Abstract
Background and Aims
Most molecular phylogenetic studies of Orchidaceae have relied heavily on DNA sequences from the plastid genome. Nuclear and mitochondrial loci have only been superficially examined for their systematic value. Since 40% of the genera within Vanilloideae are achlorophyllous mycoheterotrophs, this is an ideal group of orchids in which to evaluate non-plastid gene sequences.
Methods
Phylogenetic reconstructions for Vanilloideae were produced using independent and combined data from the nuclear 18S, 5·8S and 26S rDNA genes and the mitochondrial atpA gene and nad1b-c intron.
Key Results
These new data indicate placements for genera such as Lecanorchis and Galeola, for which plastid gene sequences have been mostly unavailable. Nuclear and mitochondrial parsimony jackknife trees are congruent with each other and previously published trees based solely on plastid data. Because of high rates of sequence divergence among vanilloid orchids, even the short 5·8S rDNA gene provides impressive levels of resolution and support.
Conclusions
Orchid systematists are encouraged to sequence nuclear and mitochondrial gene regions along with the growing number of plastid loci available.
Key words: 26S rDNA, 18S rDNA, 5·8S rDNA, atpA, nad1, orchids, plastid, Vanilla, vanilloid orchids, Vanilloideae
INTRODUCTION
With few exceptions, molecular systematic studies of Orchidaceae above the species level have relied on sequences of plastid genes or intergenic spacers (ITSs)/introns. This has also been true for the majority of angiosperm phylogenetic studies. Among the plastid loci most commonly used by orchidologists are ndhF (Neyland and Urbatsch, 1996), rbcL (Cameron et al., 1999), atpB (Cameron, 2006 ), matK (Goldman et al., 2001) and trnL-F (Kores et al., 2001). Cameron (2004) introduced psaB as an alternative plastid gene for intergeneric studies of Orchidaceae, and the plastid genes psbB and psbC were shown by Cameron and Molina (2006) to be of value in a study to determine the sister taxon of Vanilla. Others have explored the value of additional plastid regions such as rpoC1, ycf1, trnS-G and trnH-psbA for systematic studies of orchids (Dueck and Cameron, 2007; W. M. Whitten, University of Florida, pers. comm.). There is little doubt that the plastid genome holds a wealth of information for the orchid systematist. Now that the entire plastid genome sequence of Phalaenopsis is known (Chang et al., 2005), and the plastid genomes of additional orchid genera have been sequenced (N. H. Williams, University of Florida, pers. comm.), even more plastid regions are likely to be introduced.
During this same time, there has been little effort to incorporate nuclear and/or mitochondrial gene sequences into orchid phylogenetic studies. The notable exception to this observation is the use of the two highly variable nuclear rDNA ITSs (ITS1 and 2), which are nearly ubiquitous among interspecific studies of orchids (e.g. Cox et al., 1997; Gravendeel et al., 2001; Pridgeon and Chase, 2001). Only a few other published studies have looked beyond the plastid genome or nuclear ITS region. Cameron and Chase (2000) sequenced a limited number of taxa for the nuclear 18S rDNA gene in an effort to place Rhizanthella, Cyrtosia and other achlorophyllous genera of uncertain affinity within appropriate subfamilies of Orchidaceae. Likewise, Molvray et al. (2000) used 18S rDNA sequences to gain a better understanding of phylogenetic relationships among genera of Gastrodieae and Epipogieae (both Epidendroideae), all species of which are non-photosynthetic mycoheterotrophs. The only study of Orchidaceae that has sampled the mitochondrial genome has been that of Freudenstein and Chase (2001), who provided a reconstruction of the family based on nad1b-c intron sequences. Again, this study proved itself to be especially valuable in providing data for those achlorophyllous taxa of Epidendroideae for which plastid gene sequencing has proved difficult or impossible.
Subfamily Vanilloideae also contain a number of non-photosynthetic genera, and many of these have been absent in phylogenetic studies using plastid DNA sequences alone. In fact, 40% of the 15 genera of vanilloid orchids sensu Cameron (2003) are mycoheterotrophic. These are Cyrtosia, Erythrorchis, Galeola, Lecanorchis, Pogoniopsis and, to a lesser extent, Pseudovanilla. Despite this high percentage, some of these taxa retain partial or even full-length copies of some plastid genes. Cameron and co-authors in a number of studies (e.g. Cameron et al., 1999; Cameron, 2004, 2006; Cameron and Molina, 2006) have been able to place Pseudovanilla, Cyrtosia and Erythrorchis within Vanilloideae using partial sequences of rbcL, psaB, psbB, psbC and/or atpB. No plastid DNA sequences have been published for Lecanorchis, and no DNA sequences of any kind have been published for Pogoniopsis or Galeola, although Cameron (2006) stated that he had been able to amplify an intact copy of atpB from Galeola. DNA of Pogoniopsis has not been available.
Considering that two of the three plant genomes have been insufficiently sampled within Orchidaceae and that the phylogenetic relationships of Vanilloideae are still incompletely known due to missing data from achlorophyllous genera, this study was initiated to evaluate a few loci from the nuclear and mitochondrial genomes for these orchids. The loci under consideration are 18S, 5·8S and 26S rDNA, representing the tandemly repeated nuclear rDNA cistron, and the mitochondrial atpA gene and nad1b-c intron region.
