Abstract
Background and Aims
The orchid genus Dichaea, with over 100 species found throughout the neotropics, is easily recognized by distichous leaves on long stems without pseudobulbs and flowers with infrastigmatic ligules. The genus has previously been divided into four sections based primarily on presence of ovary bristles and a foliar abscission layer. The aim of this work is to use DNA sequence data to estimate phylogenetic relationships within Dichaea and map the distribution of major morphological characters that have been used to delimit subgenera/sections.
Methods
Sequence data for the nuclear ribosomal internal transcribed spacers and plastid matK, trnL intron, trnL-F spacer and ycf1 for 67 ingroup and seven outgroup operational taxonomic units were used to estimate phylogenetic relationships within Dichaea. Taxa from each of the four sections were sampled, with the greatest representation from section Dichaea, the most diverse and taxonomically puzzling group.
Key Results
Molecular data and morphology support monophyly of Dichaea. Results indicate that section Dichaeopsis is polyphyletic and based on symplesiomorphies, including deciduous leaves and smooth ovaries that are widespread in Zygopetalinae. There are at least three well-supported clades within section Dichaeopsis. Section Pseudodichaea is monophyletic and defined by setose ovaries and leaves with an abscission layer. Sections Dichaea and Dichaeastrum are monophyletic and defined by pendent habit and persistent leaves. Section Dichaeastrum, distinguished from section Dichaea primarily by a glabrous ovary, is potentially polyphyletic.
Conclusions
The leaf abscission layer was lost once, occurring only in the derived sections Dichaea and Dichaeastrum. The setose fruit is a more homoplasious character with several losses and gains within the genus. We propose an informal division of the genus based upon five well-supported clades.
Key words: Dichaea, matK, nrITS, Orchidaceae, trnL intron, trnL-F spacer, ycf1, Zygopetalinae
INTRODUCTION
Dichaea Lindl. is a rarely cultivated orchid genus, closely related to some commonly cultivated, showy genera including Zygopetalum Hook., Huntleya Bateman ex Lindl. and Pescatoria Rchb.f. With approx. 100 species, Dichaea is found throughout the neotropics, reaching peak diversity in the equatorial Andes. Dichaea is the largest genus in subtribe Zygopetalinae with about 400 species (Chase et al., 2003). Zygopetalinae form a strongly supported clade (Whitten et al., 2000) within tribe Cymbidieae (Chase et al., 2003). All members of this subtribe are part of an exclusively neotropical clade within the widespread tribe Cymbidieae that also includes Catasetinae, Coeliopsidinae, Cymbidiinae, Cyrtopodiinae, Eriopsidinae, Eulophiinae, Maxillariinae, Oncidiinae, Stanhopeinae and Vargasiellinae.
The unusual habit and floral morphology (Fig. 1) of Dichaea have made its systematic position controversial. Szlachetko (1995) placed Dichaea in the monogeneric subtribe Dichaeinae, part of a larger tribe Dichaeeae including Vargasiella C.Schweinf., Fernandezia Lindl. and Pachyphyllum Kunth, all with a similar vegetative habit. No sequence data have been published for Vargasiella, but it is now treated as the only member of subtribe Vargasiellinae (Romero and Carnevali, 1993; Pridgeon et al., 2009). Molecular data clearly place Fernandezia and Pachyphyllum in Oncidiinae (Williams et al., 2001). Although placement of Dichaea within Zygopetalinae was novel in the molecular analysis of Whitten et al. (2000), it was supported by single-flowered inflorescences, pollinarium structure and pseudobulbless stems (Dressler, 1993b).
Fig. 1.
Morphological features of Dichaea: (A) Dichaea globosa (section Pseudodichaea) (co = column, pe = petal, la = labellum, se = sepal); (B) D. panamensis (section Dichaeopsis); (C) D. glauca (section Dichaeopsis); (D) D. trulla (section Dichaeopsis); (E) D. caveroi (section Dichaeopsis); (F) D. ancoraelabia (section Dichaeopsis); (G) D. poicillantha (section Dichaea); (H) D. squarrosa (section Dichaea); (I) typical column (note the round stigma and pubescent instrastigmatic ligule; an = anther, li = infrastigmatic ligule, st = stigma, vi = viscidium); (J) pollinarium of Dichaea in natural configuration; (K) pollinarium pressed to show the elastic caudicles as the upper pollinia extend over the lower ones (ca = caudicle, po = pollinium, sp = stipe, vi = viscidium); (L) D. glauca (note erect stem and thickly glaucous leaves); (M) D. cryptarrhena; note strongly pendulous habit; (N) D. ecuadorensis (note semi-erect habit); (O) spiny fruits of section Dichaea; (P) leaves with an abscission layer between the sheath and blade (section Pseudodichaea); (Q) leaves lacking an abscission layer (section Dichaea).
Since Lindley (1833) described Dichaea, generic and subgeneric classifications have been problematic. Knowles and Westcott (1839) first addressed subgeneric categories within the genus. They erected a second genus, Epithecia Knowles & Westc., for species with articulate (deciduous) leaves. Pfitzer (1889) described another segregate genus, Dichaeopsis, to encompass species with articulate leaves, apparently ignoring the work of Knowles and Westcott. Kuntze (1904) reduced Dichaeopsis to a section within Dichaea. Cogniaux (1906) retained Dichaea as a single genus of four sections: Dichaea (as Eudichaea), Dichaeastrum Cogn., Dichaeopsis (Pfitzer) Kuntze and Pseudodichaea Cogn. Section Dichaea was distinguished by setose ovaries and non-articulate leaves, section Dichaeastrum by glabrous ovaries and non-articulate leaves, section Dichaeopsis by glabrous ovaries and articulate leaves and section Pseudodichaea by setose ovaries and articulate leaves. Although Cogniaux's treatment was limited to Brazilian Dichaea, his sections did encompass all combinations of the two characters (i.e. leaf abscission and setose ovaries). Schlechter (1914) accepted the four groups of Cogniaux but preferred the generic delimitation of Knowles and Westcott (1839). Therefore he placed sections Dichaea and Dichaeastrum in Dichaea and sections Dichaeopsis and Pseudodichaea in Epithecia. Kränzlin (1923) treated the group as a single genus with three sections: Dichaea, Dichaeopsis and Maxillariopsis Kränzl. However, the four species placed in section Maxillariopsis are currently treated as members of Maxillariella M.A.Blanco & Carnevali in subtribe Maxillariinae (Folsom, 1987, 1996). Senghas (1996), who followed Kränzlin's work closely, erected two subgenera in Dichaea. His subgenus Dichaea included section Dichaea and subgenus Epithecia included sections Dichaeopsis and Maxillariopsis.
There is no monograph, revision or even synopsis of the entire genus. Folsom (1987) monographed section Dichaea, the most taxonomically and morphologically diverse group in the genus. In addition to his monograph, he diagrammed his ideas of the relationships within section Dichaea in a non-cladistic manner. In his revision of Costa Rican Dichaea, Pupulin (2007) presented a morphological cladistic analysis (limited to Costa Rican taxa). Historically, infrageneric classifications of Dichaea were based on one to few characters without a phylogenetic framework and with conflicting results. No sectional scheme of Dichaea has ever used clearly defined apomorphic characters to circumscribe subgeneric taxa. The objective of this study was to use DNA sequence data [nuclear ribosomal internal transcribed spacers (nrITS), the plastid matK, trnL intron, trnL-F intergenic spacer and ycf1] to reconstruct phylogenetic relationships in Dichaea and map the distribution of major morphological characters that have previously been used to delimit subgenera/sections. For purposes of clarity, the classification of Cogniaux (1906), as above, will be followed.
