Abstract
Vitamin E is a mixture of eight compounds α, β, γ, δ tocopherols and α, β, γ, δ tocotrienols. Their individual role in cellular transport as antioxidants and in metabolic pathways has been highlighted in the present work. All the eight compounds have been docked with the respective metabolizing enzymes (αtocopherol transfer protein (ATTP), αtocopherol associated protein (TAP), Pglycoprotein (Pgly) and human serum albumin (HSA)) to understand molecular interactions for pharmacokinetics. These have been structurally aligned against the four human phospholipids in order to reveal their individual role in chylomicron formation and hence the mechanism of cellular transport. The study of their binding with their metabolizing enzymes provides insight to the comparative antioxidant activity of each of these isomers.
Keywords: docking data, vitamin E, enzymes, mechanism, antioxidant
Background
It has now been realized that Vitamin E, traditionally known as α tocopherol, is a mixture of eight different compounds, four tocopherols and four tocotrienols, each one being designated as α, β, γ and δ forms. The two groups differ in the hydrophobic tridecyl side chain which is saturated (phytyl) in tocopherols and unsaturated having three double bonds (geranyl) in tocotrienols. Detailed reports are available on αtocopherol. However, during the last few years it has been found that all the eight forms are biologically active and perform specific functions. Clinical research has shown that mixture of tocotrienols and tocopherols offer synergistic protective action against heart ailments and cancer that is not exclusively offered by αtocopherol. The other advantage of mixed tocopherols and tocotrienols is their role in slowing down aging. Diseases like diabetes 1 and 2, autoimmune diseases, bacterial and viral infections, Alzheimer disease, fungal (Candida) infections are prevented by these compounds. It helps in the maintenance of bones, muscles, eyes (vision), memory, sleep, lungs, infertility, skin and wrinkles. [1]
The main function of αTocopherol is to terminate a chain reaction of lipid peroxidation and thus protecting the cell membranes and LDL from oxidative disintegration. It has been reported in literature that the cell signaling activity of the enzyme protein kinase C is considerably reduced due to the presence of αtocopherol [2]. This also affects the activity of both inflammatory and immune cells [3]. αTocopherol also dilates blood vessels and interferes with aggregation of platelets. Recent reports have suggested better activity of γtocopherol VisaVis αtocopherol. γtocopherol can trap NO and other nitrogen free radicals more efficiently than αtocopherol [4]. γTocopherol is known to have a balanced effect on the transport of Na+ (sodium ions) thus affecting beneficially the uretic activity [5]. δTocopherol and its metabolites have been found to be the most effective inhibitors for the proliferation of prostate cancer cells [6].
γTcopherol has unique properties unlike alphatocopherol including the ability to neutralize certain free radicals and suppress expression of a gene (rasp21) that is known to cause cancer [7]. Stone and colleagues have reported that γtocopherol can hinder the growth of colon cancer [8]. An interesting observation has been reported elsewhere [9], that if α and γtocopherols are taken up together into the cells, the γtocopherol increases the level of αtocopherol. It has been reported that excess of αtocopherol displaces the more important gamma component in the body. The cellular uptake of tocotrienols through lipid bilayer is more due to the presence of double bonds in their side chains, enhancing their bioavailability and hydrophobicity. This makes them potentially more useful for cosmetic products. It is evident from the fact that amongst the two αcomponents, the αtocotrienol has been found to be 40 to 60 times more potent than αtocopherol in preventing lipid peroxidation and 6.5 times better at defending cytochrome P450 against oxidative damage. The efficiency of αtocotrienol in scavenging peroxyl radicals in liposomes is 1.5 fold more than αtocopherol.
It is found that δtocotrienol is uniformly distributed in cell membranes due to its hydrophobic nature which is due to its unsaturated side chain. δ tocotrienol has been shown to possess antithrombic activity and decreases platelet aggregation. δtocotrienol is the most effective form of vitamin E family in reducing the risk of developing atherosclerotic plaque. One of the most striking discoveries in tocotrienol research is their ability to clear atherosclerotic blockages (stenosis) in the carotid artery, potentially reducing the risk of stroke. These are more potent in quenching and scavenging the free radicals. Due to their molecular structure they have greater recycling activity and more effective collusion with free radicals [10].
One of the metabolite of γtocotrienol is LLUalpha which reduces the high blood pressure and congestive heart failure [11]. Tocotrienols inhibit cancer cell proliferation through apoptosis. Studies on estrogen responsive and estrogen nonresponsive human breast cancer cells have shown that tocotrienols are effective in preventing the proliferation of both [12]. Tocotrienols inhibit NF kB signaling pathway leading to suppression of antiapoptotic gene products and starts apoptosis. Tocotrienols are known to cross the bloodbrain barrier, and are potent protectors of neuron cells that may get destroyed through stroke and other neurodegenerative diseases.