MATERIALS AND METHODS
Taxon sampling and gene sequencing
Incomplete gene sequences from the nuclear ribosomal genes 26S, 18S and 5·8S rDNA were obtained for 35 taxa, representing 13 of the 15 genera of Vanilloideae (Table 1). Most of these taxa were also sampled for the mitochondrial loci atpA and nad1b-c. DNA was unavailable for two rare genera of Vanilloideae: Pogoniopsis and Dictyophyllaria. Most of the sampled taxa were field collected and vouchered, details of which are given in Table 1. Considering that tribe Pogonieae is known to be monophyletic and sister to tribe Vanilleae based on more broadly sampled studies of Orchidaceae (e.g. Cameron et al, 1999; Cameron, 2004), it was specified as the outgroup in all analyses presented here. The data matrices are available from the author upon request. All sequences have been submitted to GenBank (Table 1).
Table 1.
Species of Vanilloideae (Orchidaceae) sequenced for this study; voucher information and GenBank accession numbers are provided
| Taxon | Voucher | GenBank, 18S | GenBank, 5·8S | GenBank, 26S | GenBank, atpA | GenBank, nad1b-c |
|---|---|---|---|---|---|---|
| Tribe Vanilleae | ||||||
| Clematepistephium smilacifolium (Rchb.f.) N.Hallé | Ziesing 33 (CBG) | FJ425740 | FJ425838 | FJ425773 | – | FJ425846 |
| Cyrtosia lindleyana Hook.f. & Thomson | Cameron 2182 (WIS) | – | – | FJ425775 | FJ425807 | FJ425847 |
| Cyrtosia septentrionalis (Rchb.f.) Garay | Cameron 1048 (WIS) | FJ425742 | FJ425826 | FJ425774 | FJ425808 | FJ425848 |
| Epistephium lucidum Cogn. | Cameron 1039 (WIS) | FJ425744 | FJ425836 | FJ425780 | FJ425810 | FJ425853 |
| Epistephium parviflorum Lindl. | Cameron 1040 (WIS) | FJ425745 | FJ425828 | FJ425777 | – | FJ425850 |
| Epistephium sp. Kunth | Chase O-432 (MICH) | FJ425746 | FJ425839 | FJ425778 | FJ425811 | FJ425851 |
| Epistephium sp. Kunth | Chase O-433 (MICH) | FJ425747 | FJ425827 | FJ425779 | FJ425812 | FJ425854 |
| Epistephium subrepens Hoehne | Cameron 1037 (WIS) | FJ425748 | FJ425837 | FJ425781 | FJ425813 | FJ425852 |
| Eriaxis rigida Rchb.f. | Ziesing 5 (CBG) | FJ425749 | FJ425833 | FJ425782 | FJ425814 | – |
| Erythrorchis altissima Blume | Cameron 1029 (WIS) | FJ425750 | – | FJ425784 | FJ425815 | FJ425855 |
| Erythrorchis cassythoides (A.Cunn. ex Lindl.) Garay | Weston 1831 (WIS) | FJ425751 | FJ425841 | FJ425783 | FJ425816 | AH010950 |
| Galeola nudifolia Lour. | Cameron 1045 (WIS) | FJ425752 | – | FJ425785 | – | – |
| Lecanorchis multiflora J.J.Sm. | Cameron 1015 (WIS) | FJ425755 | FJ425831 | FJ425788 | FJ425819 | FJ425858 |
| Lecanorchis nigricans Honda | Yukawa s.n. (WIS) | FJ425756 | FJ425829 | FJ425789 | FJ425820 | FJ425859 |
| Pseudovanilla foliata (F.Muell.) Garay | Cameron 1046 (WIS) | – | – | FJ425793 | FJ425823 | FJ425863 |
| Pseudovanilla ponapensis (Kaneh. & Yamam.) Garay | Cameron s.n. (WIS) | FJ425760 | – | FJ425794 | FJ425824 | FJ425864 |
| Vanilla africana Lindl. | Chase O-584 (K) | FJ425762 | FJ425834 | FJ425798 | – | FJ425865 |
| Vanilla aphylla Blume | Cameron 1041 (WIS) | FJ425763 | AF151006 | FJ425795 | – | FJ425871 |
| Vanilla barbellata Rchb.f. | Chase O-591 (K) | FJ425764 | FJ425835 | FJ425797 | – | FJ425866 |
| Vanilla cf. planifolia Andrews | Chase O-170 (MICH) | FJ425765 | FJ425832 | FJ425800 | – | FJ425869 |
| Vanilla imperialis Kraenzl. | Chase O-587 (K) | – | FJ425830 | FJ425799 | FJ425825 | FJ425867 |
| Vanilla mexicana Mill. | McCartney s.n. | FJ425761 | – | FJ425796 | – | FJ425868 |
| Vanilla roscheri Rchb.f. | Chase O-540 (K) | FJ425766 | FJ425840 | FJ425801 | – | FJ425870 |
| Tribe Pogonieae | ||||||
| Cleistes cipoana Hoehne | Thomas 12976 (NY) | – | – | FJ425771 | FJ425802 | FJ425842 |
| Cleistes divaricata (L.) Ames | Cameron 1062 (WIS) | FJ425738 | AF151009 | FJ425767 | FJ425803 | FJ425843 |
| Cleistes rosea Lindl. | Cameron 1038 (WIS) | FJ425739 | – | FJ425769 | FJ425804 | – |
| Cleistes sp. 1 | Chase O-430 (MICH) | FJ425741 | AF151013 | FJ425772 | FJ425805 | FJ425845 |
| Cleistes sp. 2 | Jardim 2579 (NY) | – | – | FJ425768 | FJ425806 | FJ425844 |
| Cleistes sp. 3 | Thomas 12975 (NY) | – | – | FJ425770 | – | – |
| Duckeella adolphii Porto & Brade | Romero 3013 (AMES) | FJ425743 | AF151007 | FJ425776 | FJ425809 | FJ425849 |
| Isotria medeoloides Raf. | Keenan s.n. | FJ425753 | – | FJ425786 | FJ425817 | FJ425856 |
| Isotria verticillata (Muhl. ex Willd.) Raf. | Cameron 1030 (WIS) | FJ425754 | AF151008 | FJ425787 | FJ425818 | FJ425857 |