MATERIALS AND METHODS
Taxon sampling
Specimens were obtained from wild-collected and cultivated plants (Appendix). Sampling of Dichaea included 35 species, representing all four described sections of Dichaea. Outgroups included six other genera of subtribe Zygopetalinae and Heterotaxis violaceopunctata (Rchb.f.) F. Barros (subtribe Maxillariinae). Outgroups were chosen based on phylogenetic placement in previous work (Cameron, 2001, 2004; Cameron et al., 1999; Chase et al., 2003; Whitten et al., 2000).
Extractions, amplification and sequencing
All freshly collected material was preserved in silica gel (Chase and Hills, 1991). Genomic DNA was extracted using a modified 2 × CTAB (cetyl trimethylammonium bromide) technique (Doyle and Doyle, 1987), scaled to a 1-mL volume reaction. Approximately 10 mg of dried tissue were ground in 1 mL of 2 × CTAB buffer and either 8 µL of β-mercaptoethanol or 10 µL of proteinase-K. Some total DNAs were then cleaned with Qiagen QIAquick PCR purification columns to remove any inhibitory secondary compounds (e.g. species of section Pseudodichaea). Amplifications were performed using a Biometra Tgradient or an Eppendorf Mastercycler EP Gradient S thermocycler and Sigma brand reagents in 25-μL volumes with the following reaction components for ITS: 0·5–1·0 µL template DNA (approx. 10–100 ng), 11 µL water, 6·5 µL 5M betaine, 2·5 µL 10 × buffer, 3 µL MgCl2, 0·5 µL of 10 µm dNTPs, 0·5 µL each of 10 µm primers and 0·5 units Taq. For the plastid regions the following reaction components were used: 0·5–1·0 µL template DNA (approx. 10–100 ng), 16–17·5 µL water, 2·5 µL 10 × buffer, 2–4 µL MgCl2, 0·5 µL of 10 µm dNTPs, 0·5 µL each of 10 µm primers and 0·5 units Taq.
nrITS (ITS 1 + 5·8S rDNA + ITS 2)
This region was amplified with the parameters 99 °C, 10 min; 94 °C hold for Taq addition; 33 × (94 °C, 45 s; 65 °C, 1 min; 72 °C, 1 min); 72 °C, 3 min, with the primers 17SE and 26SE from Sun et al. (1994).
matK–trnK
This region includes the entire matK gene and the flanking 3′ trnK spacer. This region was amplified with the parameters 94 °C, 3 min; 33 × (94 °C, 45 s; 60 °C, 45 s; 72 °C, 2 min); 72 °C, 3 min, with primers –19F (Molvray et al., 2000) and trnK2R (Johnson and Soltis, 1994). Internal sequencing primers were 308F and 1520R (Whitten et al., 2007). Some outgroups were amplified using the primers 56F and 1520R (Whitten et al., 2000) that yielded a shorter, but nearly complete sequence of matK (missing the 3′ spacer).
trnL–trnF
This region includes both the trnL intron and the spacer between trnL and trnF (hereafter collectively referred to as trnL-F). This region was amplified with the parameters 94 °C, 3 min; 33 × (94 °C, 1 min; 58 °C, 1 min; 72 °C, 1 min, 20 s); 72 °C, 6 min, with the primers c and f from Taberlet et al. (1991). Additional primers d and e were rarely required for sequencing.
ycf1
In Phalaenopsis Blume (GenBank: AY916449), this open reading frame is nearly 6 kb in length and may be the most variable coding region within the plastid genome (M. Moore, Oberlin College, OH, USA, pers. comm.). An approx. 1500-base-pair (bp) portion from the 3' end was sequenced. This region was amplified using a ‘touchdown’ protocol with the parameters 94 °C, 3 min; 8 × (94 °C, 30 s; 60–51 °C, 1 min; 72 °C, 3 min); 30 × (94 °C, 30 s; 50 °C, 1 min; 72 °C, 3 min); 72 °C, 3 min, with primers 3720F (TAC GTA TGT AAT GAA CGA ATG G) and 5500R (GCT GTT ATT GGC ATC AAA CCA ATA GCG). Additional internal primers intF (GAT CTG GAC CAA TGC ACA TAT T) and intR (TTT GAT TGG GAT GAT CCA AGG) were also required for sequencing.
Products were cleaned with Microclean™ (The Gel Company, San Francisco, CA, USA) following the manufacturer's protocols, eluted with 50 µL of 10 mM Tris–HCl (pH 8·5) and stored at 4 °C. Purified PCR products were then cycle-sequenced using the parameters 96 °C, 10 s; 25 × (96 °C, 10 s; 50 °C, 5 s; 60 °C, 4 min), with mix of 3 µL water, 1 µL fluorescent Big Dye dideoxy terminator, 2 µL Better Buffer™ (The Gel Company), 1 µL template and 0·5 µL primer. Cycle sequencing products were cleaned using ExoSAP™ (USB Corporation, OH, USA) following the maufacturer's protocols. Purified cycle sequencing products were directly sequenced using BigDye terminator reagents on an ABI 377, 3100 or 3130 automated sequencer according to the manufacturer's protocols (Applied Biosystems, Foster City, CA, USA). Electropherograms were edited and assembled using Sequencher 4·6™ (GeneCodes, Ann Arbor, MI, USA). All sequences were deposited in GenBank (Appendix).
Data analysis
Sequence data were manually aligned using Se-Al v2.0a11 (Rambaut, 1996). No sequence data were excluded from analyses. Indels (insertions/deletions) were coded as missing. Analyses were performed using PAUP* 4.0b10 (Swofford, 1999) with Fitch parsimony (unordered characters with equal weights; Fitch, 1971). A heuristic search strategy consisted of branch swapping by tree bisection reconnection (TBR), stepwise addition with 5000 random-addition replicates holding five trees at each step, and saving multiple trees (MulTrees). Levels of support were assessed using the bootstrap (Felsenstein, 1985). Bootstrap percentages were estimated with 1000 bootstrap replicates, using TBR swapping for five random-addition replicates per bootstrap replicate. Maximum likelihood (ML) analyses were performed using the program Garli 0·95 (Zwickl, 2006), assuming a GTR + I + Γ model. Modeltest (Posada and Crandall, 1998) was used to determine the appropriate model for analysis using all combined data under the Akaike Information Criterion. However, because the results from maximum likelihood analyses are so similar to those found with parsimony searches, they are not presented here.
All analyses were performed for datasets including ITS only, plastid regions only and all data combined. Data congruence was tested using the partition homogeneity test in PAUP*4.0b10 (Swofford, 1999) as described by Johnson and Soltis (1998). Heuristic searches for the partition homogeneity tests were performed using 100 replicates and TBR branch-swapping. Probability values lower than 0·05 were used to identify data sets that were significantly different from one another.
