA sequences and phylogenetic reconstruction in Saxifragaceae s.s. Systematic Botany. 1994;19:143–156. [Google Scholar]
- Johnson LA, Soltis DE. Phylogenetic inference in Saxifragaceae sensu stricto and Gilia (Polemoniaceae) using matK sequences. Annals of the Missouri Botanical Garden. 1995;82:149–175. [Google Scholar]
- Kress WJ, Prince LM, Williams KJ. The phylogeny and a new classification of the gingers (Zigiberaceae): evidence from molecular data. American Journal of Botany. 2002;89:1682–1696. doi: 10.3732/ajb.89.10.1682. [DOI] [PubMed] [Google Scholar]
- Matias LQ, Braga PIS, Freire AG. Biologia reprodutiva de Constantia cipoensis Porto & Brade, endêmica da Serra do Cipó. Revista Brasileira de Botânica. 1996;19:119–225. [Google Scholar]
- Molvray M, Kores PJ, Chase MW. Polyphyly of mycoheterotrophic orchids and functional influences on floral and molecular characters. In: Wilson KL, Morrison DA, editors. Monocots: systematics and evolution. Melbourne: CSIRO Publishing; 2000. pp. 441–448. [Google Scholar]
- Neyland R, Urbatsch LE. Phylogeny of subfamily Epidendroideae (Orchidaceae) inferred from ndhF chloroplast gene sequences. American Journal of Botany. 1996;83:1195–1206. [Google Scholar]
- Nylander JAA. MRMODELTEST v2. Uppsala: Evolutionary Biology Centre, Uppsala University; 2004. Program distributed by the author. [Google Scholar]
- Pabst GFJ, Dungs F. Orchidaceae Brasilienses. V. 1. Hildesheim: Brücke-Verlag Kurt Schmersow; 1975. [Google Scholar]
- Pabst GFJ, Dungs F. Orchidaceae Brasilienses V. 2. Hildesheim: Brücke-Verlag Kurt Schmersow; 1977. [Google Scholar]
- Pfitzer E. Orchidaceae. II.B.13. Monandrae-Laeliinae, II.B.13.a. Monandrae-Laeliinae-Ponereae. In: Engler A, Prantl K, editors. Die Natürlichen Pflanzenfamilien Ergaenzungsheft. Leipzig: Engelmann; 1889. pp. 52–222. [Google Scholar]
- van der Pijl L, Dodson CH. Orchid flowers: their pollination and evolution. Coral Gables, FL: University of Miami Press; 1969. [Google Scholar]
- Pires MFO, Semir J, Pinna GFAM, Felix LP. Taxonomic separation of the genera Prosthechea and Encyclia (Laeliinae: Orchidaceae) using leaf and root anatomical features. Botanical Journal of the Linnean Society. 2003;143:293–303. [Google Scholar]
- Pridgeon AM, Solano R, Chase MW. Phylogenetic relationships in Pleurothallidinae (Orchidaceae): combined evidence from nuclear and plastid DNA sequences. American Journal of Botany. 2001;88:2286–2308. [PubMed] [Google Scholar]
- Ronquist F, Huelsenbeck JP. MRBAYES 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- Ronquist F, Huelsenbeck JP, van der Mark P. MRBAYES 3.1. Manual. 2005 Distributed with the program by the authors. [Google Scholar]
- Schlechter R. Das System der Orchidaceae. Notizblatt des Botanischen Gartens und Museums zu Berlin-Dahlem. 1926;9:563–591. [Google Scholar]
- Sheahan MC, Chase MW. Phylogenetic relationships within Zygophyllaceae based on DNA sequences of three plastid regions, with special emphasis on Zygophylloideae. Systematic Botany. 2000;25:371–384. [Google Scholar]
- Smidt EC, Silva-Pereira V, Borba EL. Reproductive biology of two Cattleya (Orchidaceae) species endemic to northeastern Brazil. Plant Species Biology. 2006;21:85–91. [Google Scholar]
- Soto-Arenas MA. In: Genera Orchidacearum. Pridgeon AM, Cribb PJ, Chase MW, Rasmussen FN, editors. Vol. 4. Oxford: Oxford University Press; 2005. pp. 265–271. Laelia. [Google Scholar]
- Soto-Arenas MA, Salazar G, van den Berg C. New combinations in Domingoa, Homalopetalum (Orchidaceae: Laeliinae) and Nemaconia (Orchidaceae: Ponerinae) Neodiversity. 2007;2:7–9. [Google Scholar]
- Swofford DL. PAUP* – Phylogenetic analysis using parsimony (*and other methods) version 4.0. Sunderland, MA: Sinauer Associates; 1998. [Google Scholar]
- Szlachetko DL. Systema orchidalium. Fragmenta Floristica et Geobotanica Supplementum. 1995;3:1–152. [Google Scholar]
- Taberlet P, Gielly L, Pautou G, Bouvet J. Universal primers for amplification of three non-coding regions of chloroplast DNA. Plant Molecular Biology. 1991;17:1105–1109. doi: 10.1007/BF00037152. [DOI] [PubMed] [Google Scholar]
- Tanaka R, Kamemoto H. Chromosomes in orchids: counting and numbers. In: Arditti J, editor. Orchid biology: reviews and perspectives. Vol. 3. Ithaca, NY: Comstock Publishing Associates; 1984. pp. 323–410. [Google Scholar]
- Verola CF. Brazil: Universidade Estadual de Campinas; 2008. Estudos biossistemáticos em espécies de Hoffmannseggella H.G. Jones (Orchidaceae:Laeliinae) ocorrentes nos complexos rupestres de altitude. PhD Thesis. [Google Scholar]
- Withner CL. The cattleyas and their relatives. Vol. 2. Portland, OR: Timber Press; 1990. The laelias. [Google Scholar]