| Pogonia japonica Rchb.f. | Cameron 1034 (WIS) | FJ425757 | AF151011 | FJ425790 | FJ425821 | FJ425861 |
| Pogonia minor Makino | Cameron 1033 (WIS) | FJ425758 | AF151010 | FJ425791 | FJ425822 | FJ425860 |
| Pogonia ophioglossoides (L.) Ker Gawl. | Chase O-437 (MICH) | FJ425759 | AF151012 | FJ425792 | – | FJ425862 |
All newly generated sequences were produced by automated methods, briefly described as follows. Total DNA was extracted according to the manufacturer's protocols using the DNEasy™ (Qiagen, Valencia, CA, USA) method from approx. 0·5 cm2 of dried leaf tissue. Target loci were amplified in 25 µL volumes using standard amplification protocols that typically included the addition of bovine serum albumin. Sometimes betaine was added to relax the secondary DNA structure. Amplification and double-stranded sequencing reactions were completed using primers previously published by various authors. The 18S rDNA primers are mostly based on those published by Bult et al. (1992) and modified later by others. Kuzoff et al. (1998) gave details concerning the primers for 26S rDNA sequencing. The 5·8S rDNA gene was amplified with the flanking ITS1 and 2 regions using primers that can be traced back to various publications, including Nickrent et al. (1994). The nad1b-c primers were synthesized based on information originally provided by Demesure et al. (1995). Primer sequences for atpA sequencing were taken from Davis et al. (1998). Many of these primers have been in use for over a decade, and their exact primary origins are difficult to trace. For this reason, the complete primer sequences are listed here 5'–3' as follows: for 18S, gtagtcatatgcttgtctc*, gcccttccgtcaattcctttaagtttcagc, tcctattgtgttggcctt and cgacttctccttcctcta*; for 26S, agggaagcggatgggggc*, gctatcctgagggaaacttc, cgtgcaaatcgttcgtct and acccatgtgcaagtgccgtt*; for 5·8S, tatgcttaaaytcagcgggt* and aacaaggtttccgtaggtga*; for nad1b-c, catcacctacagccctttc, gaaagggctgtaggtgatggyg, gcattacgatctgcagctca* and ggagctcgattagtttctgc*; for atpA, aagtggatgagatcggtcgag*, ttccgcgataatggaatgca, agcggctctttctaagagac and ggcattcgatcacaga*. Amplification primers are indicated with an asterisk (*). The nuclear rDNA ITS1 and 2 were amplified (with 5·8S), but the spacers could not be aligned among these genera. They were not included in these analyses. In all cases, resulting PCR products were purified using QIAquick™ spin columns (Qiagen). Cycle sequencing reactions were performed using a combination of purified PCR template, primer and BigDye™ reaction mix (Applied Biosystems, Foster City, CA, USA) for 20 cycles. To remove excess dye terminators and primer from the cycle sequencing products, Centri-Sep™ Sephadex columns (Princeton Separations, Adelphia, NJ, USA) were employed. Final purified samples were subsequently dehydrated, re-suspended in a mixture of formamide and loading dye, and pipetted onto a 5% denaturing polyacrylamide gel. Samples were analysed on an Applied Biosystems ABI 377XL automated DNA sequencer. Resulting electropherograms were edited and sequences aligned by eye using Sequencher 3·0 software (GeneCode, Ann Arbor, MI, USA).
Phylogenetic analyses
In this study the aim was to to determine whether selected nuclear and/or mitochondrial gene sequences would provide an appropriate level of variation to reconstruct phylogenetic relationships within Vanilloideae and whether they would result in parallel or conflicting topologies for intergeneric relationships within Vanilloideae when compared with previously published plastid DNA studies. Abbreviated heuristic searches were executed to calculate relative consistency index (CI) and retention index (RI) scores for each analysis, but not to find all equally parsimonious trees. Instead, parsimony jackknife consensus trees were calculated and used to address comparative issues of tree topology, resolution and support. In a few cases, a single tree was generated to highlight the variability of branch lengths and sequence divergence among taxa. In all cases, gaps were treated as missing data, and there was no attempt made to code indels. Jackknife support was calculated by performing analyses of 5000 heuristic search replicates using the TBR branching swapping algorithm and the following settings: 37% deletion, emulate ‘jac’ resampling, one random addition per replicate and saving two trees per replicate. All analyses were performed using PAUP* v. 4.0b10 (Swofford, 2002).
