Abstract
We introduce here the concept of Unique Pattern Informative Combinations (UPIC), a decision tool for the cost-effective design of DNA fingerprinting/genotyping experiments using simple-sequence/tandem repeat (SSR/STR) markers. After the first screening of SSR-markers tested on a subset of DNA samples, the user can apply UPIC to find marker combinations that maximize the genetic information obtained by a minimum or desirable number of markers. This allows a cost-effective planning of future experiments. We have developed Perl scripts to calculate all possible subset combinations of SSR markers, and determine based on unique patterns or alleles, which combinations can discriminate among all DNA samples included in a test. This makes UPIC an essential tool for optimizing resources when working with microsatellites. An example using real data from eight markers and 12 genotypes shows that UPIC detected groups of as few as three markers sufficient to discriminate all 12- DNA samples. Should markers for future experiments be chosen based only on polymorphism-information content (PIC), the necessary number of markers for discrimination of all samples cannot be determined. We also show that choosing markers using UPIC, an informative combination of four markers can provide similar information as using a combination of six markers (23 vs. 25 patterns, respectively), granting a more efficient planning of experiments. Perl scripts with documentation are also included to calculate the percentage of heterozygous loci on the DNA samples tested and to calculate three PIC values depending on the type of fertilization and allele frequency of the organism.
Availability
Perl scripts are freely available for download from http://www.ars.usda.gov/msa/jwdsrc/gbru.
Keywords: simple sequence repeats, software, best SSR markers, microsatellites, GeneMapper
Background
Repetitive DNA sequences, microsatellites or simple sequence/tandem repeats (SSR, STR) are widely spread throughout prokaryotic and eukaryotic genomes [1,2,3], and have a number of applications from marker-assisted breeding in plants [4] to detecting genetic disorders in humans [3]. Given the cost of running SSR markers, primers are usually screened on a subset of DNA samples before designing large scale experiments. Though some useful coefficients exist, such as polymorphism information content (PIC) [5] and Log10 of the likelihood ratio (LOD score) [5] to help determine which markers to use, currently, there are no available decision tools for cost-effective planning of fingerprinting or genotyping experiments.
Various PIC formulas are available in the literature, depending on whether the organisms are cross-fertilized [5], cross-fertilized and have equifrequent alleles [6], or are selffertilized [6] (Formulas S1, 1.1.1, 1.1.2 and 1.1.3). Though software is available for the calculation of PIC values, such as Cervus [7,8] or the on-line PIC calculator [9], no single site calculates the three mentioned PIC values. Other useful information when working with microsatellites is the average heterozygosity per locus (Formulas S1, 1.2.1) as a measure of the genetic variability of the population [10]. Knowing the degree of heterozygosity of the lines tested allows choosing parental lines for further studies, selecting lines with potential environmental fitness [11] or inferring ploidy of the tested DNA samples [12].
It is necessary to make a clear distinction between the polymorphism-information content (PIC) value developed by Botstein et al. (1980) [5], and the new approach presented here for choosing the best combinations of SSR markers that we now call UPIC. Whereas PIC values only indicate the information content of individual markers, UPIC calculates all possible subset combinations of markers and finds which combinations are the most informative. We introduce the concept of Unique Pattern Informative Combination (UPIC) to provide users of SSR markers with a decision tool that: (a) finds the most informative combinations of polymorphic markers based on the presence of unique patterns on the samples tested, and (b) allows the user to choose the number of markers to run depending on cost or objectives of the experiment. UPIC calculations do not require prior knowledge of genetic information of the populations to be analyzed such as genome size, ploidy or type of fertilization. In addition to UPIC values, the scripts presented here calculate percentage of heterozygous loci for each DNA sample and three PIC coefficients for self fertilized, crossfertilized, and cross-fertilized with equifrequent alleles (Formulas S1) for the user to choose from, thus representing a convenient tool for microsatellite work.
Methodology
After screening primers for developing SSR markers, a text file containing marker names, DNA samples and amplicon sizes is generated (i.e., by GeneMapper, Applied Biosystems) and used as input for the scripts. The first row in this tab or space delimited text file contains the headers for the columns, please see example in Table S1. The scripts calculate: three PIC values [5,6], percent of heterozygous loci for each line, and the UPIC values proposed here.
UPIC calculation
Allele information of eight polymorphic markers that were run on 12 lines (DNA samples) was used in our example to show the mechanics of calculating unique-pattern informative combinations (UPIC). The various allele patterns observed for each marker (fingerprint) were compared as strings of amplicon (peak) sizes (Table S2a). In our example, the possible number of combinations of 8 polymorphic markers is 255. If we assign a letter to each pattern observed for a line (Table S2b) and then convert the letters to binary values, where “0” is assigned to an allele pattern present more than once across the lines tested, and “1” is assigned to unique patterns (UP), Table S2c. Please note that UP differ in at least one allele, therefore, UP values represent unique identifiers for the DNA sample.