RESULTS
The aligned length of the ITS data set was 758 bp. Of these, 270 were potentially parsimony informative (35·6 %). Fitch parsimony analysis of the ITS region found 53 equally parsimonious trees of 896 steps [consistency index (CI) = 0·58, retention index (RI) = 0·86]. The aligned length of the combined plastid dataset (matK, trnL-F and ycf1) was 4719 bp. Of these, 413 were potentially parsimony informative (8·8 %). Fitch analysis of the combined plastid regions found 3407 equally parsimonious trees of 1054 steps (CI = 0·79, RI = 0·92). Individually (trees not shown), trnL-F had a total of 1278 characters, with 106 potentially parsimony informative, giving trees of 276 steps (CI = 0·80, RI = 0·92); matK had a total of 1813 characters, with 126 potentially parsimony informative, giving trees of 316 steps (CI = 0·83, RI = 0·93); and ycf1 had a total of 1628 characters, with 181 potentially parsimony informative, giving trees of 457 steps (CI = 0·77, RI = 0·91). Fitch parsimony analysis of all four regions found 725 equally parsimonious trees of 1982 steps (CI = 0·68, RI = 0·88). Maximum likelihood searches gave log likelihood scores (–lnL) of 6221·06, 13936·48 and 20857·29 for ITS only, plastid regions only and all five regions, respectively.
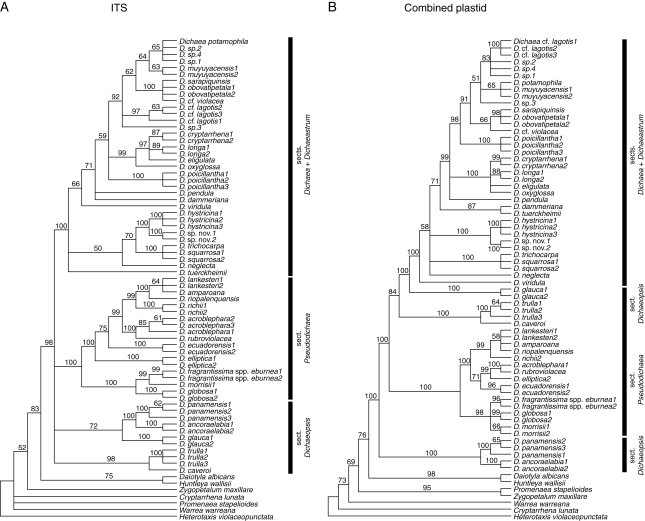
The partition homogeneity test comparing ITS and the combined plastid data showed significant incongruence compared with random partitions of the same size (P = 0·01, α = 0·05). Various combinations of each of the four of individual datasets, however, did not indicate significant incongruence (ITS/trnL-F P = 0·07; ITS/matK P = 0·16; matK/trnL-F P = 0·30; ycf1/matK P = 0·76; ycf1/trnL-F P = 0·78) except between ITS and ycf1 (P = 0·01). The combined plastid data did conflict with ITS, apparently due to the addition of ycf1. A visual comparison of bootstrap percentages between the ITS and plastid data sets shows that there are only three strongly supported cases of incongruence (e.g. the positions of D. tuerckheimii Schltr., D. glauca Lindl. and D. elliptica Dressler & Folsom; Fig. 2). Furthermore, the exclusion of ycf1 from the total combined analysis yields little support for relationships among the sections of Dichaea. Because the partition homogeneity test has been demonstrated to be overly sensitive (Graham et al., 1998; Reeves et al., 2001) and because a total evidence approach yields a highly resolved and relatively strongly supported topology, all data were combined.
Fig. 2.
Comparison of parsimony bootstrap consensus trees for (A) ITS dataset only and (B) plastid combined dataset. Bootstrap percentages are indicated above branches.
Monophyly of Dichaea and its placement within Zygopetalinae are both strongly supported by all analyses (Figs 2 and 3). The monophyly of combined sections Dichaea + Dichaeastrum is strongly supported. Also, section Pseudodichaea is strongly supported in all analyses. However, no individual or combined dataset supports the monophyly of section Dichaeopsis.
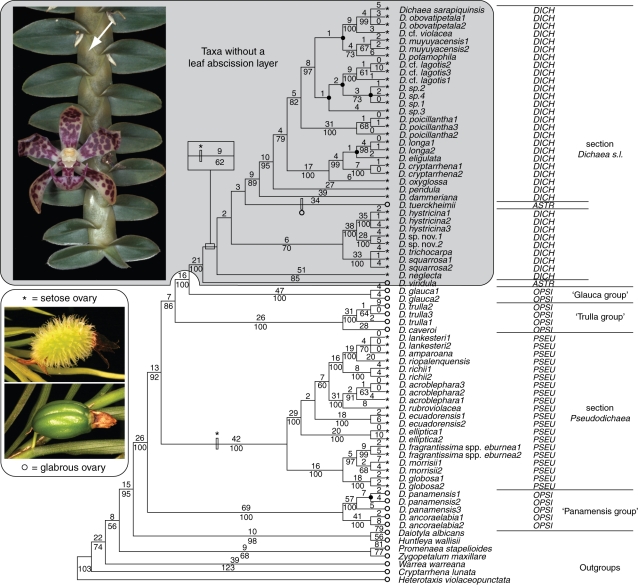
Fig. 3.
One of 725 most-parsimonious trees (MPTs) of all molecular data combined (DELTRAN optimization). Branch lengths are above the branches, and bootstrap percentages are below the branches. Branches that collapse in the strict consensus tree are indicated by closed circles. Taxa missing a leaf abscission layer are enclosed within the box. Fruit ornamentation is also shown with character states [glabrous (o) vs. setose (*)] on branch terminals, and hash-marks in the tree represent apomorphies. Traditional sectional placement for all species is listed by abbreviation (sections Dichaea, DICH; Dichaeastrum, ASTR; Dichaeopsis, OPSI; Pseudodichaea, PSEU); present circumscription is listed on the far right.
DISCUSSION
Although the placement of Dichaea within subtribe Zygopetalinae is strongly supported according to molecular data (Whitten et al., 2005), the genus does not share many vegetative or floral features with other members of the subtribe. Dichaea species lack the distinctive multi-ridged labellar callus and the short sympodial growth typical of other members of Zygopetalinae. However, Dichaea does share a characteristic pollinarium structure of four pollinia attached by four caudicles to a broad stipe terminating in a small, hyaline viscidium (Fig. 1J, K). The monophyly of Dichaea is strongly supported by both molecular and morphological data. Synapomorphies of Dichaea include elongate, monopodial stems, reduced flower size (relative to most other Zygopetalinae except Cryptarrhena R.Br.) and infrastigmatic ligules (Fig. 1I).
Plants of Dichaea appear to be monopodial, in contrast to sympodial growth found in rest of Zygopetalinae. In monopodial plants, the axillary shoot has the potential for indefinite apical growth, as in most stems of Dichaea, which continue to elongate and root at nodes (Dressler, 1993b). Roots of Dichaea are usually small, delicate and sparse. Leaves consist of a sheath and blade, although in many taxa, the abscission layer has been lost. However, no developmental studies of this unusual feature have been made, so another explanation is the loss of the blade and development of a blade-like sheath. Inflorescences of Dichaea are single-flowered, a derived condition within Zygopetalinae, but also found in some derived genera of the closely related subtribe Maxillariinae s.l. (Whitten et al., 2007). Flowers have distinctive anchor-shaped labella, which have a largely homogeneous papillose micromorphology (Davies and Stpiczynska, 2008). Fruits of Dichaea are capsules that split along two longitudinal lines of dehiscence (on only one side of the fruit), rather than having three to six lines of dehiscence observed in most orchids (Pupulin, 2007). The surface of the ovary and fruit can be glabrous, setose or spiny. Only sections Dichaea and Pseudodichaea have ornamentations on the fruit, which can become robust and prickle-like (Fig. 3).