RESULTS
Trees for individual loci
The aligned nuclear 5·8S matrix contains 191 characters of which 45 (24%) are variable and 30 (16%) potentially informative. The CI is 0·78 and the RI is 0·90. Sequences were available for 24 taxa. Only eight nodes of the tree (not shown) were supported (>50% jackknife), including clades comprised of Vanilla spp. (87%), Epistephium spp. (96%), Lecanorchis spp. (99%), Pogonia spp. (62%) and temperate Pogonieae (72%).
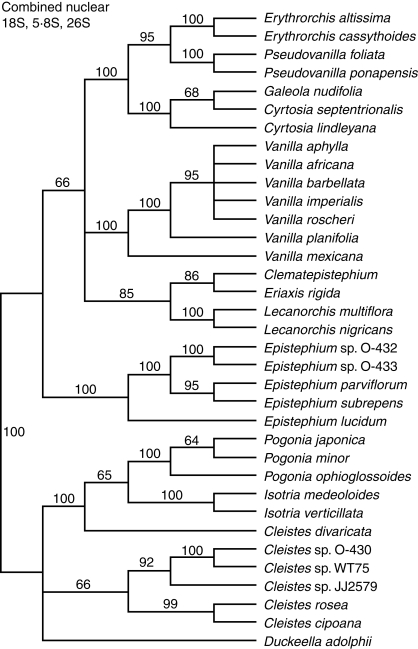
The aligned nuclear 18S matrix contains 1699 characters of which 207 (12%) are variable and 176 (10%) potentially informative. The CI for these data is 0·66 and RI 0·88. Sequences were available for 29 taxa. The parsimony jackknife tree is not shown, but see Fig. 1 for the results of the combined nuclear gene analysis.
Fig. 1.
Parsimony jackknife tree for Vanilloideae based on combined nuclear ribosomal gene sequences. Support percentages >50 are indicated above the branches.
The aligned nuclear 26S matrix contains 1098 characters of which 349 (32%) are variable and 291 (27%) potentially informative. The CI is 0·59 and RI 0·84. Sequences were available for 35 taxa. The parsimony jackknife tree is not shown, but see Fig. 1.
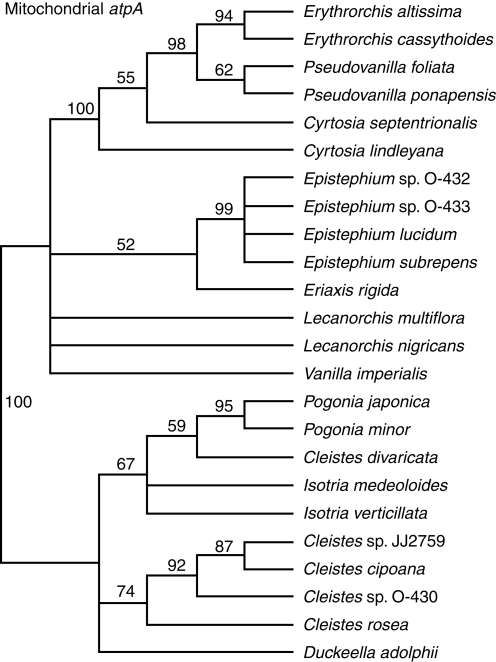
The aligned mitochondrial atpA matrix contains 1217 characters of which 76 (6%) are variable and 53 (4%) potentially informative. The CI is 0·77 and RI 0·92. Sequences were available for 24 taxa. The parsimony jackknife tree is presented (Fig. 2).
Fig. 2.
Parsimony jackknife tree for Vanilloideae based on mitochondrial atpA gene sequences. Support percentages >50 are indicated above the branches.
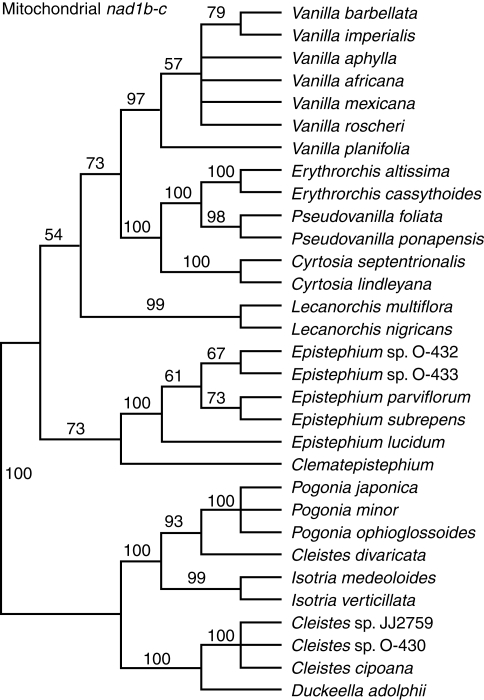
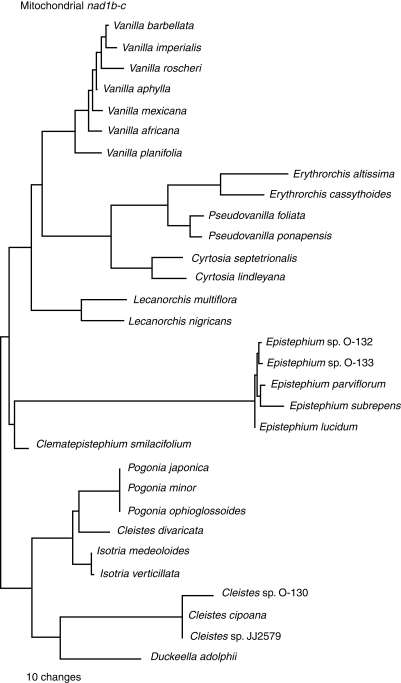
The aligned mitochondrial nad1b-c matrix contains 2412 characters of which 550 (23%) are variable and 407 (17%) potentially informative. The CI for these data is 0·84 and RI 0·92. Sequences, which ranged in size from 951 bp in the case of Clematepistephium smilacifolium to 2001 bp for Erythrorchis altissima, were available for 31 taxa. The average sequence length was approx. 1600 bp. A parsimony jackknife tree is presented as Fig. 3. Sequences from Epistephium spp. are highly divergent from all other taxa due to indels and possible inversions. To highlight this phenomenon, a single tree is depicted to show relative branch lengths (Fig. 4).
