Abstract
Summary: We describe an integrative software platform, Prequips, for comparative proteomics-based systems biology analysis that: (i) integrates all information generated from mass spectrometry (MS)-based proteomics as well as from basic proteomics data analysis tools, (ii) visualizes such information for various proteomic analyses via graphical interfaces and (iii) links peptide and protein abundances to external tools often used in systems biology studies.
Availability: http://prequips.sourceforge.net
Contact: dhhwang@postech.ac.kr
1 INTRODUCTION
Recently, liquid chromatography (LC) coupled tandem MS-based proteomics technologies have been extensively used in systems biology studies. The improved reliability and widespread use of such LC-MS/MS approaches have led to huge amounts of large datasets together with a growing number of software tools for diverse analyses of MS data (Trans-Proteomic Pipeline (TPP), Keller et al., 2005). Despite such software tools, effective integration, visualization and analysis of MS data for systems biology investigations are still complicated due to lack of (i) integration of diverse data types from different MS platforms and analytical output data types from various software tools—MSn spectra, peptides (sequences, search outputs, abundances and modifications) and proteins (coverage, abundances, modifications and significances), (ii) effective visualization of such complex integrated data for various analyses of MS data to examine characteristics and quality of data and optimize experimental conditions in comparative proteomics and (iii) automatic exchanges of such integrated information with software used in systems biology. The first problem is still challenging due to the complex relationships among data from multiple samples, despite several tools having been developed for MS data analysis (e.g. TPP). There is still a lack of tools for visualizing information from multiple samples to facilitate various analyses in comparative proteomics. Pep3D (Li et al., 2004) resolved the second problem in part, especially for a single sample. Moreover, only few tools with limited functionalities to resolve the third problem have been introduced. Therefore, there has been a need for a computational framework providing functionalities for (i) integration of complex MS data, (ii) visualization of the complex data using spectrum/chromatogram and tabular viewers, peptide feature maps and (iii) network modeling and analysis (e.g. Cytoscape and its plug-ins) and statistical analyses such as data normalization, identification of differentially expressed proteins, functional enrichment analyses and classification/clustering. Here, we present Prequips, a Java-based workbench of computational tools efficiently achieving these three tasks.
2 METHODS AND FEATURES
Prequips provides the following three key features. To demonstrate the features in Prequips, we used four datasets obtained from LC-MS/MS analysis of samples to examine changes in sub-cellular localization after chemical treatment.
2.1 Integration of multiple MS data and diverse proteomic information
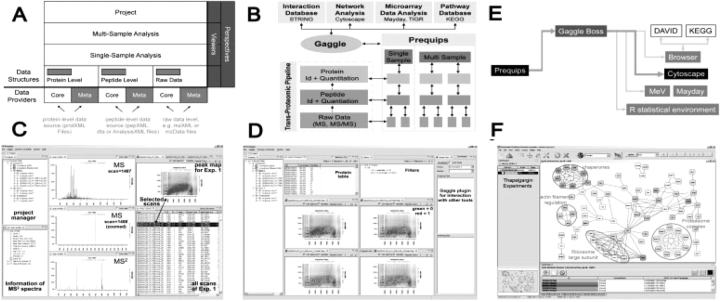
Prequips is built on a generic data model with two main parts, core data and meta information elements obtained from the data sources shown in Figure 1A. Core data refers to key information from MS experiments with hierarchical structure—spectra, peptide sequences and protein IDs (Fig. 1B). Meta information represents data generated by the basic data analysis pipeline (e.g. SEQUEST and TPP) at all stages and in various forms, which describes the core data more closely. Meta data of peptides include DB search outputs (sequences, SEQUEST outputs such as Xcorr, PeptideProphet probabilities; Keller et al., 2002), abundances and modifications, while those of proteins include sequence coverage, abundances, modifications and ProteinProphet probabilities. These core data and meta information are integrated via the data model of Prequips (Fig. 1A). In ‘single sample analysis’, both core and meta data for a single dataset are organized in a data model including spectrum-, peptide- and protein-level information (Fig. 1A). In ‘multi-sample analysis’, the core and meta information for multiple samples can be combined in the data model based on any kinds of unique proteins (Fig. 1B). A vector of values including all spectrum- and peptide-level data and related meta data over all samples can be created for each protein. To effectively load core data and meta information, Prequips employs so-called ‘data providers’, which enhances data integration capabilities (Fig. 1A). Data providers, designed in plug-in interfaces, separate Prequips from the data sources (e.g. mzXML, pepXML and protXML; Fig. 1A) and make the software independent of data formats.
Fig. 1.
Overview of Prequips' design and functionality. (A) Data model structure. (B) Schematics of data structure and exchange. (C) Spectrum, peptide feature map and table viewers in single sample analysis. (D) Peptide feature maps in multi-sample analysis. (E) Broadcasting to Cytoscape via the Gaggle interface. (F) Network analysis using broadcasted proteins in Cytoscape.