In a recent systematic revision of the Dichaea species in Costa Rica, Pupulin (2007) performed parsimony analyses of 62 morphological and anatomical characters. The phylogenetic structure obtained with morphological characters is largely similar to that produced with molecular data. Both morphology and molecular data support the monophyly of section Pseudodichaea. Monophyly of sections Dichaea and Dichaeastrum (combined) is also supported. In some morphological trees, section Dichaeastrum is sister to section Dichaea, not polyphyletic as indicated by molecular data. Most topological discrepancies between morphological and molecular data sets can be found among species groupings within section Dichaea. Morphological data also consistently support the paraphyletic topology of section Dichaeopsis, as discussed below.
Section Dichaeopsis
This section, as circumscribed in all classifications proposed to date, is not monophyletic. Cogniaux's more concise circumscription, including only those taxa with glabrous ovaries and an abscission layer in the leaves, consists of at least three clades that are not each other's closest relatives.
Dichaea panamensis Lindl. and D. ancoraelabia C. Schweinf. form a clade (‘Panamensis group’ in Fig. 3) sister to the rest of the genus with strong support. This group of at least eight species has relatively short and narrow leaves (i.e. similar to smaller species of section Pseudodichaea; see below), delicate stems, relatively thick roots, dark maroon anther caps, spotted perianth and reduced and bluntly triangular infrastigmatic ligules (Fig. 1B, F). These characters are shared by most species within this Dichaeopsis clade and represent putative synapomorphies. There are, however, many more described species not sampled in this study that are closely related to these two species, based on morphological similarity. These species include D. campanulata C.Schweinf., D. dressleri Folsom, D. hutchisonii D.E.Benn. & Christenson, D. longipedunculata D.E.Benn. & Christenson, D. peruviensis D.E.Benn. & Christenson and D. picta Rchb.f.
Another Dichaeopsis clade consists of two species sampled in this study, D. trulla Rchb.f. and D. caveroi D.E.Benn. & Christenson (‘Trulla group’ in Fig. 3). There is moderate support for this clade sister to the ‘Glauca group’ + sect. Dichaea s.l. This clade of species is well supported and easily diagnosable. The plants typically have erect or semi-erect stems with leaves that are relatively long (up to 10 cm) and narrow (many other species in the genus have leaves that are proportionally broader and 2–4 cm long). Also, the flowers typically have well-developed infrastigmatic ligules (Fig. 1D, E). Other species likely to be included in the ‘Trulla group’ are D. calyculata Poepp. & Endl., D. powellii Schltr. (likely a synonym of D. trulla) and D. benzingii Dodson.
Dichaea glauca (Sw.) Lindl. (‘Glauca group’ in Fig. 3) occurs in Central America and the Caribbean and is the tallest species in the genus. It has glaucous leaves and the thickest roots in the genus. This species also displays multiple inflorescences per stem simultaneously (Fig. 1L), an unusual feature for the genus. The ITS results place this species sister to D. panamensis and D. ancoraelabia, but with just moderate support. Dichaea panamensis also has glaucous leaves, an unusual feature in Dichaea. However, plastid results strongly support placement of D. glauca sister to sections Dichaea and Dichaeastrum, even though there is no apparent morphological basis for this relationship.
Section Pseudodichaea
Two characters define section Pseudodichaea: leaves with an abscission layer (plesiomorphic) and setose ovaries (apomorphic). Setose ovaries are also found in section Dichaea, making the character homoplasious within the genus (Fig. 3). Although circumscription of section Pseudodichaea is based on plesiomorphic and homoplasious characters, it is still monophyletic. Section Pseudodichaea is not as closely related to section Dichaea as might be supposed based on possession of setose ovaries. These two sections are separated by section Dichaeastrum (in part) and a paraphyletic grade of section Dichaeopsis.
Molecular results support two main, strongly supported sister groups in section Pseudodichaea. One group consists of at least three species: D. morrisii Fawc. & Rendl., D. fragrantissima Folsom and D. globosa Dressler & Pupulin. These have stout, more or less horizontal stems and relatively large, broad leaves with strongly ancipitous sheaths (Fig. 1P). The other group is much more species rich, with relatively small, narrow leaves (Fig. 1N). The close relationship between D. amparoana Schltr. and D. lankesteri Ames shown by molecular data was previously suggested based on morphology (Dressler, 1993a; Pupulin, 2007). Dichaea elliptica Dressler & Folsom and D. acroblephara Schltr. are similar morphologically but not closely related according to these results. Section Pseudodichaea is particularly diverse in South America, and many of these species were not sampled in this study (e.g. D. alcantarae D.E.Benn. & Christenson, D. angustisegmenta Dodson, D. chasei Dodson, D. cleistogama Dodson, D. delcastilloi D.E.Benn. & Christenson, D. galeata Dodson, D. luerorum Dodson, D. moronensis Dodson, D. sodiroi Schltr., D. suarezii Dodson, D. tamboensis Dodson and D. venezuelensis Carnevali & I. Ramírez). This section shows high levels of sequence divergence and phylogenetic resolution. Because of the high degree of endemism, biogeographic patterns could probably be determined with increased taxon sampling of this section.
Section Dichaea sensu lato (including section Dichaeastrum)
The most coherent group within Dichaea based on morphology consists of sections Dichaea and Dichaeastrum. This group has leaves lacking an abscission layer, a generally pendulous or creeping habit (Fig. 1M), stems with indeterminate growth and setose (or more rarely, glabrous) ovaries (Fig. 1O). These two sections also form a strongly supported clade with all sampled DNA regions. Pfitzer (1889) and Kuntze (1904) recognized this entire group as section Dichaea. Cogniaux (1906) and Schlechter (1914) split the group into sections Dichaea and Dichaeastrum. This study does not support monophyly of either section Dichaeastrum or Dichaea. The two species from this study representing section Dichaeastrum (D. tuerckheimii and D. viridula Pupulin) have glabrous ovaries and are relatively diminutive plants. As these two species are not sister groups and are in a poorly supported portion of the tree, section Dichaeastrum should be treated as a part of section Dichaea. There are more species belonging to this glabrous-ovary group known as section Dichaeastrum (e.g. D. escobariana Dodson, D. pumila Barb.Rodr., D. retroflexa Kränzl. and D. tenuifolia Schltr.). Species in section Dichaeastrum usually have small, thin-textured, non-articulate leaves and glabrous ovaries. Therefore, with greater taxon sampling, a larger monophyletic group of what would be recognized as section Dichaeastrum may appear. The type of section Dichaeastrum was not sampled; morphological affinity of the type species to either D. tuerckheimii or D. viridula cannot be assessed at this time.
Section Dichaea sensu stricto was monographed by Folsom (1987, 1996), who attempted to diagram the evolutionary patterns within this group, with emphasis on clusters of species complexes. However, his non-cladistic approach to phylogenetic reconstruction leaves much to interpretation as it was based on subjective ideas of relationships among species. Dichaea poicillantha Schltr. is a distinct species on a long branch in all datasets. Folsom placed it near D. schlechteri Folsom, D. cryptarrhena Rchb.f. ex Kränzl. and D. muricatoides Hamar & Garay.