Since various informative combinations (IC) with different total number of UP can be found, the UPIC script output consists of two columns, one is the total number of UPIC (i.e., 18, Table S2d) in the combination, and the other is the marker combination. All UP values of each IC, for the data in our example, are shown in the UPIC plot, Figure 1. We have written UPIC version 1.0 which calculates all possible subset combinations of markers, where the range of subsets is selected by the user. The range minimum is combinations of two and the maximum is the number of markers in the input file. Each combination subset is calculated completely before the next larger subset. Details of the calculation of UPIC are provided in Formulas S1.
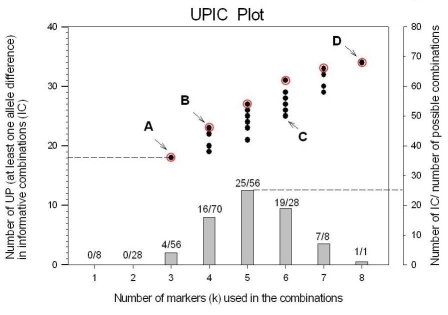
Figure 1.
Graphic representation of UPIC values for the 8 markers and 12 DNA samples in our example. •: unique patterns (UP)(y-axis,left) that allow discrimination of the 12 DNA samples tested, corresponding to informative combinations (IC) of variable number of polymorphic markers (x-axis). Ο: optimum UPIC values for different number of markers in the combination. A: minimum number of markers (3) in an IC that can discriminate the 12 DNA samples, the 3 markers can detect up to 18 unique patterns (UP) or alleles; B and C: point to IC of 4 and 6 markers (B, C) respectively, both providing similar amount of information in UP values; D: shows the maximum number of UP (34) detectable by all 8 markers. Numbers on top of the histogram are the actual number of IC for K number of markers used in the combinations, i.e., for combinations of 5 markers, there are 25 IC out of 70 possible combinations.
Details on Input/Output files and Scripts
The input file for UPIC needs to contain four columns, please see example in Table S1. The first column (in GeneMapper exported data corresponds to dye and amplicon/peak order) is not used by the scripts. Columns 2, 3 and 4 correspond to marker, DNA sample and amplicon size (peaks) respectively, these are the columns used by the scripts. An example of the output file for the calculation of UPIC values is shown in supplementary Table S3. The output shows the number of markers in the group, then the first column corresponds to the number of unique patterns (UP) observed for that combination of markers. An example of the output file for the calculation of percent of heterozygous loci and polymorphic information content (PIC) values is shown in supplementary Table S4, where the first column is for the name of the DNA sample (or line), and the second column is the percent of heterozygous loci. In the same output file there are another 5 columns that correspond to name of the marker, square of the allele frequencies, PIC value of selffertilization, PIC value for equifrequent alleles and PIC values for cross-fertilized organisms. The user needs to select the PIC value that applies for his/her biological system. In order to run the script for UPIC calculation the user must install the Math::Combinatorics and Array::Compare, and Benchmark::Stopwatch Perl modules. The approximate computer time required to run UPIC version 1.0 script for calculating 2 to 8 combinations of 120 polymorphic markers across 6 DNA samples using a Dell Optiplex GX745 2.66 GHz dual-core Intel processor with 3.25 GB of RAM is ca. 5 min. Perl scripts for the calculations of UPIC, PIC and heterozygosity are available from the authors upon request. Each line of Perl script is either clearly self evident as to its function or is preceded by an explanatory comment. The user will receive a self extracting Zip file including test data and a README file with instructions for installation and use. UPIC Perl scripts can be downloaded from http://www.ars.usda.gov/msa/jwdsrc/gbru under Products and Sevices/Bioinformatics Tools.
Discussion
When working with microsatellites, the size of the experiments that can be conducted in terms of number of samples and number of SSR markers to run is often limited by cost. The general recommendation is to run more markers with greater numbers of polymorphism or high PIC values [5]. However, no specific number of markers to run per experiment can be extracted from PIC values. Although PIC value gives a good estimation of the informativeness of a marker, the PIC value only refers to a particular marker, whereas UPIC analyzes all the markers in relation to each other and in the context of all samples evaluated, and provides the user with the most informative marker combinations to choose from. Another useful tool to choose markers is the LOD [5], however, this is used for known pedigrees and known genome sizes, and this information is not always available when working with diverse species and populations.