2.2 Visualization of integrated core data and meta information
Prequips supports interactive data exploration through a graphical user interface (GUI) permitting to efficiently interact with the data through graphical viewers and data filters. Figure 1C shows summary windows, spectrum viewers, peptide feature maps and a tabular viewer showing integrated core and meta information for ‘single sample analysis’: (i) the project manager allows users to select a particular sample (e.g. a chemical-treated sample) or a group of samples (e.g. control samples); (ii) a spectrum summary window showing core and meta information (peak numbers, retention time, etc.) for the MS2 scan being displayed in the spectrum viewer; (iii) MS/MS2 spectrum viewers; (iv) chromatogram viewers (not shown); and (v) peptide feature map. Note that the data being displayed are all connected across the viewers through the integration of core and meta information in the data model (see the linked selection in Fig. 1C). Also, a sophisticated filtering framework has been implemented in Prequips, based on Boolean operators AND, OR and XOR (exclusive-or), by which a group of objects can be selected using multiple data filters. For instance, users can select peptides whose PeptideProphet probabilities are larger than 0.95. Once those peptides were selected in the tabular viewer, the corresponding peptides and scans are highlighted in the other viewers. Prequips enables users to open a virtually arbitrary number of viewers to investigate the data based on the Model-View-Controller architecture. Figure 1D shows four peptide feature map viewers from four MS datasets, as an example of a ‘multi-sample analysis’. Exploring multiple MS datasets using the viewers and data filters assist users to examine characteristics and quality of data, thus aiding optimizing experimental conditions to improve particular aspects of results (e.g. increase the detected proteome size).
2.3 Mapping proteomic information into systems biology tools
Besides data integration and visualization tools for data exploration, Prequips also provides extensive support for systems biology analyses of MS-based proteomic data. Prequips employs the Gaggle (Shannon et al., 2006) to exchange core and meta data of interest (e.g. a list of proteins detected from multiple experiments) with external software tools (Fig. 1B) being used for network analysis (Cytoscape, Biotapestry and String) and statistical analyses (TIGR MeV; Saeed et al., 2003). Prequips is able to broadcast both lists of names and matrices via the Gaggle plug-in. Figure 1E schematically shows broadcasting protein IDs from Prequips to Cytoscape via the Gaggle. As shown in Figure 1F, we can reconstruct a network model for the broadcasted proteins using protein–protein interactions obtained by Cytoscape plug-ins. The network shows cellular processes (see circled network modules) enriched by the proteins whose sub-cellular locations were changed after the chemical treatment. The Gaggle in Prequips also passes protein IDs via Firefox to web-based tools (DAVID) for functional enrichment analysis and also to R-package as inputs to R-scripts for sophisticated analyses.
3 IMPLEMENTATION
Prequips is open-source software implemented in Java based on the Eclipse Rich Client Platform and released under the GNU GPL. The modularity of Prequips enables third party developers to adapt it into their specific software environment or add their own functionalities to Prequips as plug-ins (e.g. the Inclusion List Builder; Schmidt et al., 2008). The use of Java renders the software platform-independent. To efficiently handle large datasets, the data providers were designed to dynamically load data from data sources into memory: Prequips could load a 589 MB mzXML in 1.5 min on an iMac with 2.16 GHz and 2 GB RAM.
Conflict of Interest: none declared.
Funding: GAES (grant DAMD17-03-1-0321, DAMD17-03-1-0425); NIH (grant NS41997); NHLBI (grant N01-HV-28179); DOD (grant DAMD17-3-1-0321); Korean MEST (grant FPR08A1-050, SC-2120); BioGreen21 (grant 20080401-034-041-008-02-00).
REFERENCES
- Keller A, et al. Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal. Chem. 2002;74:5383–5392. doi: 10.1021/ac025747h. [DOI] [PubMed] [Google Scholar]
- Keller A, et al. A uniform proteomics MS/MS analysis platform utilizing open XML formats. Mol. Syst. Biol. 2005;1:1–8. doi: 10.1038/msb4100024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X-J, et al. A tool to visualize and evaluate data obtained by liquid chromatography-electrospray ionization-mass spectroscopy. Anal. Chem. 2004;76:3856–3860. doi: 10.1021/ac035375s. [DOI] [PubMed] [Google Scholar]
- Saeed A, et al. TM4: a free, open-source system for microarray data management and analysis. Biotechniques. 2003;34:374–378. doi: 10.2144/03342mt01. [DOI] [PubMed] [Google Scholar]
- Shannon PT, et al. The gaggle: an open-source software system for integrating bioinformatics software and data sources. BMC Bioinformatics. 2006;7:176. doi: 10.1186/1471-2105-7-176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt A, et al. An integrated, directed mass spectrometric approach for in-depth characterization of complex peptide mixtures. Mol. Cell. Proteomics. 2008;7:2138–2150. doi: 10.1074/mcp.M700498-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]