Fig. 3.
Parsimony jackknife tree for Vanilloideae based on mitochondrial nad1b-c intron sequences. Support percentages >50 are indicated above the branches.
Fig. 4.
A single tree depicting relative branch lengths for one of the most equally parsimonious trees for Vanilloideae based on mitochondrial nad1b-c intron sequences.
Trees for combined loci
The three nuclear genes, 26S, 5·8S and 18S rDNA, were combined into a single matrix containing 2988 characters, of which 497 are potentially informative. The CI is 0·63 and RI 0·84. A parsimony jackknife tree is presented (Fig. 1).
Likewise, the two mitochondrial loci, atpA and nad1b-c, were combined into a single matrix containing 3629 characters, of which 459 are potentially informative. The CI is 0·83 and RI 0·92 (tree not shown).
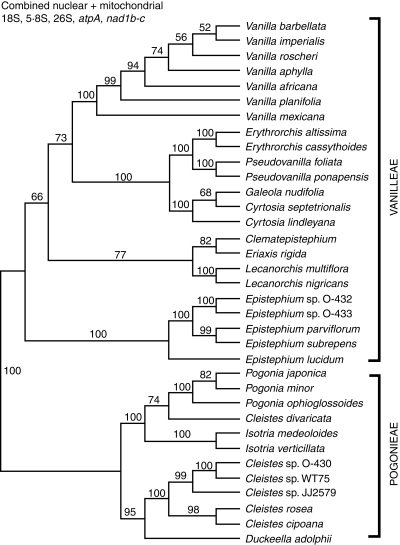
The three nuclear and two mitochondrial loci were combined into a five-locus matrix containing 6617 characters. The matrix contains 35 taxa. The CI is 0·71 and RI 0·88. A parsimony jackknife tree is presented (Fig. 5). From this it can be seen that every node of the tree is supported by jackknife percentages >50%, and all but three are supported by >70%. Vanilla is monophyletic and sister to a clade containing mostly mycoheterotrophic vines. A second lineage of mycoheterotrophs, Lecanorchis spp., is sister to the pair of monotypic genera endemic to New Caledonia, Eriaxis and Clematepistephium. Sister to all of the rest of Vanilleae is Epistephium. Relationships within Pogonieae are highly supported. Pogonia is sister to Cleistes divaricata, and this pair is sister to Isotria. These orchids form a clade of temperate Pogonieae. Sister to the temperate clade are tropical Pogonieae, represented by five accessions of Cleistes, with Duckeella as their sister.
Fig. 5.
Parsimony jackknife tree for Vanilloideae based on combined nuclear 26S, 5·8S and 18S rDNA gene sequences plus the mitochondrial atpA gene and nad1b-c intron sequences. Support percentages >50 are indicated above the branches.
DISCUSSION
The value of non-plastid gene sequences within Vanilloideae
The greatest value of the data presented here lies in the positioning of those achlorophyllous taxa of Vanilloideae for which only partial, functionless and/or divergent plastid sequences have been obtained; in some cases no plastid gene sequences have been available. For all five loci considered in this rerport, complete sequences of Lecanorchis multiflora and L. nigricans were successfully amplified and sequenced. To date, no sequences from any species of this mycoheterotrophic genus have been obtained from the plastid genome, despite repeated efforts to do so. Similarly, there are no published sequences of the genus Galeola. This may, in part, be due to the fact that only a degraded DNA sample has been available. Nevertheless, quality sequences of nuclear 18S and 26S rDNA were obtained from this sample. The other non-photosynthetic members of the subfamily for which samples are available were also successfully sequenced for both mitochondrial loci and most of the nuclear loci. These included two species each of Cyrtosia, Erythrorchis and Pseudovanilla. As stated in the Introduction, plastid gene or pseudogene sequences have been recovered from these plants in the past, and so there are existing phylogenetic hypotheses with which to compare these nuclear and mitochondrial results.
It is also worth commenting on the unexpected utility of the nuclear 5·8S rDNA gene for reconstructing relationships within Vanilloideae. This short gene (<200 bp) is flanked by the ITS1 and ITS2 regions, and is generally considered to be so highly conserved as to be of little value for systematic studies. However, Vanilloideae are known for their exceptional rates of molecular evolution in all three genomic compartments, and hypervariable regions such as nuclear ITS rDNA or the plastid trnL-F intron/spacer are almost impossible to align among them. On the other hand, genes typically considered useful primarily for interfamilial studies (i.e. many of those considered herein) are of value within Vanilloideae. Thus, it is not surprising that there is a relatively large amount of phylogenetic information in the 5·8S rDNA sequences for this set of taxa.