Dichaea oxyglossa Schltr., D. eligulata Folsom, D. longa Schltr. and D. cryptarrhena form a strongly supported complex of species in the present analyses. This group does not entirely agree with Folsom's diagram. He placed D. oxyglossa and D. eligulata near D. obovatipetala Folsom, D. sarapiquinsis Folsom and D. retroflexiligula Folsom. Dichaea retroflexiligula was not sampled in this study. Dichaea obovatipetala and D. sarapiquinsis are not closely related to this group, as postulated by Folsom (1987, 1996).
Dichaea obovatipetala and D. sarapiquinsis are strongly supported and part of a strongly supported clade of primarily South American taxa (including D. cf. lagotis Rchb.f., D. muyuyacensis Dodson, D. potamophila Folsom and D. cf. violacea Folsom). Folsom (1987, 1996) suspected that D. obovatipetala and D. sarapiquinsis, species endemic to Central America, were closely related to South American taxa based on morphology. Dichaea obovatipetala and D. sarapiquinsis lack clear cross-venation. Although some accessions sampled from this clade are unidentified, they do represent the South American group, most of which have clear cross-venation.
Folsom (1987, 1996) ignored some taxa that should properly be placed within section Dichaea. He referred to one such group as the ‘Dichaea hystricina complex,’ including D. hystricina Rchb.f., D. ciliolata Rolfe and other unspecified species. This complex is monophyletic and distinguished by ciliate leaf margins. The exclusion of this complex from section Dichaea would make the latter paraphyletic. Pupulin (2005) studied the D. hystricina complex and showed that variation in vegetative morphology in Costa Rican specimens corresponded to a single species. The type of D. ciliolata is from Costa Rica, and Pupulin suggested that it is a synonym of D. hystricina, which is further corroborated by these data. Molecular and morphological data support the existence in Ecuador of a distinct, undescribed species that is sister to D. hystricina. However, more data need to be collected before it should be formally described.
Another distinct clade included D. trichocarpa (Sw.) Lindl. and D. squarrosa Lindl. Dichaea intermedia Ames & Correll is sometimes considered to be a hybrid between D. trichocarpa and D. squarrosa (Ames and Correll, 1985; Folsom, 1987). Taxon sampling was insufficient to determine the distinctiveness of these three species individually, but they form a clade. Among these taxa in section Dichaea, D. neglecta Schltr. was poorly supported as sister to the rest of section Dichaea with the exclusion of D. viridula (Fig. 3). Folsom also cited a few aberrant species that were not closely related to any other species. Species such as D. pendula (Aubl.) Cogn. and D. dammeriana Kraenzl. were sampled in this study and shown to be part of a paraphyletic grade relative to the core group of section Dichaea.
Evolution of leaves
According to the present results, the leaf abscission layer was lost once within the genus Dichaea. The absence of the leaf abscission layer is a synapomorphy for section Dichaea sensu lato, with no apparent reversions. The lack of an abscission layer has no obvious adaptive value. This condition (marcescent leaves) is uncommon in tropical Orchidaceae, but is found in many Arecaceae and some temperate trees, such as Fagus L. Some studies of marcescent leaves in alpine tropical plants have shown that marcescent leaves protect the stem against tissue damage by freezing and desiccation (Smith, 1979). Species of Dichaea never occur at such high elevations (Pupulin, 2007), so this feature is unlikely to be an adaptation to freezing. The occurrence of marcescent leaves in sections Dichaea and Dichaeastrum is associated with a creeping or pendent habit. In these groups, the leaf bases are often twisted 90 ° so that their adaxial surfaces all face the same direction, making the stem complanate. This condition presumably maximizes the photosynthetic area exposed to light. However, the association among these leaf characters needs further study.
Evolution of fruits
Compared with other orchids, Dichaea is unusual in having setose or muricate fruits. Some members of the orchid subtribes Pleurothallidinae (e.g. Pleurothallis R.Br. subgenera Kraenzlinella Kuntze and Aenigma Luer; Luer, 1994), Laeliinae (e.g. Pygmaeorchis Brade; Pridgeon et al., 2005) and Oncidiinae (e.g. Saundersia Rchb.f.) also have muricate ovaries, although these taxa are not closely related to Dichaea. Like almost all other orchid fruits, those of Dichaea are dehiscent capsules with wind-dispersed seeds.
It is possible that the spines on the fruits are used in dispersal by adhering to animals. Hooked spines that aid in dispersal are fairly common in many families of flowering plants. However, the setae of Dichaea fruits are not strongly uncinate, so it is unlikely that the fruits are exozoochorous. Also, Dichaea species have ‘dust seeds’ that are readily dispersed by wind, so exozoochory of fruits seems superfluous.
Many plants have setose or spiny fruits, which presumably protect seeds against herbivory. Spininess has been shown to inhibit herbivory by small mammals (Cooper and Ginnett, 1998) and by larger animals (Young and Augustine, 2007). However, the fruit setae of many species of sections Dichaea and Pseudodichaea are soft and could hardly inhibit predation by large or medium-sized herbivores. No assessment of the adaptive value, if any, of these setae has ever been performed. More puzzling than their apparent lack of adaptive value is the polyphyletic nature of their occurrence. Setose fruits appear to have evolved at least twice (Fig. 3), and the groups in Dichaea with setose fruits are also the most diverse in terms of species richness.
CONCLUSIONS
DNA sequence data were used to investigate the circumscription of sectional classification and species relationships of Dichaea. Characters used to separate the sections within the genus have been misleading. Section Dichaeopsis was found to be polyphyletic whereas section Pseudodichaea is monophyletic. We suggested a broader circumscription of section Dichaea, including section Dichaeastrum, because Dichaeastrum is polyphyletic and Dichaea and Dichaeastrum are similar vegetatively and together are easily distinguished from other sections. The marcescent leaves of section Dichaea sensu lato have been attained only once, whereas setae on fruits have been attained at least twice within the genus. However, there is no obvious adaptive value to the plants of these unique morphological features. Further investigation into the anatomical structure of the leaves in section Dichaea could address the developmental and evolutionary significance of the lack of an abscission layer between sheath and blade. Careful in situ observations of orchid taxa with fruit setae may yield more meaningful phylogenetic interpretations of this character. The question of how such a wide range of morphological diversity was shaped and the ecological role behind these features requires further probing in this group of plants.
ACKNOWLEDGEMENTS
We thank Jardín Botánico Lankester (Universidad de Costa Rica) for contributing vouchered specimens and tissue. We are grateful to the Portillas of Ecuagenera Ltd in Ecuador for generous access to their collections. Walter Judd gave much taxonomic advice. Bruce Holst facilitated sampling of herbarium specimens for DNA at Marie Selby Botanical Gardens. We thank Mario Blanco, Alec Pridgeon, Mark Chase and one anonymous reviewer for helpful comments on this manuscript. Barbara Sue Carlsward provided technical support. Robert Dressler and Calaway Dodson have helped with identification of specimens. We also thank Michael Moore for his suggestion to use the ycf1 gene region for phylogenetic analyses, J. Richard Abbott for the use of photographs and Savita Shanker and Patrick Thimote at the Interdisciplinary Center for Biotechnology Research at UF. Portions of this research were funded by the Lewis and Varina Vaughn Fellowship in Orchid Biology and the American Orchid Society's 11th World Orchid Conference Fellowship to K. Neubig, and the US National Science Foundation grant No. DEB-234064 to N. H. Williams and W. M. Whitten.