The reports discussed thus far about the enhanced activity of other members of the vitamin E family besides the mostly studied αtocopherol, motivated us to study the structureactivity relationship of the whole group. It is known that αtocopherol is taken up together with dietary lipids and bile in the proximal part of the intestine. The tocopherols get assembled together with triglycerides, cholesterol, phospholipids, and apolipoproteins into chylomicrons. The exact mechanism of this process of chylomicron formation is not very well known. In this paper we have made an effort to structurally align all the four mammalian phospholipids viz. phosphatidyl choline, phosphatidyl ethanolamine, phophatidyl inositol, lysophosphatidyl inositol , present in the human liver with tocopherols and tocotrienols to establish their mechanism of action.
Methodology
ACD/CHEMSKETCH
2D structures of all the eight compounds of vitamin E were developed using ACD/chemsketch software (to draw molecules, reactions and schematic diagrams and calculate their properties).
CHIMERA
3D structures of all the eight isomers of vitamin E were obtained from chimera (python based software generates three dimensional atomic coordinates from 2D structure of a molecule).
AUTODOCK
Docking was performed using AUTODOCK version 3.0 software package running on PC Intelbased Pentium 4, running Linux operating system.
Protein receptors for docking
The following four target proteins were selected for the study,
Alphatocopherol transfer protein (ATTP)
Alphatocopherol (αT) transfer protein (ATTP) is a member of the family of lipidbinding proteins containing two CRALTRIO domains, pfam03765 (residues 1183) and pfam00650 (residues 89275). The corresponding structure with PDB ID 1R51 is downloaded from PDB.
Alphatocopherol associated protein (TAP)
It is tocopherol binding protein with a molecular mass of 46 kDa present in the cytosol of bovine liver. This is referred as alpha tocopherolassociated protein (TAP). The corresponding structure with PDB ID 1OLM is downloaded from PDB.
Pglycoprotein (Pgp)
3D structure of pglycoprotein was downloaded from PDB (PDB ID 2GHI). Pglycoprotein is found in the gut, gonads, kidneys, biliary system, brain and other organs. It belongs to the family of efflux transporter. Pgp is also called ABCB1, ATPbinding cassette subfamily B member 1, MDR1, and PGY1. It is a 170 kDa encoded by the MDR (multidrug resistance) gene(s) that are highly conserved across species.
Human serum albumin (HSA)
3D structure of human serum was downloaded from PDB (PDB ID: 1E7H). The protein is a helical monomer of 66 kDa containing three homologous domains (IIII) each of which is composed of A and B subdomains.
Structural Alignment
We used the VMD software for structural alignment of the phospholipid molecules present in human liver with all the eight forms of tocopherols and tocotrienols. The lipid structures were first downloaded from lipid database, and thereafter the 2D structures were drawn using the Chemsketch software. Root mean square deviation was then calculated using appropriate tools in Chemsketch.
Molecular docking
ATTP
The PDB ID used for ATTP is 1R51. The crystal structure of human alphatocopherol transfer protein bound to its ligand αtocopherol is used. The bound tocopherol was removed and the active site of the ligand was analyzed with the help of CASTP server. Volume of the active site is 1030 Å3 and surface area is 727 Å2. This active site is made of residues TYR100 to ILE222. This is tested for all the eight isomers.
TAP
The PDB ID used for TAP is 1OLM. The structure is protein complex with rrralphatocopherylquinone. The active site has volume 10262 Å3 and surface area 6402.2 Å2.
HSA
The PDB ID used for HSA is 1E7H. HSA forms complex with palmitic acid. The volume of the active site (LYS413 LYS538) 88.7 Å3 and the surface area is 90.2 Å2.
Pgp
The PDB ID used for Pgp is 2GHI. Pgp has four chains with ATP binding sites. The volume of the active site (LYS37ILE231) is 10.1 Å3 and the surface area is 18.9 Å2.
Docking procedure
The active sites are located in each of the target proteins before being loaded to AUTODOCK. Polar hydrogens were then added using the protonate utility available in AUTODOCK. The ligand is made flexible by providing rotation to bonds. AUTOGRID (using Kollman united atom charges and salvation parameters) is then used to map grid to active sites of target protein. The grid dimensions are [1.344, 63.053, 14.596] for TTP, [-20.33, 19.575, 87.289] for TAP, [-3.188, 1.003, 26.114] for HSA and [45.57, 14.8, -17.9] for Pgp. The genetic search algorithm with 100 iterative runs was selected for dockings.