In general, the resulting trees presented here are congruent with one another and with previously published cladograms. Both nuclear and mitochondrial sequences, in this case, are worthy of further consideration as more taxa are collected and sampled. At the same time, however, caution must be exercised when using these regions, particularly those derived from the mitochondrial genome. Others have commented on the perils and pitfalls of analysing mitochondrial sequences in plants (Palmer et al., 2000; Adams et al., 2002; Peterson et al., 2006), and some of the same anomalies of molecular evolution seen in other plant lineages are evident in these data as well. For example, among these nad1 intron data are seen extremes in length variation ranging as much as 1500 bp between Clematepistephium and Erythrorchis. Numerous indels are present, making alignment a challenge, and putative inversions and/or highly divergent stretches of nucleotides relative to the other genera are evident within Epistephium, thereby placing the genus on an especially long branch (Fig. 4).
Relationships among Pogonieae
These data corroborate relationships among taxa of Pogonieae uncovered by earlier studies (e.g. Cameron et al., 1999; Cameron, 2004; Cameron and Molina, 2006). In all cases, the three species of Pogonia form a clade, with the two Asian species sister to each other in most trees. The two species of Isotria are always sister to each other, and Cleistes divaricata is always part of a clade of temperate taxa. This renders the genus polyphyletic since the tropical species sampled almost always form an unrelated clade. Cleistes divaricata should be treated as a species of Pogonia or as a distinct, possibly monotypic genus [Smith et al. (2004) showed there to be two genetic lineages in North American Cleistes, but the division did not follow the current division into C. bifaria and C. divaricata, with some populations of C. bifaria being genetically closer to C. divaricata than to the other populations of C. bifaria]. Identifying morphological characters to support the generic status for C. divaricata s.l. separate from Pogonia or Cleistes has proved difficult.
The only taxa that vary in their placement within Pogonieae among the individual trees are Duckeella and C. divaricata. In the plastid and ITS analyses published to date, Duckeella is sister to all Pogonieae, but that position is not supported here. Instead, Duckeella is either sister to temperate Pogonieae (26S, 69% jackknife support; 5·8S, 50% jackknife support), sister to tropical Cleistes species (18S, 60% jackknife support; nad1, 100% jackknife support) or unresolved (atpA, <50% jackknife support). The conflicting positions are mostly poorly supported and may simply be due to sampling error since there are no taxa included in these analyses from other orchid subfamilies. In any case, combination of data from all three genomic compartments positions Duckeella sister to all Pogonieae with 100% jackknife support (tree not shown), and this would seem to be its proper placement. It is morphologically distinct from all other taxa in the tribe (see Cameron and Chase, 1999).
The position of C. divaricata from North America also varies among trees. In some cases it is sister to Pogonia; in other cases it is sister to Pogonia plus Isotria, and in other cases its position is unresolved within temperate Pogonieae. Among nuclear and mitochondrial gene trees, only nad1b-c provides high jackknife support (93%) for the position of C. divaricata, and this is sister to Pogonia. That same relationship is recovered when all data are combined, including plastid and morphological data (e.g. Cameron and Chase, 1999; Cameron et al., 1999; Cameron, 2006).
Relationships among Vanilleae
Unlike its sister clade, relationships among the major lineages of Vanilleae have been less certain in previous studies. Clear molecular and morphological evidence has been accumulating over the past decade to support the monophyly of Epistephium, Vanilla, the pair of New Caledonian endemics (Clematepistephium and Eriaxis) and the clade of mostly achlorophyllous mycoheterotrophic genera related to Pseudovanilla. These new data confirm those findings. Also, Cameron and Molina (2006) presented well-supported evidence for a sister relationship between Vanilla and the mycoheterotrophic Pseudovanilla clade based on plastid gene sequences. That sister relationship is also supported by the nuclear and mitochondrial gene trees depicted here.
What remains unclear is the exact relationship among Epistephium, the New Caledonian sister genera and Lecanorchis. Each individual jackknife consensus tree calculated for the separate nuclear and mitochondrial gene matrices places Lecanorchis in an unresolved (or at least unsupported) position among Vanilleae. In combination, however, the three nuclear genes place Lecanorchis sister to the New Caledonian genera with 85% jackknife support (see Fig. 1). That relationship is not supported by the mitochondrial data, but holds up with 77% jackknife support in the combined nuclear + mitochondrial gene trees (Fig. 5). This is a surprising relationship. Lecanorchis has seeds unlike those of the New Caledonian genera (or any other orchid, for that matter), Lecanorchis spp. are distributed throughout Southeast Asia but not in Australia or the Pacific Islands, and the floral morphology of Lecanorchis is more similar to that of some Vanilla spp. than to anything else. However, Lecanorchis does share with Eriaxis and Clematepistephium the phenomenon of placental intrusion into the ovary resulting in a pseudo-trilocular condition. Lecanorchis is such an enigmatic genus of orchids, and with such reduced vegetative morphology, that its placement as sister to the New Caledonian taxa is just as believeable as a position anywhere else in Vanilleae. However, all three genera are relatively phylogenetically isolated, and undoubtedly there have been numerous extinction events of perhaps entire lineages throughout Vanilloideae that could confound reconstruction of phylogenetic relationships of the extant relictual members. More systematic data for Lecanorchis are needed before we can have confidence in the result uncovered in this study, but there is now a hypothesis in hand to be further evaluated. No plastid data from Lecanorchis exist for making further comparisons.