We have introduced here the concept of UPIC, a decision tool for the cost-effective design of DNA fingerprinting/genotyping experiments using polymorphic simple-sequence/tandem repeat (SSR/STR) markers. UPIC is a set of Perl scripts the user can apply to find the highest number of unique patterns (UP) or alleles on the best informative combination (IC) of polymorphic markers to use in an experiment. UPIC calculations consider the information of all markers and samples used in preliminary screening, and do not require having genetic information of the populations to be analyzed such as genome size, ploidy or type of fertilization. To the best of our knowledge, there is no program available that can assist in choosing the number of polymorphic markers to use as well as determine which combination of markers will provide the maximum discrimination among the DNA samples for fingerprinting or genotyping.
The UPIC plot in Figure 1 represents the number of UP obtainable with IC of polymorphic markers for our example of 8 markers and 12 DNA samples. From our example, the benefits of UPIC calculation are: 1) Not all combinations of polymorphic markers are IC, only those that allow discrimination among all samples; in our example, only 72 IC were found out of 255 possible subset combinations of 8 polymorphic markers (histogram, Figure 1). 2) UPIC calculations identified a single combination of three markers (Figure 1A) that can discriminate all the DNA samples tested. 3) If using an IC of 4 markers, the amount of information (UP value) can vary from 19 to 23 (Figure 1B), so the user can choose the most informative one. 4) Running an IC of 4 markers provided almost the same information as running 6 markers (UP = 23,25; Figure 1B, C), therefore, the user could maximize information and minimize costs. 5) The scripts presented here also calculate three PIC values (for various fertilization types and allele frequencies) and the percent of heterozygous loci as additional decision tools. The flow diagram for the scripts is shown on Figure 2.
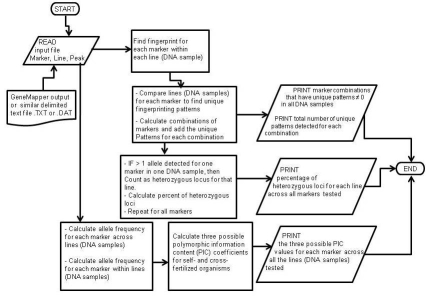
Figure 2.
Flow diagram of Perl scripts for calculation of three PIC values, percentage of heterozygous loci and UPIC values.
Conclusion
We believe that UPIC values will become a very useful tool for planning cost-effective studies using SSR markers. UPIC will minimize the cost of experiments while maximizing the information obtained by polymorphic SSR markers. The users will also be able to choose the number of markers to run based on the obtainable information. In addition to UPIC values, the scripts presented here calculate the percent of heterozygosity of the samples and PIC values for various types of fertilization in populations. Having this information available at a single location in a user-friendly format will also facilitate research with microsatellites.
Supplementary material
Footnotes
Citation:Arias et al. Bioinformation 3(8): 352-360 (2009)
References
- 1.Mrázek J, et al. Proc Natl Acad Sci. 2007;104(20):8472–8477. doi: 10.1073/pnas.0702412104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anwar T, Khan AU. Bioinformation. 2005;1(1):64–68. doi: 10.6026/97320630001064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Richard GF, et al. Microbiol Mol Biol Rev. 2008;72(4):686–727. doi: 10.1128/MMBR.00011-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Varshney RK, et al. Trends Plant Sci. 2005;10(120):621–630. doi: 10.1016/j.tplants.2005.10.004. [DOI] [PubMed] [Google Scholar]
- 5.Botstein D, et al. Am J Hum Genet. 1980;32:314–331. [PMC free article] [PubMed] [Google Scholar]
- 6.Shete S, et al. Theor Popul Biol. 2000;57:265–271. doi: 10.1006/tpbi.2000.1452. [DOI] [PubMed] [Google Scholar]
- 7. Cervus http://www.fieldgenetics.com/pages/aboutCervus_Functions.jsp.
- 8.Marshall TC, et al. Mol Ecol. 1998;7:639–655. doi: 10.1046/j.1365-294x.1998.00374.x. [DOI] [PubMed] [Google Scholar]
- 9. PIC calculator, http://www.liv.ac.uk/~kempsj/pic.html.
- 10.Nei M, Roychoudhury AK. Genetics. 1974;76:379–390. doi: 10.1093/genetics/76.2.379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hansson B, Westerberg L. Mol Ecol. 2002;11:2467–2474. doi: 10.1046/j.1365-294x.2002.01644.x. [DOI] [PubMed] [Google Scholar]
- 12.Bruvo R, et al. Mol Ecol. 2004;13:2101–2106. doi: 10.1111/j.1365-294X.2004.02209.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.