Discussion
We aligned all the structures of eight tocopherols and tocotrienols with four human phospholipids to elucidate the exact binding mode in the formation of chylomicrons before reaching the intestine [13]. Literature reports suggest that αtocopherol along with lipids is packed into chylomicrons and transported to the liver [14]. They appearance in plasma is only after passing through the liver. Most of the ingested β, γ, and δ tocopherols are secreted into bile are not taken up and hence excreted in the feces. This is true for all the other compounds used in this study. However, results show that αtocopherol and αtocotrienol are more structurally similar to phophatidyl inositol. β tocopherol is similar to phosphatidyl choline and betatocotrienol is aligned with phosphotidyl ethanolamine. δ and γ forms are more similar to lysophosphatidyl ethanolamine. Nonetheless, all the four phospholipids are associated with the transportation process. The binding mode of these eight components of vitamin E with different proteins in metabolism, transport and absorption is of interest. The binding energy of these compounds with four metabolizing enzymes is given in Table 1.
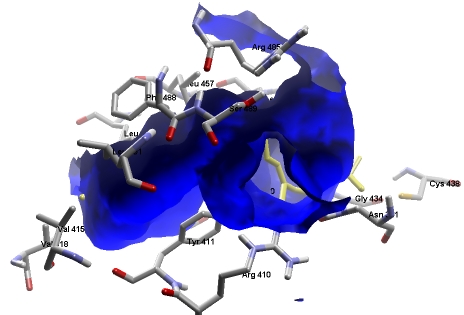
?lphatocopherol transfer protein (αTTP) present in liver is selectively involved in retension of α tocopherol from dietary vitamin E (a mixture of α, β, γ, δ tocopherols and corresponding tocotrienols). The components of vitamin E are taken up along with emulsified lipid molecules, extracted from food, in equal amounts in an unspecific manner. These are transported from intestine packed in chylomicrons and subsequently found as remnants in the liver. The cytosolic protein αTTP is then released into circulation from the liver. This enzyme is responsible for the stereoselective transfer of α tocopherol to VLDL. In vitro and in vivo assays have shown that α TTP preferentially binds to the αtocopherol. However, our docked complex shows that αtocotrienol shows the highest binding affinity among all the eight isomers tested for the protein. The other three tocotrienols (β, γ, δ) have appreciably high binding energy as compared to the corresponding tocopherols. Data also show that the four tocopherols have almost equivalent binding energy. We then selected the specific active sites for docking of all the eight isomers. The docked conformation of αtocotrienol with α TTP has two hydrogen bonded interaction with αTTP (Figure 1). The interaction is with ser140 of intensity -0.28 and with ser136 of intensity -1.26.
Figure 1.
The docked conformation of αtocotrienol with α TTP has two hydrogen bonded interaction with α TTP.
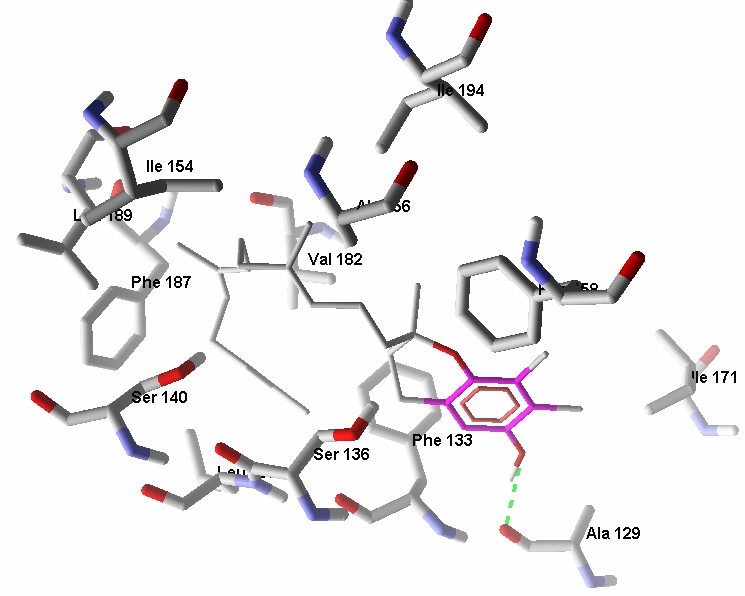
Tocopherolassociated protein (TAP) is also been documented extensively as a metabolizing enzyme. It is present in brain, liver and prostate gland. It increases the uptake and absorption of Vitamin E and hence facilitates the anti proliferation effect in prostate cancer cells. TAP also functions like a tumor suppressor gene to control cancer cell viability through a nonvitamin E functional mechanism. Therefore, TAP is a new prophetic marker for prostate cancer progression. TAP is a vitaminbinding protein and it is involved in transport of Vitamin components specifically tocopherols. Docking data suggests that γtocotrienol binds better than others. Three tocotrienols except δtocotrienol have comparable binding energies higher affinity than tocopherols. They are easily transported between the membranes due to unsaturated side chains. Thus, trienols are potential tumor suppressors for prostate cancer. Docking data shows that gamma tocotrienol have four hydrogen bond interactions with active site residues of TAP (Figure 2). The interactions are with Ile80 of intensity -0.899, Leu84 of intensity -0.4962, Leu84 of intensity -0.30 and Asn259 of intensity -2.203.
