Abstract
The AllergenPro database has developed a web-based system that will provide information about allergen in microbes, animals and plants. The database has three major parts and functions:(i) database list; (ii) allergen search; and (iii) allergenicity prediction. The database contains 2,434 allergens related information readily available in the database such as on allergens in rice microbes (712 records), animals (617 records) and plants (1,105 records). Furthermore, this database provides bioinformatics tools for allergenicity prediction. Users can search for specific allergens by various methods and can run tools for allergenicity prediction using three different methods.
Availability
The database is available for free at http://www.niab.go.kr/nabic/
Keywords: allergen database, allergenicity prediction, AllergenPro
Background
Allergy and other hypersensitivity reactions have become major causes of chronic health problems in developed countries. With advances in genomic technologies, there is a rapid increase in allergy-related data, including allergen sequences, allergic cross-reactivity and clinical measurements [1]. Starting from the first release of the International Union of Immunological Societies (IUIS) allergen database [2], several institutions have started to develop databases of various functions on allergens [3]. Various researches performed different algorithms to try to predict allergenicity [4]. Since then, a variety of Webbased bioinformatics tools for protein allergenicity have been developed [5]. Generally, allergen databases contain at least a list of allergens, bioinformatics tools for sequence search and for performing allergenicity prediction. IUIS Allergens [6] is the official web site for allergen nomenclature. ADFS [7] is a database containing information on allergen structures with computational tools for defining allergenicity. The SWISS-PROT is an annotated protein sequence database and SDAP is a database of allergenic proteins with various computational tools [8–9]. ALLERDB database contains sequences of allergens and focuses on the analysis of allergenicity and allergic cross-reactivity of clinically relevant protein allergens [10]. AllergenOnline [11] provides access to a peer reviewed allergen list and sequence searchable database intended for identifying proteins that may present potential risks of allergenic cross-reactivity. These allergen databases will be important in updating the allergen reference information on a regular basis as new allergens are identified in order to improve the performance of allergenicity prediction.
Methodology
Dataset
The allergen, iso-allergen and epitopes information were collected from the IUIS allergens, SDAP, SWISS-PROT and NCBI PubMed database. At present, the allergens' information are categorized with their accession numbers under microbes, animals and plants. The current version of AllergenPro contains a total of 2,434 allergen data including 712 records on allergens in microbes (fungi 213, bacteria 499), 617 records on allergens in animals (insects 175, foods 79, mites 148, rodents 39, mammals 27, worms 66, others 83) and 1,105 records on allergens in plants (foods 600, latex 40, pollen 461, others 4). Additional information on allergen structure and epitopes were connected with UniProtKB/TrEMBL [8] and Protein Data Bank (PDB) database.
Development
The platform was developed using MYSQL and JAVA languages. The data were stored in Oracle relational database management system (RDBMS). Using the collected allergen information, we have developed an allergen database with three main features: (i) allergen list with structure and epitopes; (ii) searching of allergen by keyword and sequence information; and (iii) computing methods for allergenicity prediction.
Implementation and Features
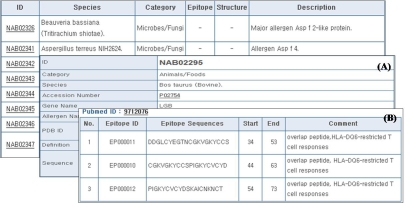
The AllergenPro has developed a web-based system that will provide information about allergen in microbes, animals and plants. The database has three major parts and functions: a database list, allergen search and allergenicity prediction. Users can search for specific allergens by keywords, sequence and allergen list of categories. To identify potential cross-reactivity with known allergens, users can run tools for allergenicity prediction using three different methods such as the FAO/WHO, motif-based and epitope-based methods. The individual data tables are linked to each other through their respective hypertexts as shown in Figure 1. In browsing the result table (Figure 1), a user can access the individual characterization tables by clicking a specific accession number on the linked hypertext. An ’ID‘ field provides a detailed table (Figure 1A), which has records about category, species, accession number, gene name, allergen name, PDB ID, definition and sequence. In addition, clicking the hypertext displays the characterization information of the epitope, if it exists (Figure 1B).
Figure 1.
A snap shot of the database. The individual view table shows samples of search results such as the (A) allergen information table (ex: NAB02295) by clicking a specific accession number in the left area and the (B) epitope information view table.
Discussion
AllergenPro database provides allergen characterization information which includes the structure and epitope of allergens in microbes, animals and plants. The database contains 2434 specific allergens related information readily available in the database such as on allergens in rice microbes (712 records), animals (617 records) and plants (1,105 records). Furthermore, this database provides bioinformatics tools for allergenicity prediction. Users can search for specific allergens using various methods and can run tools for allergenicity prediction using three different methods.
Footnotes
Citation:Kim et al., Bioinformation 3(8): 344-345 (2009)
References
- 1.Tong JC, et al. Front Biosci. 2008;13:4882. doi: 10.2741/3047. [DOI] [PubMed] [Google Scholar]
- 2.King TP, et al. J Allergy Clin Immunol. 1995;96:5. [Google Scholar]
- 3.Adriano M, et al. Cellular Immunology. 2006;244(2):97. doi: 10.1016/j.cellimm.2007.02.012. [DOI] [PubMed] [Google Scholar]
- 4.Stadler MB, et al. FASEB J. 2003;17:1141. doi: 10.1096/fj.02-1052fje. [DOI] [PubMed] [Google Scholar]
- 5.Saha S, et al. Nucleic Acids Res. 2006;34:202. doi: 10.1093/nar/gkl343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. http://www.allergen.org.
- 7. http://allergen.nihs.go.jp/ADFS/
- 8.David FP, et al. BMC Bioinformatics. 2008;23:391. doi: 10.1186/1471-2105-9-391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ivanciuc O, et al. Nucleic Acids Res. 2003;31:359. doi: 10.1093/nar/gkg010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang ZH, et al. Cell Immunol. 2006;244(2):90. doi: 10.1016/j.cellimm.2007.01.007. [DOI] [PubMed] [Google Scholar]
- 11. http://www.allergenonline.com/