APPENDIX
Voucher information and GenBank accession numbers for all taxa used in this study
| Species | Country | Voucher information (herbarium) | ITS | matK + trnK | trnL-F | ycf1 |
|---|---|---|---|---|---|---|
| Cryptarrhena lunata R. Br. | Costa Rica | Whitten 98000 (FLAS) | AY870081 | AY869982 | AY869894 | EU123733 |
| Daiotyla albicans (Rolfe) Dressler | Panama | Whitten 1932 (FLAS) | AY870016 | AY869917 | AY869831 | EU123734 |
| Dichaea acroblephara Schltr. 1 | Costa Rica | Pupulin 4795 (USJ-L) | EU123545 | EU123611 | EU123677 | EU123735 |
| D. acroblephara Schltr. 2 | Panama | Whitten 2669 (FLAS) | EU123546 | NA | NA | NA |
| D. acroblephara Schltr. 3 | Panama | Blanco 2994 (FLAS) | EU123547 | NA | NA | NA |
| D. amparoana Schltr. | Costa Rica | Bogarín 679 (USJ-L) | EU123548 | EU123612 | EU123678 | NA |
| D. ancoraelabia C.Schweinf. 1 | Ecuador | Neubig 1–2004 (FLAS) | EU123549 | EU123613 | NA | EU123736 |
| D. ancoraelabia C.Schweinf. 2 | Ecuador | Whitten 2542 (FLAS) | EU123550 | EU123614 | EU123679 | EU123737 |
| D. caveroi D.E. Benn. & Christenson | Ecuador | Whitten 2417 (FLAS) | EU123551 | EU123615 | EU123680 | EU123738 |
| D. cryptarrhena Rchb.f. ex Kraenzl. 1 | Costa Rica | Pupulin 4436 (USJ-L) | EU123556 | EU123620 | EU123685 | EU123743 |
| D. cryptarrhena Rchb.f. ex Kraenzl. 2 | Panama | Whitten 2610 (FLAS) | EU123557 | EU123621 | EU123686 | EU123744 |
| D. dammeriana Kraenzl. | Costa Rica | Bogarín & León-Páez 197 (USJ-L) | EU123558 | EU123622 | EU123687 | EU123745 |
| D. ecuadorensis Schltr. 1 | Ecuador | Whitten 1799 (FLAS) | EU123559 | EU123623 | NA | EU123746 |
| D. ecuadorensis Schltr. 2 | Ecuador | Whitten 2416 (FLAS) | EU123560 | EU123624 | EU123688 | NA |
| D. eligulata Folsom | Costa Rica | Pupulin 1094 (USJ-L) | EU123561 | EU123625 | EU123689 | EU123747 |
| D. elliptica Dressler & Folsom 1 | Costa Rica | Pupulin 5133 (USJ-L) | EU123562 | NA | NA | NA |
| D. elliptica Dressler & Folsom 2 | Costa Rica | Pupulin 4945 (USJ-L) | EU123563 | EU123626 | EU123690 | EU123748 |
| D. fragrantissima Folsom ssp. eburnea Dressler & Pupulin 1 | Costa Rica | Blanco 513 (USJ-L) | EU123564 | EU123627 | NA | EU123749 |
| D. fragrantissima Folsom ssp. eburnea Dressler & Pupulin 2 | Costa Rica | Pupulin 4601 (USJ-L) | EU123565 | EU123628 | EU123691 | EU123750 |
| D. glauca (Sw.) Lindl. 1 | Costa Rica | Pupulin 4734 (USJ-L) | EU123566 | EU123629 | EU123692 | EU123751 |
| D. glauca (Sw.) Lindl. 2 | Mexico | Neubig 9–2006 (FLAS) | EU123567 | EU123630 | EU123693 | EU123752 |
| D. globosa Dressler & Pupulin 1 | Costa Rica | Pupulin 4517 (USJ-L) | EU123568 | EU123631 | EU123694 | EU123753 |
| D. globosa Dressler & Pupulin 2 | Panama | Neubig 2–2005 (FLAS) | EU123569 | EU123632 | EU123695 | EU123754 |
| D. hystricina Rchb.f. 1 | Costa Rica | Pupulin 3925 (USJ-L) | EU123570 | EU123633 | EU123696 | EU123755 |
| D. hystricina Rchb.f. 2 | Costa Rica | Pupulin 2925 (USJ-L) | EU123571 | EU123634 | EU123697 | EU123756 |
| D. hystricina Rchb.f. 3 | Costa Rica | Pupulin 4320 (USJ-L) | EU123572 | EU123635 | EU123698 | EU123757 |
| D. cf. lagotis Rchb.f. 1 | Ecuador | Whitten 1801 (FLAS) | EU123552 | EU123616 | EU123681 | EU123739 |
| D. cf. lagotis Rchb.f. 2 | Ecuador | Whitten 2477 (QCA) | EU123553 | EU123617 | EU123682 | EU123740 |
| D. cf. lagotis Rchb.f. 3 | Ecuador | Whitten 2523 (QCA) | EU123554 | EU123618 | EU123683 | EU123741 |
| D. lankesteri Ames 1 | Panama | Blanco 2993 (FLAS) | EU123573 | EU123636 | NA | EU123758 |
| D. lankesteri Ames 2 | Costa Rica | Pupulin 3030 (USJ-L) | EU123574 | EU123637 | EU123699 | EU123759 |
| D. longa Schltr. 1 | Ecuador | Whitten 2684 (FLAS) | EU123575 | EU123638 | NA | EU123760 |
| D. longa Schltr. 2 | Ecuador | Whitten 2685 (FLAS) | EU123576 | EU123639 | EU123700 | EU123761 |
| D. morrisii Fawc. & Rendl. 1 | Panama | Neubig 3–2004 (FLAS) | EU123577 | EU123640 | EU123701 | EU123762 |
| D. morrisii Fawc. & Rendl. 2 | Ecuador | Neubig 8–2006 (FLAS) | NA | EU123641 | EU123702 | EU123763 |
| D. muyuyacensis Dodson 1 | Panama | Neubig 5–2004 (FLAS) | EU123578 | EU123642 | EU123703 | EU123764 |
| D. muyuyacensis Dodson 2 | Ecuador | Whitten 1512 (FLAS) | EU123579 | EU123643 | NA | EU123765 |
| D. neglecta Schltr. | Mexico | Higgins 1005 (FLAS) | EU123580 | EU123644 | EU123704 | EU123766 |
| D. obovatipetala Folsom 1 | Costa Rica | Pupulin 5023 (USJ-L) | EU123581 | EU123645 | EU123705 | EU123767 |
| D. obovatipetala Folsom 2 | Costa Rica | Pupulin 4202 (USJ-L) | EU123582 | EU123646 | EU123706 | EU123768 |
| D. oxyglossa Schltr. | Costa Rica | Bogarín & León-Páez 186 (USJ-L) | EU123583 | EU123647 | EU123707 | EU123769 |
| D. panamensis Lindl. 1 | Costa Rica | Pupulin 3667 (USJ-L) | EU123584 | EU123648 | EU123708 | EU123770 |
| D. panamensis Lindl. 2 | Ecuador | Whitten 2348 (FLAS) | EU123585 | EU123649 | EU123709 | EU123771 |
| D. panamensis Lindl. 3 | Panama | Whitten 2556 (FLAS) | EU123586 | EU123650 | NA | EU123772 |
| D. pendula (Aubl.) Cogn. | Costa Rica | Pupulin 3024 (USJ-L) | EU123587 | EU123651 | EU123710 | EU123773 |
| D. poicillantha Schltr. 1 | Panama | Blanco 2981 (FLAS) | EU123588 | EU123652 | EU123711 | EU123774 |
| D. poicillantha Schltr. 2 | Costa Rica | Pupulin 4662 (USJ-L) | EU123589 | EU123653 | EU123712 | EU123775 |
| D. poicillantha Schltr. 3 | Panama | Whitten 2557 (FLAS) | EU123590 | EU123654 | EU123713 | EU123776 |
| D. potamophila Folsom | Peru | Neubig 4–2004 (FLAS) | EU123591 | EU123655 | EU123714 | EU123777 |
| D. richii Dodson 1 | Ecuador | Whitten 1526 (FLAS) | EU123592 | NA | NA | NA |
| D. richii Dodson 2 | Ecuador | Whitten 2429 (QCA) | EU123593 | EU123656 | EU123715 | NA |