The position of Galeola in these trees must be addressed because it renders Cyrtosia paraphyletic. This is probably an anomaly related to taxon and gene sampling. Sequences for all three species in question, Galeola nudifolia, Cyrtosia septentrionalis and Cyrtosia lindleyana, are available only for 26S rDNA. That gene provides just 67% support for Galeola being sister to C. septentrionalis; the same relationship is seen in the combined tree. For all other single locus matrices, Galeola and/or C. lindleyana are missing. There is little doubt that the two genera are closely related since they have similar floral morphology, but their seeds, fruits and habit are different. Galeola species are climbing vines with dry dehiscent fruits and winged seeds; Cyrtosia species are erect herbs with fleshy indehiscent fruits and crustose seeds. Further sampling of data and taxa may resolve each as monophyletic.
Future molecular systematics studies among Vanilloideae
This study demonstrates the potential value of sampling nuclear and mitochondrial gene regions (not only plastid) for reconstructing the phylogeny of Orchidaceae, especially for lineages that include mycoheterotrophic species. Most relationships supported by these new data confirm and even strengthen those uncovered with plastid gene data alone (results not shown). At the same time they remind us that issues related to incomplete sampling (of data and/or taxa) can cloud our interpretation of phylogeny. There are still no molecular data for Pogoniopsis, and this genus could be important for understanding the evolutionary history of vanilloid orchids, assuming that it even belongs within the subfamily. This achlorophyllous genus from Brazil shares reproductive characters with some Pogonieae and Vanilleae. Placing it among these taxa could affect current tree topologies in a manner that we have no way to predict. Even more enigmatic is Dictyophyllaria, an orchid that is known only from the type specimen and a line drawing. It shares morphological features with both Vanilla and Epistephium, but may well be extinct. If this is the case, then it serves to remind us that extant Vanilloideae, a line of orchids perhaps >76 million years old (Ramirez et al., 2007), are relicts of what may have been an even more diverse clade of Orchidaceae with a complex history of evolution.
Our understanding of the fundamental biology of Pogonia ophioglossoides, Cleistes divaricata and Vanilla mexicana has come a long way since Linnaeus (1753) described them in Species plantarum. It has advanced most noticeably during the past decade, thanks in part to early molecular systematics studies, but, like all scientific knowledge, these are incomplete. Such studies of Vanilloideae must be expanded and refined.
ACKNOWLEDGEMENTS
Special thanks are extended to Ken Wurdack and Susan Pell for their assistance in the laboratory. The Lewis B. and Dorothy Cullman Foundation as well as the USA National Science Foundation (grant DEB-0108100) are acknowledged for their financial support. This paper was presented as part of the Linnaean Society's tercentenary celebration, ‘Orchid Evolutionary Biology and Conservation – from Linnaeus to the 21st Century’, held at the Royal Botanic Gardens, Kew. Gratitude is extended to the Society and the organizers for their invitation to participate in this special event.
LITERATURE CITED
- Adams KL, Qiu YL, Stoutemyer M, Palmer JD. Punctuated evolution of mitochondrial gene content: high and variable rates of mitochondrial gene loss and transfer to the nucleus during angiosperm evolution. Proceedings of the National Acadamy of Sciences, USA. 2002;99:9905–9912. doi: 10.1073/pnas.042694899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bult C, Kallersjo M, Suh Y. Amplification and sequencing of 16/18S rDNA from gel-purified total plant DNA. Plant Molecular Biology Reporter. 1992;10:273–284. [Google Scholar]
- Cameron KM. Vanilloideae. In: Pridgeon A, Cribb P, Chase M, Rasmussen F, editors. Genera orchidacearum vol 3. Oxford: Oxford University Press; 2003. pp. 281–334. [Google Scholar]
- Cameron KM. Utility of plastid psaB gene sequences for investigating intrafamilial relationships within Orchidaceae. Molecular Phylogenetics and Evolution. 2004;31:1157–1180. doi: 10.1016/j.ympev.2003.10.010. [DOI] [PubMed] [Google Scholar]
- Cameron KM. A comparison of plastid atpB and rbcL gene sequences for inferring phylogenetic relationships within Orchidaceae. Aliso. 2006;22:447–464. [Google Scholar]
- Cameron KM, Chase MW. Phylogenetic relationships of Pogoniinae (Vanilloideae, Orchidaceae): an herbaceous example of the eastern North America–eastern Asia phytogeographic disjunction. Journal of Plant Research. 1999;112:317–329. [Google Scholar]
- Cameron KM, Chase MW. Nuclear 18S rDNA sequences of Orchidaceae confirm the subfamilial status and circumscription of Vanilloideae. In: Wilson KL, Morrison DA, editors. Monocots, systematics and evolution. Collingwood: CSIRO; 2000. pp. 457–464. [Google Scholar]