Figure 2.
Docking data shows that gamma tocotrienol have four hydrogen bond interactions with active site residues of TAP
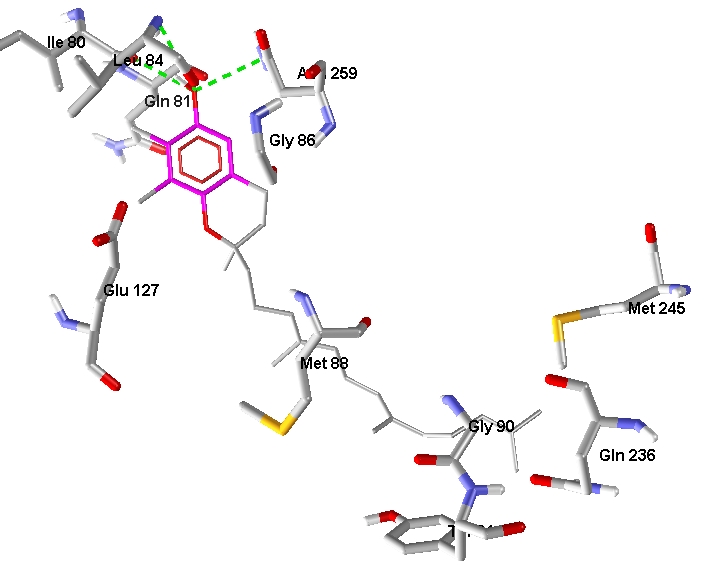
HSA is the target protein for circulation of tocopherols and tocotrienols through blood. This protein has an ability to bind variety of hydrophobic small molecules. These include fatty acids, bilirubin, thyroxin, bile acids, steroids and a group of other bio active molecules. HSA can solubilize the ligands to facilitate transport and buffering effect in free concentration. It is documented that HSA binds a wide variety of drugs in two primary sites which overlap with the binding locations of endogenous ligands [15]. We then selected a site specific for palmitic acid. Data suggest that γ tocotrienol and δtocotrienol have highest binding energy with HSA as compared to other members of the family. Overall the binding of tocotrienols with HSA is comparatively higher than the four tocopherols. The docked conformation of γ tocotrienol with the highest binding energy.-11.34 and docking energy is -14.24 kcal/mol. Figure 3 shows the electrostatic interactions of γ tocotrienol with the aminoacid of HAS. This shows the better absorption and transport potential of tocotrienols.
Figure 3.
The electrostatic interactions of γ tocotrienol with the aminoacid of HSA.
Pglycoproteins are involved in active transport of various ligands across cell membranes through ATP mediated processes. These are either localized in the plasma membrane or intracellular and are responsible for efflux of various foreign substances including drugs. This protects the body against toxic xenobiotics and drugs. It prevents their accretion in sensitive organs like brain, placenta and gonads by removing these compounds into bile, urine, and the intestinal lumen. The role played by Pgp efflux transporters in assaying the overall bioavailability of drugs is emphasized in recent years. The involvement of Pgp in drug metabolism has immense pharmacokinetic importance. The role of efflux transporters in determining the permeability and overall bioavailability of drugs has gained considerable attention. Pgp efflux pump is localized in a wide range of tissues, including enterocytes of the GI tract. The interaction of Pgp with vitamin E components highlights their bioavailability and pharmacokinetics for distribution in organs/tissues. The docked conformation of αtocotrienol with Pgp shows a single hydrogen bond interaction (Figure 4). The interaction is with Gln100 of interaction energy -0.781 kcal/mol, binding energy -9.06 kcal/mol and docking energy -11.91 kcal/mol.
Figure 4.