| D. riopalenquensis Dodson | Ecuador | Whitten 2731 (FLAS) | EU123594 | EU123657 | EU123716 | EU123778 |
| D. rubroviolacea Dodson | Ecuador | Whitten 2945 (FLAS) | EU123595 | EU123658 | EU123717 | EU123779 |
| D. sarapiquinsis Folsom | Costa Rica | Pupulin 4856 (USJ-L) | EU123596 | EU123659 | EU123718 | EU123780 |
| D. squarrosa Lindl. 1 | Costa Rica | Pupulin 5127 (USJ-L) | EU123604 | EU123667 | EU123726 | EU123788 |
| D. squarrosa Lindl. 2 | Mexico | Neubig 4–2006 (FLAS) | EU123603 | EU123666 | EU123725 | EU123787 |
| D. trichocarpa (Sw.) Lindl. | Costa Rica | Bogarín 173 (USJ-L) | EU123605 | EU123668 | EU123727 | EU123789 |
| D. trulla Rchb.f. 1 | Costa Rica | Whitten 2096 (USJ-L) | EU123607 | EU123670 | EU123729 | EU123791 |
| D. trulla Rchb.f. 2 | Ecuador | Whitten 2474 (QCA) | EU123608 | EU123671 | EU123730 | EU123792 |
| D. trulla Rchb.f. 3 | Ecuador | Whitten 2475 (FLAS) | EU123606 | EU123669 | EU123728 | EU123790 |
| D. tuerckheimii Schltr. | Costa Rica | Whitten 2097 (USJ-L) | EU123609 | EU123672 | EU123731 | EU123793 |
| D. cf. violacea Folsom | Panama | Neubig 6–2004 (FLAS) | EU123555 | EU123619 | EU123684 | EU123742 |
| D. viridula Pupulin | Costa Rica | Pupulin 4752 (USJ-L) | EU123610 | EU123673 | EU123732 | EU123794 |
| D. sp. 1 | Ecuador | Whitten 2709 (FLAS) | EU123597 | EU123660 | EU123719 | EU123781 |
| D. sp. 2 | Ecuador | Whitten 2434 (QCA) | EU123598 | EU123661 | EU123720 | EU123782 |
| D. sp. 3 | Ecuador | Whitten 2435 (QCA) | EU123599 | EU123662 | EU123721 | EU123783 |
| D. sp. 4 | Ecuador | Whitten 2476 (QCA) | EU123600 | EU123663 | EU123722 | EU123784 |
| D. sp. nov. 1 | Ecuador | Neubig 6–2006 (FLAS) | EU123601 | EU123664 | EU123723 | EU123785 |
| D. sp. nov. 2 | Ecuador | Whitten 2329 (FLAS) | EU123602 | EU123665 | EU123724 | EU123786 |
| Heterotaxis violaceopunctata (Rchb.f.) F. Barros | Brazil | Whitten 2294 (FLAS) | DQ210308 | DQ210807 | NA | EU123795 |
| H. violaceopunctata (Rchb.f.) F. Barros | Suriname | Whitten 1980 (FLAS) | NA | NA | AY869911 | NA |
| Huntleya wallisii (Rchb.f) Rolfe | Ecuador | Whitten 88026 (FLAS) | AY870074 | EU123674 | AY869887 | EU123796 |
| Promenaea stapelioides (Link & Otto) Lindl. | Brazil | Whitten 94102 (FLAS) | AY870101 | AY870002 | AY869905 | EU123797 |
| Warrea warreana (Lodd. ex Lindl.) C. Schweinf. | Costa Rica | Whitten 1752 (FLAS) | AF239321 | EU123675 | AF239513 | EU123798 |
| Zygopetalum maxillare Lodd. | Brazil | Whitten 94103 (FLAS) | AY870095 | EU123676 | AY869899 | EU123799 |
Vouchers are deposited at FLAS (Florida Museum of Natural History Herbarium), USJ-L (University of Costa Rica, San José, Lankester Botanical Garden) and QCA (Pontifica Universidad Católica del Ecuador, Quito).
LITERATURE CITED
- Ames O, Correll DS. Orchids of Guatemala and Belize. New York, NY: Dover Publications; 1985. [Google Scholar]
- Cameron KM. An expanded phylogenetic analysis of Orchidaceae using three plastid genes: rbcL, atpB, and psaB. American Journal of Botany. 2001;88:104. [Google Scholar]
- Cameron KM. Utility of plastid psaB gene sequences for investigating intrafamilial relationships within Orchidaceae. Molecular Phylogenetics and Evolution. 2004;31:1157–1180. doi: 10.1016/j.ympev.2003.10.010. [DOI] [PubMed] [Google Scholar]
- Cameron KM, Chase MW, Whitten WM, et al. A phylogenetic analysis of the Orchidaceae: evidence from rbcL nucleotide sequences. American Journal of Botany. 1999;86:208–224. [PubMed] [Google Scholar]
- Chase MW, Hills HG. Silica gel: an ideal material for field preservation of leaf samples for DNA studies. Taxon. 1991;40:215–220. [Google Scholar]
- Chase MW, Freudenstein JV, Cameron KM, Barrett RL. DNA data and Orchidaceae systematics: a new phylogenetic classification. In: Dixon KW, Kell SP, Barrett RL, Cribb PJ, editors. Orchid conservation. Kota Kinabalu: Natural History Publications; 2003. pp. 69–89. [Google Scholar]
- Cogniaux A. Orchidaceae III. In: de Martius CFP, editor. Flora Brasiliensis. Koenigstein: Otto Koeltz Science Publishers; 1906. pp. 484–504. [Google Scholar]
- Cooper SM, Ginnett TF. Spines protect plants against browsing by small climbing mammals. Oecologia. 1998;113:219–221. doi: 10.1007/s004420050371. [DOI] [PubMed] [Google Scholar]
- Davies KL, Stpiczynska M. Labellar micromorphology of two euglossine-pollinated orchid genera; Scuticaria Lindl. and Dichaea Lindl. Annals of Botany. 2008;102:805–824. doi: 10.1093/aob/mcn155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle JJ, Doyle JL. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochemical Bulletin, Botanical Society of America. 1987;19:11–15. [Google Scholar]
- Dressler RL. Field guide to the orchids of Costa Rica and Panama. Ithaca, NY: Comstock Publishing Associates; 1993a. [Google Scholar]
- Dressler RL. Phylogeny and classification of the orchid family. Portland, OR: Dioscorides Press; 1993b. [Google Scholar]
- Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- Fitch WM. Toward defining the course of evolution: minimum change for a specific tree topology. Systematic Zoology. 1971;20:406–416. [Google Scholar]
- Folsom JP. Austin: University of Texas; 1987. A systematic monograph of Dichaea section Dichaea (Orchidaceae) PhD Thesis. [Google Scholar]
- Folsom JP. An introduction to the genus Dichaea and a synopsis of section Dichaea. Orchid Digest. 1996;60:148–155. [Google Scholar]
- Graham SW, Kohn JR, Morton BR, Eckenwalder JE, Barrett SCH. Phylogenetic congruence and discordance among one morphological and three molecular data sets from Pontederiaceae. Systematic Biology. 1998;47:545–567. doi: 10.1080/106351598260572. [DOI] [PubMed] [Google Scholar]