- Cameron KM, Molina MC. Photosystem II gene sequences of psbB and psbC clarify the phylogenetic position of Vanilla (Vanilloideae, Orchidaceae) Cladistics. 2006;22:239–248. [Google Scholar]
- Cameron K, Chase M, Whitten M, et al. A phylogenetic analysis of the Orchidaceae, evidence from rbcL nucleotide sequences. American Journal of Botany. 1999;86:208–224. [PubMed] [Google Scholar]
- Chang CC, Lin HC, Lin IP, et al. The chloroplast genome of Phalaenopsis aphrodite (Orchidaceae): comparative analysis of evolutionary rate with that of grasses and its phylogenetic implications. Molecular Biology and Evolution. 2005;23:279–291. doi: 10.1093/molbev/msj029. [DOI] [PubMed] [Google Scholar]
- Cox AV, Pridgeon AM, Albert VA, Chase MW. Phylogenetics of the slipper orchids (Cypripedioideae, Orchidaceae): nuclear rDNA ITS sequences. Plant Systematics and Evolution. 1997;208:197–223. [Google Scholar]
- Davis JI, Simmons MP, Stevenson DW, Wendel JF. Data decisiveness, data quality, and incongruence in phylogenetic analysis: an example from monocotyledons using mitochondrial atpA sequences. Systematic Biology. 1998;47:282–310. doi: 10.1080/106351598260923. [DOI] [PubMed] [Google Scholar]
- Demesure B, Sodzi N, Petit RJ. A set of universal primers for amplification of polymorphic non-coding regions of mitochondrial and chloroplast DNA in plants. Molecular Ecology. 1995;4:129–131. doi: 10.1111/j.1365-294x.1995.tb00201.x. [DOI] [PubMed] [Google Scholar]
- Dueck LA, Cameron KM. Sequencing re-defines Spiranthes relationships, with implications for rare and endangered taxa. Lankesteriana. 2007;7:190–195. [Google Scholar]
- Freudenstein JV, Chase MW. Analysis of mitochondrial nad1b-c intron sequences in Orchidaceae: utility and coding of length-change characters. Systematic Botany. 2001;26:643–657. [Google Scholar]
- Goldman D, Freudenstein J, Kores P, et al. Phylogenetics of Arethuseae (Orchidaceae) based on plastid matK and rbcL sequences. Systematic Botany. 2001;26:670–695. [Google Scholar]
- Gravendeel B, Chase MW, de Vogel EF, Roos MC, Mes THM, Bachmann K. Molecular phylogeny of Coelogyne (Epidendroideae; Orchidaceae) based on plastid RFLPs, matK, and nuclear ribosomal ITS sequences: evidence for polyphyly. American Journal of Botany. 2001;88:1915–1927. [PubMed] [Google Scholar]
- Kores PJ, Molvray M, Weston PH, et al. A phylogenetic analysis of Diurideae (Orchidaceae) based on plastid DNA sequence data. American Journal of Botany. 2001;88:1903–1914. [PubMed] [Google Scholar]
- Kuzoff RK, Sweere JA, Soltis DE, Soltis PS, Zimmer EA. The phylogenetic potential of entire 26S rDNA sequences in plants. Molecular Biology and Evolution. 1998;15:251–263. doi: 10.1093/oxfordjournals.molbev.a025922. [DOI] [PubMed] [Google Scholar]
- Linnaeus C. Species plantarum. 2 vols. Stockholm: L. Salvii; 1753. (Facsim. ed. 1957–1959, London.) [Google Scholar]
- Molvray M, Kores PJ, Chase MW. Polyphyly of mycoheterotrophic orchids and functional influences on floral and molecular characters. In: Wilson KL, Morrison DA, editors. Monocots: systematics and evolution. Collingwood: CSIRO; 2000. pp. 441–448. [Google Scholar]
- Neyland R, Urbatsch L. Evolution in the number and position of fertile anthers in Orchidaceae inferred from ndhF chloroplast gene sequences. Lindleyana. 1996;11:47–53. [Google Scholar]
- Nickrent DL, Schuette KP, Starr EM. A molecular phylogeny of Arceuthobium based upon rDNA internal transcribed spacer sequences. American Journal of Botany. 1994;81:1149–1160. [Google Scholar]
- Palmer JD, Adams KL, Cho Y, Parkinson CL, Song K. Dynamic evolution of plant mitochondrial genomes: mobile genes and introns and highly variable mutation rates. Proceedings of the National Acadamy of Sciences, USA. 2000;97:6960–6966. doi: 10.1073/pnas.97.13.6960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen G, Seberg O, Davis JI, et al. Mitochondrial data in monocot phylogenetics. Aliso. 2006;22:52–62. [Google Scholar]
- Pridgeon A, Solano R, Chase MW. Phylogenetic relationships in Pleurothallidinae (Orchidaceae): combined evidence from nuclear and plastid DNA sequences. American Journal of Botany. 2001;88:2286–2308. [PubMed] [Google Scholar]
- Ramirez SR, Gravendeel B, Singer RB, Marshall CR, Pierce NE. Dating the origin of the Orchidaceae from a fossil orchid with its pollinator. Nature. 2007;448:1042–1045. doi: 10.1038/nature06039. [DOI] [PubMed] [Google Scholar]
- Smith SD, Cowan RS, Gregg KB, Chase MW, Maxted N, Fay MF. Genetic discontinuities among populations of Cleistes in North America (Orchidaceae, Vanilloideae) Botanical Journal of the Linnean Society. 2004;147:87–95. [Google Scholar]
- Swofford DL. PAUP* 4.0b10, Phylogenetic analysis using parsimony (* and other methods) Sunderland, MA: Sinauer Associates; 2002. [Google Scholar]