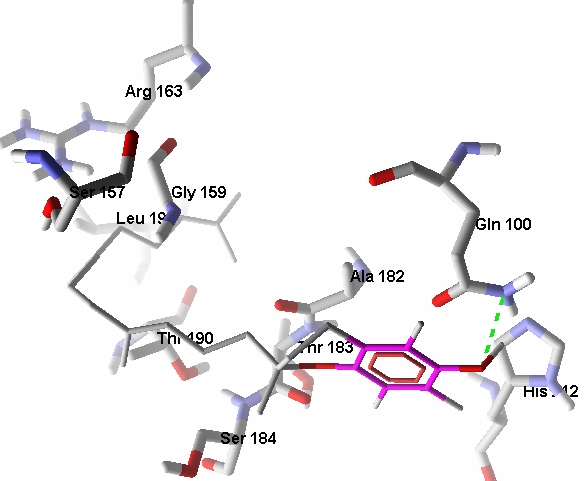
The docked conformation of αtocotrienol with Pgp shows a single hydrogen bonded interaction
We selected the ATP binding site for docking of all the eight components of vitamin E in order to analyze their efflux potential. When binding intensity is high efflux is low resulting in more bioavailability. In this study, data shows that α and δtocotrienols show highest binding affinity at the ATP site. Therefore, it can stop the efflux of all other components more efficiently and thus, enhancing their bioavailability. The other two tocotrienols (β and γ) have comparable activity. Tocotrienols are potent candidate as antioxidant and anticancer agent and thus it is important to increase their bioavailability. The bioavailability of all the components of vitamin E is increased when taken with compounds (eg. piperine) that bind strongly at the ATP site of Pglycoprotein which enhances the bioavailability of many antioxidants like curcumin and vitamin C.
Conclusion
A study to understand the mechanism of cellular uptake of phospholipids through the formation of chylomicron with lipids is of interest. The eight components of vitamin E show similar mechanism of cellular uptake (chylomicron formation with lipids). The docked complexes of ATTP, TAP, HSA and Pgp with tocotrienols and tocopherols provide insight to their binding mode during metabolism, absorption, transport and efflux. Data shows that the tocotrienols with unsaturated side chain show better activity than tocopherols due to high binding with the corresponding metabolizing enzymes. This suggests tocotrienols as potential antioxidants and tumor suppressors than tocopherols. Nonetheless, it should be noted these docked data should be verified using appropriate experiments.
Supplementary material
Footnotes
Citation:Upadhyay & Misra, Bioinformation 3(8): 326-331 (2009)
References
- 1.Azzi A, Stocker R. Prog Lipid Res. 2000;39:231. doi: 10.1016/s0163-7827(00)00006-0. [DOI] [PubMed] [Google Scholar]
- 2.Freedman JE, et al. Circulation. 1996;94:2434. doi: 10.1161/01.cir.94.10.2434. [DOI] [PubMed] [Google Scholar]
- 3.Azzi A, et al. Biofactors. 1998;7:3. doi: 10.1002/biof.5520070102. [DOI] [PubMed] [Google Scholar]
- 4.Cooney R, et al. Proc Natl Acad Sci U S A. 1993;90:1771. doi: 10.1073/pnas.90.5.1771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wechter WJ, et al. Proc Natl Acad Sci USA. 1996;93:6002. [Google Scholar]
- 6.Jiang Q, et al. Proc Natl Acad Sci USA. 2004;101:17825. doi: 10.1073/pnas.0408340102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stone WL, et al. Cancer Detect Prev. 2002;26:78. doi: 10.1016/s0361-090x(02)00003-x. [DOI] [PubMed] [Google Scholar]
- 8.Stone WL, et al. Ann NY Acad Sci. 2004;1031:223. doi: 10.1196/annals.1331.022. [DOI] [PubMed] [Google Scholar]
- 9.Le Magezine American Medical Association. 2005 [Google Scholar]
- 10.Packer L, et al. J Nutr. 2001;131:369. doi: 10.1093/jn/131.2.369S. [DOI] [PubMed] [Google Scholar]
- 11.Wechter WJ, et al. Proc Natl Acad Sci USA. 1996;93:6002. [Google Scholar]
- 12.Nesaretnam K, et al. Lipids. 1995;12:1139. doi: 10.1007/BF02536615. [DOI] [PubMed] [Google Scholar]
- 13.Traber MG, Kayden HJ. Am J Clin Nutr. 1989;49:517. doi: 10.1093/ajcn/49.3.517. [DOI] [PubMed] [Google Scholar]
- 14.Rigotti A. Molecular Aspects of Medicine. 2007;28:423. doi: 10.1016/j.mam.2007.01.002. [DOI] [PubMed] [Google Scholar]
- 15.Sugio S, et al. Protein Eng. 1999;12:439. doi: 10.1093/protein/12.6.439. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.