- Johnson LA, Soltis DE. matK DNA sequences and phylogenetic reconstruction in Saxifragaceae s.st. Systematic Botany. 1994;19:143–156. [Google Scholar]
- Johnson LA, Soltis DE. Assessing congruence: empirical examples from molecular data. In: Soltis DE, Soltis PS, Doyle JJ, editors. Molecular systematics of plants II: DNA sequencing. Boston, MA: Kluwer Academic Publishers; 1998. pp. 297–348. [Google Scholar]
- Knowles G, Westcott F. Dichaea. Floral Cabinet. 1839;2:167. [Google Scholar]
- Kränzlin F. Orchidaceae-Monandrae-Pseudomonopodiales. In: Engler A, editor. Das Pflanzenreich. Leipzig: Engelmann; 1923. pp. 33–59. [Google Scholar]
- Kuntze CEO. Revision of Dichaea. In: von Post TE, editor. Lexicon Generum Phanerogamarum. Stuttgart: Deutsche Verlags-Anstalt; 1904. p. 171. [Google Scholar]
- Lindley J. The genera and species of orchidaceous plants. London: Ridgways; 1833. [Google Scholar]
- Luer CA. Icones pleurothallidinarum. XI. Systematics of Lepanthes subgenus Brachycladium, and Pleurothallis subgenus Aenigma, subgenus Elongatia, and subgenus Kraenzlinella (Orchidaceae) Monographs in Systematic Botany from the Missouri Botanical Garden, St Louis. 1994;52:1–50. [Google Scholar]
- Molvray M, Kores PJ, Chase MW. Polyphyly of mycoheterotrophic orchids and functional influences of floral and molecular characters. In: Wilson KL, Morrison DA, editors. Monocots: systematics and evolution. Collingswood: CSIRO Publishing; 2000. pp. 441–448. [Google Scholar]
- Pfitzer EH. Orchidaceae. In: Engler A, Prantl K, editors. Die Naturlichen Pflanzenfamilien. 6 edn. 1889. pp. 206–207. [Google Scholar]
- Posada D, Crandall KA. Modeltest: tesing the model of DNA substitution. Bioinformatics. 1998;14:817–818. doi: 10.1093/bioinformatics/14.9.817. [DOI] [PubMed] [Google Scholar]
- Pridgeon AM, Cribb PJ, Chase MW, Rasmussen FN. Genera orchidacearum. Oxford: Oxford University Press; 2005. Vol. 4. Epidendroideae (part one) [Google Scholar]
- Pridgeon AM, Cribb PJ, Chase MW, Rasmussen FN. Genera orchidacearum. Oxford: Oxford University Press.; 2009. Vol. 5. Epidendroideae (part two) [Google Scholar]
- Pupulin F. Dichaea hystricina and Dichaea ciliolata: two species in one and an interesting variation. Orchids. 2005;74:678–683. [Google Scholar]
- Pupulin F. Contributions toward a reassessment of Costa Rican Zygopetalinae (Orchidaceae). 3. A systematic revision of Dichaea in Costa Rica. Journal of the Arnold Arboretum. 2007;12:15–153. [Google Scholar]
- Rambaut A. Se-Al: Sequence alignment editor, v2.0a11. Oxford, UK: University of Oxford; 1996. Available at website, http://evolve.zoo.ox.ac.uk/ , last accessed 8 August 2002. [Google Scholar]
- Reeves G, Chase MW, Goldblatt P, et al. Molecular systematics of Iridaceae: evidence from four plastid regions. American Journal of Botany. 2001;88:2074–2087. [PubMed] [Google Scholar]
- Romero G, Carnevali G. Reappraisal of subtribe Vargasiellinae (Maxillarieae, Orchidaceae) Novon. 1993;3:79–80. [Google Scholar]
- Schlechter R. Die Orchideen-Gruppe Dichaeinae Pfitzers. Orchis. 1914;8:1–8. [Google Scholar]
- Senghas K. Dichaeinae. In: Schlechter R, editor. Die Orchideen. Berlin: Blackwell Wissenschafts-Verlag; 1996. pp. 1853–1861. [Google Scholar]
- Smith AP. Function of dead leaves in Espeletia schultzii (Compositae), an Andean caulescent rosette species. Biotropica. 1979;11:43–47. [Google Scholar]
- Sun Y, Skinner DZ, Liang GH, Hulbert SH. Phylogenetic analysis of Sorghum and related taxa using internal transcribed spacers of nuclear ribosomal DNA. Theoretical and Applied Genetics. 1994;89:26–32. doi: 10.1007/BF00226978. [DOI] [PubMed] [Google Scholar]
- Swofford DL. PAUP*: phylogenetic analysis using parsimony (*and other methods), version 4.0b10. Sunderland, MA: Sinauer Associates; 1999. [Google Scholar]
- Szlachetko DL. Systema orchidalium. Fragmenta Floristica et Geobotanica Supplementum. 1995;3:1–152. [Google Scholar]
- Taberlet P, Gielly L, Pautou G, Bouvet J. Universal primers for amplification of three non-coding regions of chloroplast DNA. Plant Molecular Biology. 1991;17:1105–1109. doi: 10.1007/BF00037152. [DOI] [PubMed] [Google Scholar]
- Whitten MW, Williams NH, Chase MW. Subtribal and generic relationships of Maxillarieae (Orchidaceae) with emphasis on Stanhopeinae: combined molecular evidence. American Journal of Botany. 2000;87:1842–1856. [PubMed] [Google Scholar]
- Whitten WM, Williams NH, Dressler RL, Gerlach G, Pupulin F. Generic relationships of Zygopetalinae (Orchidaceae: Cymbidieae): combined molecular evidence. Lankesteriana. 2005;5:87–107. [Google Scholar]
- Whitten WM, Blanco MA, Williams NH, et al. Molecular phylogenetics of Maxillaria and related genera (Orchidaceae: Cymbidieae) based on combined molecular data sets. American Journal of Botany. 2007;94:1860–1889. doi: 10.3732/ajb.94.11.1860. [DOI] [PubMed] [Google Scholar]
- Williams NH, Chase MW, Fulcher T, Whitten WM. Molecular systematics of the Oncidiinae based on evidence from four DNA sequence regions: expanded circumscriptions of Cyrtochilum, Erycina, Otoglossum, and Trichocentrum and a new genus (Orchidaceae) Lindleyana. 2001;16:113–139. [Google Scholar]
- Young TP, Augustine DJ. Interspecific variation in the reproductive response of Acacia species to protection from large mammalian herbivores. Biotropica. 2007;39:559–561. [Google Scholar]
- Zwickl DJ. Genetic algorithm approaches for the phylogenetic analysis of large biological sequence datasets under the maximum likelihood criterion. Austin: University of Texas; 2006. PhD Thesis. [Google Scholar]